Abstract
Leucine-rich repeat containing protein 10 (LRRC10) is a heart-specific factor whose function remains unknown. Examination of the intracellular location of the gene products is a critical step in determining the biological functions of the protein. Our expression analyses in mice indicate that LRRC10 is exclusively expressed from the precardiac region in early embryos to the adult heart. LRRC10 expression is markedly elevated upon birth, suggesting its role in the embryonic as well as adult hearts. Of interest, LRRC10 exhibits dynamic intracellular expression patterns in cardiomyocytes. Cardiomyocytes from embryos and newborns show diffuse cytoplasmic and nuclear staining of LRRC10. In contrast, striking striations are observed in adult cardiomyocytes, which are colocalized with the markers for the Z-line, sarcoplasmic reticulum (SR), and transverse (T)-tubule by double immunostaining. Further investigation by electron micrographs places LRRC10 in a diad region where the SR interacts with the T-tubule that locates along the Z-line.
Keywords: leucine-rich repeat containing protein 10, LRRC10, cardiac-specific gene, heart development, subcellular localization, striations in cardiomyocytes
INTRODUCTION
Cardiac development is a complex biological process requiring the integration of cell specification, differentiation, and morphogenesis (for reviews, see Chien, 2000; Srivastava and Olson, 2000; Olson and Schneider, 2003; Gruber and Epstein, 2004; Olson, 2004). Many factors have been implicated in this process on the basis of their spatial and temporal expression patterns or their phenotypic effects when they are functionally inactivated in various animal models (Brannan et al., 1994; Lyons et al., 1995; Kuo et al., 1997; Sanford et al., 1997; Schott et al., 1998; Lee et al., 2000; Berdougo et al., 2003; Schneider et al., 2003; Krause et al., 2004; Toyofuku et al., 2004; Jung et al., 2005b; Bendig et al., 2006; Rottbauer et al., 2006; Walton et al., 2006). However, the exact molecular mechanisms of embryonic cardiac development remain to be elucidated.
Therefore, we reasoned that there are yet unidentified cardiac factors that play critical roles in normal heart development as well as in maintaining normal cardiac function in adults. To identify novel cardiac-specific genes that are exclusively expressed in the heart, we performed a virtual subtractive screen (in silico approach) as described previously (Passier et al., 2000; Wang et al., 2001). Mouse expression sequence tag (EST) databases from embryonic day (E) 13 heart were searched using the National Center for Biotechnology (NCBI) database. By subtracting genes that are expressed in other organs, we identified a list of putative cardiac-specific genes. These genes were subsequently subjected to in situ hybridization and Northern blot analyses to examine their expression patterns. From this expression screening, we have identified a gene encoding leucine-rich repeat containing protein 10 (LRRC10), whose molecular and cellular functions remain unknown.
Leucine-rich repeat (LRR) motifs are present in an increasing number of proteins with diverse functions such as enzyme inhibition, cell growth, cell adhesion, signal transduction, regulation of gene expression, apoptosis, and development (Inohara et al., 1999; Bilder and Perrimon, 2000; Mutai et al., 2000; Tong et al., 2000; Wu et al., 2000; Kobe and Kajava, 2001; Linhoff et al., 2001). There are over 2000 LRR motif-containing proteins present in organisms ranging from viruses to eukaryotes. LRRs are 20- to 29-residue sequence motifs, and the repeat number ranges from 2−45. The primary function of these motifs is believed to provide a structural framework for protein–protein interactions. The LRRC10 protein is conserved in most vertebrates, including mouse, rat, human, chicken, and frog, suggesting an important function in an evolutionally conserved manner. For example, mouse, zebrafish, and human LRRC10 contain seven LRR motifs and do not contain any N-terminal or C-terminal domains that are often present in other LRRC proteins. These additional domains seem to confer specific functions to LRRC proteins such as a kinase domain or a membrane spanning domain (Buchanan and Gay, 1996). Therefore, LRRC10 represents a unique member of the LRRC protein family, which contains only the leucine-rich repeat motifs.
LRRC10 was isolated by other laboratories, which was named as HRLRRP or SERDIN1 (Nakane et al., 2004; Adameyko et al., 2005) while we were characterizing this factor. Herein, we refer to this gene as LRRC10 in accordance with the NCBI nomenclature. Although mouse LRRC10 has been shown to exhibit heart-restricted or -specific expression patterns, there is controversy about the intracellular expression pattern of LRRC10 in cardiomyocytes during development and in the adult. It has been reported that overexpressed LRRC10 in noncardiac cells localized in the nucleus of some cells and in the mitochondria in most cells, but endogenous LRRC10 was detected in the nuclei of the adult mouse heart tissue (Nakane et al., 2004). However, the antibody used in this study recognized multiple bands on a Western blot and, therefore, was not suitable for immunostaining experiments to examine the endogenous expression patterns. Diffuse localization (both nuclei and cytoplasm) has been reported in cells that overexpress LRRC10, whereas cardiomyocytes from differentiated P19 cells and embryos showed nuclear localization of LRRC10 (Adameyko et al., 2005). In adult cardiac tissue, LRRC10 was detected as a doublet in the I bands flanking the Z-line of sarcomeres, which conflicts with the reported nuclear staining of LRRC10 in the adult heart (Nakane et al., 2004).
Sarcomeres are the smallest contractile units of striated muscle, which are conventionally believed to be the most regular macromolecular assemblies in biology. However, recent studies have revealed a dynamic nature of sarcomere proteins and their interacting proteins in response to various stimuli (Lange et al., 2006). The Z-line represents a critical link between the transverse (T) -tubule network and cytoskeleton of cardiac cells with a role in anchoring structural proteins, ion channels, and signaling molecules. T-tubules are the periodic deep membrane invaginations of ventricular myocytes. T-tubules form a series of bracelets that wrap around each myofibril at every Z-line. Each T-tubule is associated with a single cistern of sarcoplasmic reticulum (SR) to form a diad at the Z-line. Increasing evidence indicates that many cytoplasmic and membrane spanning proteins in the sarcolemma or SR exhibit striation patterns in adult cardiac muscle, which colocalize with Z-discs. In addition, the functional significance of the specific subcellular localization of various proteins has been reported (Puri et al., 1997; Robu et al., 2003; Sullivan et al., 2003; Singh et al., 2004; Kang and Walker, 2005; Robia et al., 2005; Balijepalli et al., 2006; Bendig et al., 2006).
