Summary
The catalytic pentad of tyrosine recombinases, that assists the tyrosine nucleophile, includes a conserved histidine/tryptophan (His/Trp-III). Flp and Cre harbor tryptophan at this position; most of their kin recombinases display histidine. Contrary to the conservation rule, Flp(W330F) is a much stronger recombinase than Flp(W330H). The hydrophobicity of Trp-330 or Phe-330 is utilized in correctly positioning Tyr-343 during the strand cleavage step of recombination. Why then is phenylalanine almost never encountered in the recombinase family at this conserved position? Using exogenous nucleophiles and synthetic methylphosphonate or 5'-thiolate substrates, we decipher that Trp-330 also assists in the activation of the scissile phosphate and the departure of the 5'-hydroxyl leaving group. These two functions are consistent with the hydrogen bonding property of Trp-330 as well as its location in structures of the Flp recombination complexes. However, van der Waals contact between Trp-330 and Arg-308 may also be important for the phosphate activation step. A structure based suppression strategy permits the inactive variant Flp(W330A) to be rescued by a second site mutation A339M. Modeling alanine and methionine at positions 330 and 339, respectively, in the Flp crystal structure suggests a plausible mechanism for active site restoration. Successful suppression suggests the possibility of evolving, by design, new active site configurations for tyrosine recombination.
Keywords: Flp, Cre, methylphosphonate, site-specific recombination, tyrosine family, 5'-thiolate
Introduction
Tyrosine family site-specific recombinases employ four subunits and utilize a type IB topoisomerase mechanism to bring about recombination between two DNA partners.1-3 Each core target site is organized as two recombinase binding elements (each a little over one turn of DNA; 11-14 bp) in head-to-head orientation flanking the strand exchange region (or spacer) that is 6-8 bp long. The reaction is carried out in two steps of single strand exchanges. The first results in the formation of a Holliday junction intermediate, and the second mediates its resolution into reciprocal recombinant products. As implied by their family name, these recombinases employ a tyrosine nucleophile to bring about DNA cleavage, which results in the covalent attachment of tyrosine to the 3'-phosphate at the strand break point. The other product of the reaction is a 5'-hydroxyl end, the nucleophile for the subsequent strand joining step. The attack by the 5'-hydroxyl from one DNA partner on the 3'-phosphotyrosyl linkage formed on the other results in a recombinant strand. Overall, one round of recombination is the sum of four mechanistically identical ‘cleavage and joining’ events. This basic reaction scheme applies to the ‘simple’ members of the family such as Flp (coded for by the Saccharomyces cerevisiae 2 micron plasmid) and Cre (from phage P1) as well as its more ‘complex’ members such as phage λ Int and XerC/XerD (from Escherichia coli). However, depending on the substrate contexts, some of the complex members have to rely on accessory protein factors and their cognate binding sites to bring about recombination.4, 5
In addition to the invariant active site nucleophile, a conserved catalytic pentad, consisting of two arginines (Arg-I, Arg-II), a lysine (Lys-β), a histidine (His-II) and a histidine/tryptophan (His/Trp-III), can be recognized among the tyrosine recombinases.3,6,7 A similar cluster is also present among the type I topoisomerases, although a lysine residue replaces His-II, and the His/Trp-III is represented by histidine.8,9 Based on structural data and extensive biochemical analyses carried out particularly on the vaccinia topoisomerase IB system,8-12 the individual contributions of a subset of these amino acids have become apparent. The arginines, hydrogen bonded to the non-bridging oxygen atoms of the scissile phosphate, not only facilitate its correct positioning but may also stabilize the negative charge built up in the transition state. Lys-β has been suggested to function as the general acid that stabilizes the 5'-hydroxyl leaving group during the strand cleavage step. Studies with the vaccinia topoisomerase implicate Arg-I to be also involved in this step either by assisting Lys-β through a proton relay mechanism or perhaps by acting as the primary general acid.9;10 The His-II residue in the tyrosine recombinases Flp and Cre are well positioned to serve as general base to activate the tyrosine nucleophile.13,14 However, mutations of this residue in Flp and Cre to leucine and alanine, respectively, do not eliminate the cleavage reaction. Whereas the Flp mutant accumulates large amounts of the cleaved product due to a strong defect in the joining reaction, the Cre mutant shows an approximately 150-fold reduction in the rate of cleavage. 15,16 As described below, the role of the His/Trp-III position in catalysis, which is the primary focus of this study, may not be uniform among tyrosine recombinases and type IB topoisomerases.