Because the intracellular location of LRRC10 is inconclusive, it is necessary to determine detailed expression profiles and precise subcellular localizations. We generated anti-LRRC10 antibodies that are highly specific to LRRC10 to thoroughly examine the expression profile of endogenous LRRC10. Our expression study indicates that LRRC10 is specifically expressed in the precardiac crescent region of early mouse embryos. LRRC10 is readily detectable throughout mouse embryonic development followed by a marked increase upon birth. The high level of expression is maintained in the adult mouse heart. Intracellularly, LRRC10 shows a diffuse staining pattern in both the cytoplasm and nucleus of embryonic and neonatal primary cultured cardiomyocytes. LRRC10 shows a striated pattern as the heart matures, and colocalizes with sarcomeric Z-line markers or other markers colocalizing with the Z-line by immuno-staining. Interestingly, electron micrographs of adult mouse heart using immunogold labeling techniques show localization of LRRC10 near the diad region where the SR interacts with the T-tubule. LRRC10 is not observed on the Z-line. The present study provides a critical foundation to further investigate the biological roles of LRRC10 in the developing and mature heart.
RESULTS
Expression Pattern of Lrrc10 During Development
Information on the detailed expression patterns of a gene provides the important basis of understanding the biological roles of the gene product. Therefore, we investigated the temporal and spatial expression patterns of LRRC10 in mouse, human, and zebrafish by various methods. Although it has been reported that LRRC10 shows cardiac-restricted or -specific expression patterns in mice (Nakane et al., 2004; Adameyko et al., 2005), its intracellular distribution is controversial and a detailed expression pattern during development has not been reported. We first investigated the developmental expression pattern of LRRC10 in the mouse. Whole-mount in situ hybridization showed that Lrrc10 was detected specifically in the cardiac crescent region of embryonic day (E) 7.5 mouse embryos with an antisense Lrrc10 probe, but not with a sense Lrrc10 probe (compare Fig. 1A with 1C). The heart founders located bilaterally in the anterior–lateral plate mesoderm have been designated the primary heart fields (for reviews, see Kelly and Buckingham, 2002; Abu-Issa et al., 2004). The cranial-most parts of the heart fields are drawn to the midline creating the cardiac crescent, a horseshoe-shaped field composed of the bilateral heart fields joined in the midline. The secondary heart field is a recently described developmental field that contributes to the developing outflow tract (Mjaatvedt et al., 2001; Waldo et al., 2001) and right ventricle (Kelly et al., 2001). The Nkx2.5 antisense probe was used as an early cardiac marker (Fig. 1B), which is expressed in the primary and secondary heart fields (Waldo et al., 2005). Lrrc10 was detected in a restricted region of the precardiac region, which appears to be the primary heart field as compared with a broader expression pattern of Nkx2.5. However, it is possible that this may be due to a lower expression level of Lrrc10. Upon the completion of the looping process, the heart showed cardiac-specific expression of Lrrc10 at E9.5 (Fig. 1D). The Lrrc10 expression domain again seems to be restricted compared with the Nkx2.5 expression especially at the proximal region of the arterial and venous poles and pharyngeal floor (Fig. 1E; Lints et al., 1993).
Fig. 1.

Cardiac-specific expression of Lrrc10 during development. A–D: Mouse embryos (embryonic day [E] 7.5, and E9.5) were subjected to whole-mount in situ hybridization using digoxigenin-labeled Lrrc10 antisense cRNA probes. G–K: Section in situ hybridization was performed on transverse sections of E12 mouse embryos using 35S-labeled probes. B,E,H,K: Nkx2.5 antisense cRNA probe was used for positive control to visualize the heart. C,F,I: Negative control was hybridized with sense Lrrc10 probe. J and K are shown at a higher magnification of G and H, respectively. To show the endocardial cushion, brightness and contrast were enhanced in J and K. Arrows and red arrowheads indicate cardiac region and endocardial cushion, respectively. SP, spinal cord; A, atrium; V, ventricle; SV, superior vena cava valve; AS, atrial septum.
At E12, section in situ hybridization was performed to examine the expression profile within the mouse heart. Lrrc10 was observed in the myocardium of the atrium and ventricle (Fig. 1G). In the ventricle, Lrrc10 was observed in the compact and trabecular layers, and in the interventricular septum. In the atrium, Lrrc10 was detected in the atrial wall, the developing atrial septum, and the superior vena cava valve (Fig. 1G) where Nkx2.5 was expressed (Fig. 1H). Neither Lrrc10 nor Nkx2.5 was detected in the endocardial cushion that is indicated by red arrowhead (Fig. 1J,K). To visualize the endocardial cushion, contrast was increased in Figure 1J,K. These expression studies indicate that Lrrc10 is expressed from the precardiac region and its expression largely overlaps with that of Nkx2.5, a cardiomyocyte marker.
Next, we investigated whether the level of Lrrc10 expression is regulated during development. To examine the level of Lrrc10 expression quantitatively, Northern blot analyses were performed using the RNA from mouse hearts from different developmental stages (Fig. 2A). Lrrc10 expression was readily detectable in the heart at E12.5, and a similar level of expression was maintained during embryonic development. Lrrc10 expression was markedly elevated in the neonatal heart (approximately fourfold increase by NIH Image J analysis), and a high level of expression was maintained in the adult mouse heart. Next, we investigated whether LRRC10 is also cardiac-specific in other vertebrates including human and zebrafish, because orthologous genes may have different expression patterns. Human LRRC10 encodes 277 amino acids, which is 95% homologous to mouse LRRC10. Human LRRC10 transcripts of an expected size of 2.3 kb were detected only in the adult heart by Northern blot analyses, confirming cardiac-specific expression of LRRC10 in humans (Fig. 2B). Zebrafish embryos and adults also showed remarkably cardiac-specific expression of lrrc10 by whole-mount in situ hybridization and reverse transcriptase-polymerase chain reaction (data not shown).
Fig. 2.

The expression patterns of Lrrc10. A: The level of Lrrc10 expression in the heart increases upon birth. Total RNA extracted from the mouse heart (20 μg/lane) was subjected to Northern blot analysis using 32P-labeled Lrrc10 probe. The Gapdh probe is a loading control. Lanes 1 to 5 indicate the heart from embryonic day (E) 12.5, E14.5, E16.5, newborn, and adult mice. B: Cardiac-specific expression of LRRC10 in human. Human RNA blot (Clontech) was subjected to Northern blot analysis using 32P-labeled LRRC10 probe. The β-actin probe is a loading control. Lanes 1 to 8 indicate the heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas.
Thus far, we have demonstrated that Lrrc10 is exclusively expressed in the heart region from early embryos to adult in vertebrates. Lrrc10 expression in the mouse is markedly elevated upon birth when increased cardiac function is required to support increased physical activities, suggesting a critical role of LRRC10 in the developing and adult heart.
Subcellular Localization of LRRC10 in Transfected Cells
Because reports of the subcellular localization of LRRC10 conflict, we conducted a thorough investigation into the intracellular location of LRRC10. To detect endogenous LRRC10 protein, we generated an antibody that is highly specific to mouse LRRC10. The LRRC10 antibody was characterized by Western blot analysis (Fig. 3A). One band at the expected size of 32 kDa was detected in the heart and primary cardiomyocyte extracts (lanes 2 and 5, respectively), but not in the liver (lane 1). This band comigrated with the protein in 10T1/2 cells transfected with pcDNA3.1-Lrrc10 (lane 3), which was absent in control 10T1/2 cells (lane 4), confirming that the band is indeed LRRC10. These results demonstrate that this antibody is highly specific to LRRC10, which was used for characterizing the endogenous expression patterns of LRRC10.
Fig. 3.

Detection of the leucine-rich repeat containing protein 10 (LRRC10) protein. A: Western blot analysis of LRRC10. Various extracts were subjected to Western blot analyses using affinity purified anti-LRRC10 antibodies. LRRC10 was detected as a 32-kDa band. Lanes 1 to 5 indicate the cell extract from the adult liver, heart, 10T1/2 transfected with Lrrc10-pcDNA3.1, 10T1/2, and neonatal primary cultured cardiomyocytes, respectively. B: Intracellular localization of LRRC10 in transfected fibroblasts. 10T1/2 fibroblast cells were transfected with Lrrc10-pEGFP-N1. LRRC10-GFP was detected in the nucleus (a) or cytoplasm (b) or in both (c) in transfected cells. Nuclei were visualized by Hoechst staining.
To determine the intracellular localization of cells overexpressing the protein, LRRC10 fused to green fluorescent protein (GFP) was expressed in 10T1/2 cells. LRRC10-GFP was detected in the cytoplasm or both in the nucleus and cytoplasm of most transfected cells (Fig. 3Bb, or c, respectively). Some transfected cells showed the nuclear localization of LRRC10-GFP (Fig. 3a). GFP alone showed the typical diffuse pattern both in the cytoplasm and nucleus (data not shown). To exclude the possibility of cell-specific or tag-dependent artifacts, the Lrrc10 plasmid with or without a myc tag was transfected into 10T1/2 or Cos cells followed by immunostaining with LRRC10 or myc-antibodies. In all cases, LRRC10 showed a similar intracellular distribution to LRRC10-GFP (data not shown).
It has been reported that the cytoplasmic LRRC10 colocalized with mitochondria (Nakane et al., 2004). However, our data showed that the cytoplasmic LRRC10 was not colocalized with a mitochondrial marker (MitoTracker, Molecular Probes) both in transfected cells and primary cultured neonatal mouse cardiomyocytes (data not shown).
Subcellular Localization of LRRC10 in Developing Cardiomyocytes
We next set out to determine the intracellular localization of endogenous LRRC10 during development by immunohistochemical staining. Primary cultured cardiomyocytes prepared from neonatal mice were immunostained with anti-LRRC10 antibody. The MF20 antibody that recognizes sarcomeric myosin heavy chain (MHC) was used for a cardiomyocyte marker, because the culture contains noncardiomyocytes such as fibroblasts. As Figure 4 shows, diffuse LRRC10 staining was observed only in cardiomyocytes (indicated by arrowheads) and not in fibroblasts. LRRC10 was detected both in the nucleus and cytoplasm of some cardiomyocytes (Fig. 4A), but only in the cytoplasm of other cardiomyocytes (Fig. 4B). Primary cardiomyocytes from embryonic heart at E14 showed the same LRRC10 staining pattern as neonatal cardiomyocytes (data not shown). These expression patterns suggest that LRRC10 is distributed both in the cytoplasm and nucleus in neonatal or embryonic cardiomyocytes, suggesting it shuttles between the cytoplasm and nucleus.
Fig. 4.
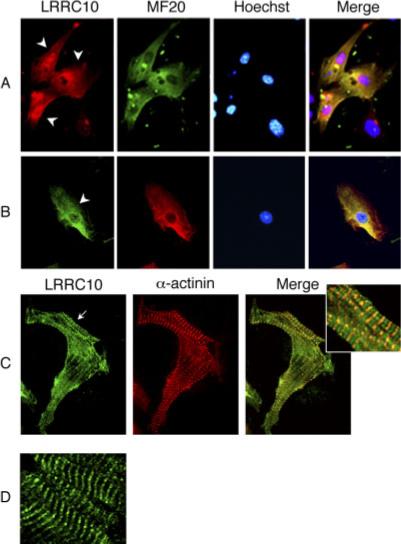
Intracellular localization of endogenous leucine-rich repeat containing protein 10 (LRRC10) in cardiomyocytes. A–C: Immunostaining of primary cultured cardiomyocytes. Neonatal mouse cardiomyocytes grown on cover slips for 2 days (A,B) or 7 days (C) were fixed in 100% methanol. Fixed cells were incubated with anti-LRRC10 and MF20 or cardiac α-actinin antibodies followed by secondary antibodies coupled to Alexa 488 or 546. D: The striated expression pattern of LRRC10 in adult heart sections. Frozen mouse heart sections were immunostained with LRRC10 antibodies followed by anti-rabbit immunoglobulin G–Alexa 488. Arrowheads indicate cardiomyocytes that are MF20-positive. Arrow points the striated pattern of LRRC10 in neonatal cardiomyocytes.
Of interest, a small percentage of neonatal cardiomyocytes grown in our culture condition for 1 or 2 days showed LRRC10 striations in the cytoplasm (Fig. 4C), and this number was increased when cultured for a longer period of time. These observations are consistent with Adameyko et al. (2005). To identify the subcellular localization of LRRC10, co-immunostaining experiments were performed using confocal microscopy. Of interest, LRRC10 was reproducibly colocalized with cardiac α-actinin, a Z-line marker (Fig. 4C). We also observed inconsistently an additional band between the Z-lines in immature cardiomyocytes. It is possible that the localization of LRRC10 near the Z-line is stable under our culture and immunostaining conditions, but the localization between the Z-lines is either labile or transient in nature because these bands were not readily observed in mature cardiomyocytes (see Fig. 5). We next examined the expression pattern of LRRC10 in the adult heart. Frozen adult mouse heart was sectioned for immunostaining experiments, which showed striking striations (Fig. 4D).
Fig. 5.

Leucine-rich repeat containing protein 10 (LRRC10) is localized near the Z-line in adult cardiomyocytes. Isolated adult mouse cardiomyocytes were double immunostained with anti-LRRC10 (A,D,G) with cardiac α-actinin, a Z-line marker (B), ryanodine receptor (RyR) in the sarcoplasmic reticulum (E), or creatine kinase (CK) MM, an M-line marker (H) antibody. Cells were incubated with the secondary antibodies coupled to Alexa 488 or 546 and examined using confocal microscope. The right column shows striations in the indicated square at a higher magnification.
Subcellular Localization of LRRC10 in Adult Cardiomyocytes
T-tubules are always in contact with the SR, forming a diad at the Z-line. Therefore, our results so far suggest that LRRC10 is associated with the Z-line in the sarcomere or the SR, or the T-tubule as cardiomyocytes become mature. Because the T-tubules are not formed until 2 weeks after birth in mice, initial striations of LRRC10 observed in some embryonic and neonatal cardiomyocytes do not seem to be dependent on the formation of the T-tubule.
To determine the detailed subcellular localization of LRRC10 in mature cardiomyocytes, a series of co-immunostaining experiments were performed using various subcellular markers that show striations (Fig. 5). Adult cardiomyocytes isolated from the mouse heart were subjected to immunostaining and examined by confocal microscopy. LRRC10 colocalized with the Z-line associated protein cardiac α-actinin (Fig. 5A–C), and with the SR membrane protein the ryanodine receptor (Fig. 5D–F). The ryanodine receptor is a Ca2+-dependent Ca2+ release pump in the SR membrane, which is concentrated in the region of SR that interacts with the T-tubule. Therefore, ryanodine receptors show striations colocalizing with a Z-line marker (Benkusky et al., 2004). To further confirm the subcellular localization, we co-immunostained cardiomyocytes with a creatine kinase-MM antibody, an M-line marker located in the middle of the A band (Fig. 5G–I). LRRC10 showed an alternate expression pattern with the M-line marker, indicating LRRC10 is not localized on the M-line or A band. LRRC10 was detected only in the cytoplasm of most adult cardiomyocytes, as shown in Figure 5. Negative controls using nonspecific rabbit or mouse IgG did not show any specific staining (data not shown).
These results so far demonstrate that LRRC10 is colocalized with Z-line markers, which suggest that LRRC10 is localized either on the Z-line or closely associated with a diad region where the SR membrane interacts with the T-tubule. To determine whether LRRC10 is closely associated with the T-tubule, the sarcolemma was removed by detergent (skinned cardiomyocytes) to remove the T-tubule (Wang et al., 2006). Removal of the T-tubule was confirmed by staining with wheat germ agglutinin (WGA) that recognizes glycosylated membrane components (compare b and f in Fig. 6A). As shown in Figure 6Ab, WGA marked the T-tubule, which is colocalized with LRRC10 (Fig. 6Ad). When T-tubules were removed (Fig. 6Af), the striated pattern of LRRC10 was also abolished (Fig. 6Ae). In our skinned cardiomyocytes, the SR membrane or sarcomere structure was intact as evidenced by the striated pattern of ryanodine receptor of SR or cardiac α-actinin, respectively (Fig. 6B or C). Intense staining of LRRC10 in some skinned cardiomyocytes was observed at former intercalated discs at the ends of cardiomyocytes and in cell membrane (see Fig. 6Ae). However, this staining pattern appears to be an artifact of skinning, because we have not observed this pattern in normal isolated cardiomyocytes (Figs. 4, 5) or in intact myocardium (data not shown).
Fig. 6.

Examination of leucine-rich repeat containing protein 10 (LRRC10) localization using skinned adult cardiomyocyte. A–C: Intact or skinned cardiomyocytes isolated from adult rats were co-immunostained with anti-LRRC10 and wheat germ agglutinin (WGA) -conjugated with Alexa Flour 488, T-tubule marker (A) or RyR, sarcoplasmic reticulum marker (B) or cardiac α-actinin, sarcomeric Z-line marker (C) antibodies. D: C2C12 myoblasts transfected with Lrrc10-pcDNA3.1-myc (left column) were differentiated into myotubes (right column) and immunostained with anti-LRRC10 antibody. E: C2C12 myotubes were co-immunostained with anti-myc and α-actinin (sarcomeric Z-line marker) or myomesin (sarcomeric M-line marker) antibodies and shown at a higher magnification. Cells were incubated with the secondary antibodies coupled to Alexa 488 or 546 and examined under confocal microscope. Nuclei were visualized by Hoechst staining.
Cardiac and skeletal muscles share many contractile proteins and the same basic sarcomeric structure. To examine whether the periodic sarcomeric structure is sufficient for LRRC10 striations, we examined the subcellular expression pattern of LRRC10 in skeletal myoblasts and myotubes. Because LRRC10 is not expressed in skeletal muscle, LRRC10 was transfected into C2C12 skeletal myoblasts, which were then induced to differentiate to form myotubes. In both C2C12 myoblasts and myotubes, LRRC10 showed a diffuse staining pattern mainly in the cytoplasm (Fig. 6D, left and right columns, respectively). When C2C12 cells were differentiated into mature multinuclear myotubes, they exhibited organized sarcomere structures as evidenced by striations of cardiac α-actinin (Z-line marker) and myomesin (M-line marker; Fig. 6E). LRRC10 still showed the diffuse staining pattern in the cytoplasm of mature myotubes, indicating that the sarcomere structure alone is not sufficient for assembling LRRC10 striations.
In summary, our data indicate a dynamic intracellular expression profile of LRRC10. The diffuse expression pattern of LRRC10 in developing cardiomyocytes becomes a striated pattern in the adult heart, which colocalizes with the SR and T-tubule markers, independent from the formation of the sarcomeric Z-line.
Electron Microscopic Examination
To define the precise subcellular location of LRRC10, transmission electron microscopic (TEM) analyses were performed on adult mouse heart sections. Heart sections were incubated with anti-LRRC10 antibodies and gold-labeled secondary antibodies. LRRC10 was detected mainly at the diad region where the T-tubule interacts closely with the SR, but neither on the Z-line nor mitochondria (Fig. 7A,B). The negative control using nonspecific rabbit IgG did not show any specific immunogold labeling (Fig. 7C,D). These results correlated well with the co-immunostaining results presented in Figures 5 and 6.
Fig. 7.
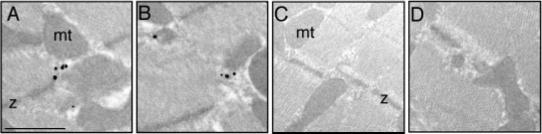
Electron micrographs showing immunogold labeling of leucine-rich repeat containing protein 10 (LRRC10). Adult mouse heart tissue was incubated with an affinity-purified LRRC10 antibodies followed by gold-labeled secondary anti-rabbit antibodies. A,B: Black dots represent the LRRC10 localization at the diad region where the sarcoplasmic reticulum (SR) interacts with the T-tubule. C,D: Negative controls were performed with nonspecific rabbit immunoglobulin G instead of anit-LRRC10 antibodies followed by gold labeled secondary antibodies. The representative pictures are shown from three independent experiments. z, Z-line; mt, mitochondria. Scale bar = 1 μm.
DISCUSSION
In this study, we determined the subcellular distribution of the LRRC10 protein during development and in the adult heart. LRRC10 was detected both in the cytoplasm and nucleus of embryonic and neonatal primary cultured cardiomyocytes. LRRC10 may shuttle between two compartments and have different functions depending on the subcellular location. It is plausible that cytoplasmic LRRC10 interacts with signal transduction pathways, metabolic enzymes, or Ca2+ handling proteins or a developing contractile apparatus. In the nucleus, LRRC10 may regulate transcriptional networks involved in cardiac-specific gene expression by interacting with other nuclear factors. We cannot exclude the possibility that LRRC10 may be passively translocated to the nucleus due to a relatively small molecular weight of 32 kDa. The nuclear pore exclusion limit is generally believed to be approximately 20−40 kDa, without considering the shapes or net charges of the protein (Ribbeck and Gorlich, 2001). However, this is unlikely because LRRC10-GFP, with an estimated molecular weight of 59 kDa, also showed the cytoplasmic and nuclear distribution. Although there is a putative nuclear localization like signal (NLS) in mouse LRRC10 (aa 249−255), this sequence is not well conserved in other species. This putative NLS is either absent in pig, chicken, and frog or completely substituted in zebrafish.
LRRC10 showed a striated pattern in isolated adult cardiomyocytes and frozen heart sections using our LRRC10 antibodies. Co-immunostaining analyses indicated that LRRC10 was colocalized with the Z-line markers, including cardiac α-actinin, the ryanodine receptor in the SR, and the T-tubule that wraps each myofibril at every Z-disc. In skinned cardiomyocytes where the T-tubule was removed, the striated pattern of LRRC10 was abolished. Moreover, TEM analyses indicated that LRRC10 was located at the diad region where the SR interacts with the T-tubules. Because LRRC10 lacks any membrane-spanning domain, LRRC10 is likely to be located in the cytosolic fraction at the diad region to interact with the components of the SR membrane, T-tubules, or cytoplasmic proteins, which are concentrated in this region.
The contractile proteins show striated patterns indicating the Z-line, M-line, A band, and I band. The Z-line represents a critical link between the T-tubule network and cytoskeleton of cardiac cells with a role in anchoring structural proteins, ion channels, and signaling molecules. For example, protein kinase C-epsilon regulates cardiac excitability, cardiac protection, and growth, possibly as a consequence of translocation to the Z-line/T-tubule region (Kang and Walker, 2005; Robia et al., 2005). Integrin-linked kinase (ILK), a serine/threonine protein kinase, is expressed at high levels in cardiac and skeletal muscle. ILK specifically localizes to sarcomeric Z-discs and controls contractility in the zebrafish heart (Bendig et al., 2006). It has been reported that the presence of active glycolytic enzymes in the cell is not sufficient for muscle function; colocalization of the enzymes in the sarcomere is required for muscle contraction (Sullivan et al., 2003). In addition, some transmembrane components, such as the endothelin receptor and L-type Ca2+ channels in sarcolemma are preferentially localized in the T-tubule (Puri et al., 1997; Robu et al., 2003). Ca2+/calmodulin-dependent protein kinase (CaMKIIβ) in muscle exists in complex with enzymes involved in glycolysis at the SR membrane. The functional significance of the SR membrane bound CaMKIIβ may be to target the glycolytic machinery to the SR and modulate the local levels of NADH and ATP in response to calcium signaling (Singh et al., 2004).
In contrast to our findings, LRRC10 has been reported to be a doublet in the I band flanking the Z-line of sarcomeres (Adameyko et al., 2005), or in the nucleus in adult cardiac tissue (Nakane et al., 2004). Reasons for this discrepancy are not clear, but it may be due to different characteristics of antibodies, different sample preparation, and different fixing methods. There is a possibility that LRRC10 binds to more than one site with different affinities, leading to either stable or labile interactions. A critical future direction will be to identify physical interacting factors of LRRC10 to determine the molecular/cellular functions of LRRC10. The present study describing the precise subcellular localization of LRRC10 in the developing and adult heart will provide an important basis to further investigate the biological roles of LRRC10. While this manuscript was in press, we have described the critical role of Lrrc10 in heart development and function in zebrafish (Kim et al., 2007).
EXPERIMENTAL PROCEDURES
Cloning of Lrrc10
We identified EST clones encoding Lrrc10 as a putative cardiac-specific gene by searching a mouse E13 embryonic heart EST library in the NCBI database. The full-length cDNA consisting of the entire coding sequence, and 5′ and 3′ untranslated regions in pDONR201 vector was obtained from Open Biosystems (GenBank accession no. CA460331.1) and sequenced. The nucleotide sequence of Lrrc10 is identical to the reported cDNA sequence (gi: 113930741). The cDNA was subcloned into pCRII-TOPO vector (Invitrogen) to examine the expression pattern by in situ hybridization and Northern blotting. Subsequently, various mammalian expression plasmids were constructed by standard methods: Lrrc10-pcDNA3.1, Lrrc10-pcDNA3.1-myc, and Lrrc10-pEGFP-N1 (Invitrogen). The sequence was confirmed by diagnostic restriction enzyme digestion and sequencing.
Anti-LRRC10 Antibody Generation, Cell Culture, and Transfections
Polyclonal anti-LRRC10 antibodies were generated in rabbit by injecting a synthesized peptide by standard methods (Covance, Inc.). The peptide [H]–CRKARRYALAKEE-[OH], corresponding to amino acids 250−261 in mouse LRRC10 was selected as an immunogen, which is highly antigenic and hydrophilic using the Kyte-Doolittle software program (UWGCG). Antiserum was first screened by an enzyme-linked immunosorbent assay against the immunogen peptide and characterized by Western blotting and immunostaining. The antiserum was subsequently affinity purified by a peptide column, which is highly specific to LRRC10 (Fig. 3A).
10T1/2 cells were cultured in a medium consisting of 10% fetal bovine serum in Dulbecco's modified Eagle's medium (DMEM) supplemented with penicillin and streptomycin (Invitrogen). C2C12 myoblasts were maintained in 20% fetal bovine serum (FBS). To differentiate C2C12 myoblasts to myotubes, differentiation medium consisting of 10% heat-inactivated horse serum was used, which was replenished every 2 days. Transient transfections of Cos, 10T1/2, and C2C12 myoblasts with various plasmids encoding Lrrc10 were performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
In Situ Hybridization and Northern and Western Blot Analyses
Whole-mount in situ hybridization of mouse embryos was performed as previously described (Sun et al., 2002). To examine the expression of Lrrc10 mRNA in mouse embryos, the antisense cRNA probe for full-length Lrrc10 were labeled by digoxigenin-11-UTP (Roche Diagnostics Corp). For section in situ hybridization, 35S-labeled cRNA probes were used as previously described (Lee et al., 2000; Micales and Lyons, 2001). The sense probe for Lrrc10 was synthesized as a negative control, and the antisense probe for full-length Nkx2.5 was synthesized as a cardiac marker.
Northern blot analyses were performed as described elsewhere (Kim et al., 2005). Total RNA isolated from the mouse heart on the fixed membrane was hybridized with a 32P-labeled probe by using Prime-a-Gene random prime labeling system (Promega) followed by autoradiography. To verify consistent loading, the membrane was stripped and reprobed for glyceraldehyde-3-phosphate dehydrogenase (Gapdh) or β-actin.
Western blot analyses were performed as previously described (Kim et al., 2003). Briefly, total protein extracts from the adult mouse liver and heart and whole cell lysates from neonatal primary cultured cardiomyocytes and 10T1/2 cells were loaded (30 μg protein /lane) onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred onto a polyvinylidene difluoride membrane. The membrane was incubated with purified anti-LRRC10 antibody (1.7 μg/ml), and the protein was detected with an ECL kit (Amersham Biosciences).
Isolation of Cardiomyocytes and Immunohistochemistry
Neonatal mouse cardiomyocytes were prepared as described previously (Jung et al., 2005a). Briefly, the isolated cells were plated in DMEM supplemented with 10% FBS. The next day, the medium was replaced by DMEM supplemented with 5% horse serum. Neonatal cardiomyocytes were also cultured on laminin-coated cover-slips for immunostaining experiments. Adult mouse and rat cardiomyocytes were enzymatically isolated as previously described (Kang and Walker, 2005; Balijepalli et al., 2006). Cardiomyocytes were skinned by incubation for 10 min in relaxing solution (100 mM KCl, 1 mM MgCl2, 2 mM ethyleneglycoltetraacetic acid, 4.5 mM ATP, and 10 mM imidazole pH 7.0) with 1% Triton X-100 (Wang et al., 2006).
Immunostaining was performed as previously described (Kim et al., 2003). Briefly, cells fixed with cold methanol were blocked for 1 hr in phosphate buffered saline (PBS), 1% bovine serum albumin, 5% FBS, 10% goat serum, and 0.05% Triton X-100. Next, the cells were incubated in primary antibodies diluted in PBS, 5% goat serum, and 0.2% Triton X-100 for 2 hr followed by secondary antibodies for 1 hr. Cells were stained with Hoechst 33258 (Molecular Probes) to visualize nuclei and mounted with Prolong mounting medium (Invitrogen), and examined using confocal microscope (Bio-Rad radiance 2100 MP Rainbow, W.M. Keck Laboratory at University of Wisconsin). The following antibodies were used as primary antibodies: affinity purified rabbit anti-LRRC10 antibody (17 μg/ml), mouse anti-MF20 antibody (Developmental Studies Hybridoma Bank, IA, 1:200 dilution), mouse anti-α-actinin antibody (Sigma, 1:3,000 dilution), mouse anti-ryanodine receptor antibody (Affinity BioReagents, 1:500 dilution), rabbit anti-myomesin antibody (Atlas Antibodies, 1:500 dilution), mouse or rabbit anti-myc antibody (Santa Cruz Biotechnology, 1:200 dilution), and mouse anti–creatine kinase-MM antibody (Fitzgerald, 1:100 dilution). Secondary antibodies were goat anti-rabbit Alexa Fluor 488, or 546 antibodies, and goat anti-mouse Alexa Fluor 488, or 546 antibodies (Molecular Probes). WGA conjugated to Alexa Fluor 488 (Invitrogen, 10 μg/ml) was used to detect the sarcolemma including T-tubules.
Immunoelectron Microscopy
Transmission electron microscopic analyses were performed at the Electron Microscopic Core Facility (University of Wisconsin) as described elsewhere (Massion et al., 2004). Small blocks (0.5 mm3) from hearts fixed with 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M PBS were dehydrated in alcohol and embedded into soft grade LR Gold resin (Sigma). Rehydrated ultrathin sections collected on nickel grids were pretreated with glycine (0.05 mol/L), blocked with AURION blocking solution (Hatfield, Pa), and incubated with affinity purified anti-LRRC10 antibody (1.7 μg/ml). Sections were incubated with Ultra Small gold-conjugated anti-rabbit antibodies (Aurion, 1:100 dilution), counterstained with uranyl acetate and lead citrate, and examined with a Philips CM120 STEM.
ACKNOWLEDGMENTS
We thank Drs. Timothy R. Kamp and Jeffrey W. Walker for valuable discussions. We thank Dr. Marion L. Greaser for generously providing antibodies. We also thank Ka Young Chung and Jing Wang for preparation of adult cardiomyocytes and Matthew R. Mysliwiec for critical reading of this manuscript. Y.L. was funded by the National Institutes of Health, and G.E.L. by American Heart Association GIA 0650032Z.
Grant sponsor: NIH; Grant number: HL67050.
REFERENCES
- Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–285. doi: 10.1016/j.ydbio.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Adameyko II, Mudry RE, Houston-Cummings NR, Veselov AP, Gregorio CC, Tevosian SG. Expression and regulation of mouse SERDIN1, a highly conserved cardiac-specific leucine-rich repeat protein. Dev Dyn. 2005;233:540–552. doi: 10.1002/dvdy.20368. [DOI] [PubMed] [Google Scholar]
- Balijepalli RC, Foell JD, Hall DD, Hell JW, Kamp TJ. Localization of cardiac L-type Ca(2+) channels to a caveolar macromolecular signaling complex is required for beta(2)-adrenergic regulation. Proc Natl Acad Sci U S A. 2006;103:7500–7505. doi: 10.1073/pnas.0503465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, Trano N, Katus HA, Fishman MC, Rottbauer W. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006;20:2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkusky NA, Farrell EF, Valdivia HH. Ryanodine receptor channelopathies. Biochem Biophys Res Commun. 2004;322:1280–1285. doi: 10.1016/j.bbrc.2004.08.033. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403:676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Buchanan SG, Gay NJ. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature. 2000;407:227–232. doi: 10.1038/35025196. [DOI] [PubMed] [Google Scholar]
- Gruber PJ, Epstein JA. Development gone awry: congenital heart disease. Circ Res. 2004;94:273–283. doi: 10.1161/01.RES.0000116144.43797.3B. [DOI] [PubMed] [Google Scholar]
- Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- Jung J, Kim TG, Lyons GE, Kim HR, Lee Y. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J Biol Chem. 2005a;280:30916–30923. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn. 2005b;232:21–32. doi: 10.1002/dvdy.20204. [DOI] [PubMed] [Google Scholar]
- Kang M, Walker JW. Protein kinase C delta and epsilon mediate positive inotropy in adult ventricular myocytes. J Mol Cell Cardiol. 2005;38:753–764. doi: 10.1016/j.yjmcc.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 2002;18:210–216. doi: 10.1016/s0168-9525(02)02642-2. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Kim K, Antkiewicz DS, Yan L, Eliceriri KW, Heideman W, Peterson RE, Lee Y. Lrrc10 is required for early heart development and function in zebrafish. Dev Biol. 2007a doi: 10.1016/j.ydbio.2007.06.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278:42247–42255. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- Kim TG, Jung J, Mysliwiec MR, Kang S, Lee Y. Jumonji represses alpha-cardiac myosin heavy chain expression via inhibiting MEF2 activity. Biochem Biophys Res Commun. 2005;329:544–553. doi: 10.1016/j.bbrc.2005.01.154. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–120. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Lange S, Ehler E, Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Linhoff MW, Harton JA, Cressman DE, Martin BK, Ting JP. Two distinct domains within CIITA mediate self-association: involvement of the GTP-binding and leucine-rich repeat domains. Mol Cell Biol. 2001;21:3001–3011. doi: 10.1128/MCB.21.9.3001-3011.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Massion PB, Dessy C, Desjardins F, Pelat M, Havaux X, Belge C, Moulin P, Guiot Y, Feron O, Janssens S, Balligand JL. Cardiomyocyte-restricted overexpression of endothelial nitric oxide synthase (NOS3) attenuates beta-adrenergic stimulation and reinforces vagal inhibition of cardiac contraction. Circulation. 2004;110:2666–2672. doi: 10.1161/01.CIR.0000145608.80855.BC. [DOI] [PubMed] [Google Scholar]
- Micales BK, Lyons GE. In situ hybridization: use of 35S-labeled probes on paraffin tissue sections. Methods. 2001;23:313–323. doi: 10.1006/meth.2000.1143. [DOI] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Mutai H, Toyoshima Y, Sun W, Hattori N, Tanaka S, Shiota K. PAL31, a novel nuclear protein, expressed in the developing brain. Biochem Biophys Res Commun. 2000;274:427–433. doi: 10.1006/bbrc.2000.3133. [DOI] [PubMed] [Google Scholar]
- Nakane T, Satoh T, Inada Y, Nakayama J, Itoh F, Chiba S. Molecular cloning and expression of HRLRRP, a novel heart-restricted leucine-rich repeat protein. Biochem Biophys Res Commun. 2004;314:1086–1092. doi: 10.1016/j.bbrc.2003.12.202. [DOI] [PubMed] [Google Scholar]
- Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10:467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev. 2003;17:1937–1956. doi: 10.1101/gad.1110103. [DOI] [PubMed] [Google Scholar]
- Passier R, Richardson JA, Olson EN. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev. 2000;92:277–284. doi: 10.1016/s0925-4773(99)00330-5. [DOI] [PubMed] [Google Scholar]
- Puri TS, Gerhardstein BL, Zhao XL, Ladner MB, Hosey MM. Differential effects of subunit interactions on protein kinase A- and C-mediated phosphorylation of L-type calcium channels. Biochemistry. 1997;36:9605–9615. doi: 10.1021/bi970500d. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robia SL, Kang M, Walker JW. Novel determinant of PKC-epsilon anchoring at cardiac Z-lines. Am J Physiol Heart Circ Physiol. 2005;289:H1941–H1950. doi: 10.1152/ajpheart.01111.2004. [DOI] [PubMed] [Google Scholar]
- Robu VG, Pfeiffer ES, Robia SL, Balijepalli RC, Pi Y, Kamp TJ, Walker JW. Localization of functional endothelin receptor signaling complexes in cardiac transverse tubules. J Biol Chem. 2003;278:48154–48161. doi: 10.1074/jbc.M304396200. [DOI] [PubMed] [Google Scholar]
- Rottbauer W, Wessels G, Dahme T, Just S, Trano N, Hassel D, Burns CG, Katus HA, Fishman MC. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99:323–331. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MD, Gaussin V, Lyons KM. Tempting fate: BMP signals for cardiac morphogenesis. Cytokine Growth Factor Rev. 2003;14:1–4. doi: 10.1016/s1359-6101(02)00053-9. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Singh P, Salih M, Leddy JJ, Tuana BS. The muscle-specific calmodulin-dependent protein kinase assembles with the glycolytic enzyme complex at the sarcoplasmic reticulum and modulates the activity of glyceraldehyde-3-phosphate dehydrogenase in a Ca2+/calmodulin-dependent manner. J Biol Chem. 2004;279:35176–35182. doi: 10.1074/jbc.M402282200. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Sullivan DT, MacIntyre R, Fuda N, Fiori J, Barrilla J, Ramizel L. Analysis of glycolytic enzyme co-localization in Drosophila flight muscle. J Exp Biol. 2003;206:2031–2038. doi: 10.1242/jeb.00367. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418:501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- Tong ZB, Nelson LM, Dean J. Mater encodes a maternal protein in mice with a leucine-rich repeat domain homologous to porcine ribonuclease inhibitor. Mamm Genome. 2000;11:281–287. doi: 10.1007/s003350010053. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Zhang H, Kumanogoh A, Takegahara N, Yabuki M, Harada K, Hori M, Kikutani H. Guidance of myocardial patterning in cardiac development by Sema6D reverse signalling. Nat Cell Biol. 2004;6:1204–1211. doi: 10.1038/ncb1193. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Walton RZ, Bruce AE, Olivey HE, Najib K, Johnson V, Earley JU, Ho RK, Svensson EC. Fog1 is required for cardiac looping in zebrafish. Dev Biol. 2006;289:482–493. doi: 10.1016/j.ydbio.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Grant JE, Doede CM, Sadayappan S, Robbins J, Walker JW. PKC-betaII sensitizes cardiac myofilaments to Ca2+ by phosphorylating troponin I on threonine-144. J Mol Cell Cardiol. 2006;41:823–833. doi: 10.1016/j.yjmcc.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Wu H, Maciejewski MW, Marintchev A, Benashski SE, Mullen GP, King SM. Solution structure of a dynein motor domain associated light chain. Nat Struct Biol. 2000;7:575–579. doi: 10.1038/76804. [DOI] [PubMed] [Google Scholar]