The conserved His/Trp-III corresponds to His-265 and His-632 in vaccinia and human topoisomerases, respectively, Trp-330 in Flp and Trp-315 in Cre. Structural or biochemical evidence from the topoisomerases and Cre supports the formation of a hydrogen bond between His/Trp-III and one of the non-bridging oxygen atoms of the scissile phosphate14;,17-19 In structures of the Flp-DNA complex, the positions of Trp-330 are quite interesting.13;20 In one case, within an ‘active’ Flp monomer (meaning that its active site is in the chemically competent state for strand cleavage), Trp-330 is within hydrogen bonding distance of the 5'-hydroxyl leaving group. In a second case, Trp-330 is located almost equidistant between the scissile phosphate and the leaving group. Taken together, these observations suggest that Trp-330 could potentially be involved in dynamic interactions during strand cleavage, by hydrogen bonding initially to the phosphate group and subsequently to the 5'-hydroxyl group. Thus, the hydrogen bonding character of His/Trp-III may, depending on the particular recombination system, assist phosphate positioning or leaving group departure or both. Contrary to expectation, however, replacement of Trp-330 in Flp by histidine results in a nearly inactive recombinase, whereas that by phenylalanine produces a reasonably active enzyme.20 Thus, at least in Flp, it appears that it is the hydrophobic character of Trp-330 that is pertinent to recombination. Consistent with this explanation, the crystal structure of Flp(W330F) reveals that the phenylalanine moiety from one Flp monomer interacts with several residues, including the cleavage nucleophile Tyr-343, from the M helix of its neighboring monomer. Why then is the conservation His/Trp-III, and not His/Trp/Phe-III?
We now describe appropriately designed nucleophile and substrate contexts that unveil catalytic contributions by Trp-330 that were not evident previously. The hydrophobic role of Trp-330 in orienting the Tyr-343 nucleophile does not fully account for its function in recombination. Evidence from the present study suggests that Trp-330 assists strand cleavage by also helping to balance the developing negative charge within the transition state and by promoting the stabilization of the 5'-hydroxyl leaving group. As revealed by structures of Flp recombination complexes, the hydrogen bonding potential of Trp-330, and perhaps its van der Waals contact with Arg-308, can account for these functions. By directed mutagenesis of residues selected on the basis of structure, it is possible to construct a functional Flp active site in which Trp-330 is replaced by alanine. We suggest that structure driven directed evolutionary schemes may reveal yet more novel catalytic constellations that can perform the phosphoryl transfer steps of tyrosine family recombination.
Results
Differential effects of Trp-330 mutations on strand cleavage and joining by Flp
As noted earlier, the cleavage reaction during tyrosine family recombination generates a 3'-phosphate covalently linked to the catalytic tyrosine, plus a hydroxyl group at the adjacent 5' terminus of the cleaved strand. The strand joining reaction involves the attack by the 5'-hydroxyl from one DNA substrate on the 3'-O-phosphotyrosyl bond formed on its recombination partner. Since the DNA-bonded tyrosine is more or less fixed in its position in the active site for the joining reaction, the relative importance of Trp-330 may not be the same during the cleavage and joining steps of Flp recombination.
The half-site substrate used for strand cleavage is so designed that the short trinucleotide product of the reaction (TTT with its 5'-hydroxyl end) diffuses away, and has little chance to partake in a joining reaction (Fig. 2A). Furthermore, the 5'-hydroxyl of the bottom strand is blocked from the joining reaction by phosphorylation. In the strand joining substrate, the top strand mimics the cleaved intermediate by having its 3'-phosphate end linked to a tyrosine mimic: the p-nitrophenolate moiety (Fig. 2B). The 5'-hydroxyl from the bottom strand carries out the nucleophilic attack to generate a hairpin product that is refractory to cleavage. Note that the joining reaction is independent of the cleavage nucleophile: Tyr-343; it can be carried out efficiently by Flp(Y343F).
Fig. 2.
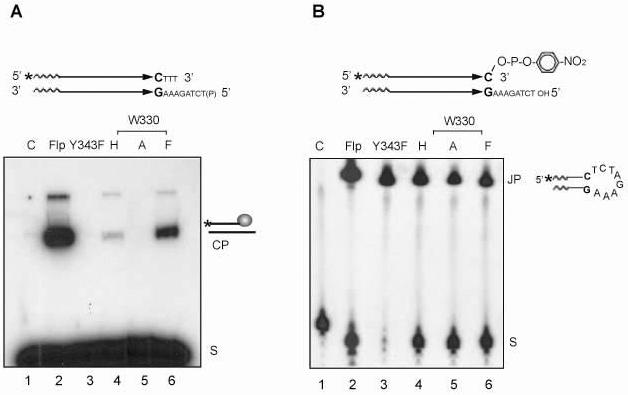
Strand cleavage and joining by Flp mutants altered at Trp-330. A. In the strand cleavage substrate, schematically diagrammed at the top, the parallel arrows ending in the CG bp represent the Flp binding element. The asterisk denotes the 32P label at the 5' end. Strand cleavage by Flp would result in the loss of the trinucleotide 5'HOTTT3' by diffusion. The 5'-hydroxyl group of the bottom strand was blocked by phosphorylation. The substrate (S) and the cleaved product covalently linked to Flp (CP) were separated by SDS-polyacrylamide gel electrophoresis. B. The 5' end-labeled substrate contained a p-nitrophenolate moiety linked to the 3'-phosphate end. The joined product (JP) was fractionated away from the substrate (S) by electrophoresis in a denaturing polyacrylamide gel. Note that some intermolecular joining reaction could also occur under the reaction conditions. In the denaturing gel, this product was not distinguishable from the hairpin product diagrammed. Reactions in A and B were carried out at 30°C for 30min. The lanes marked ‘C’ here and in other reactions (Fig. 3-6 and Fig. 8) are ‘substrate-alone’ controls.
In reactions with Flp and its three Trp-330 variants, the cleavage product was readily detected with Flp(W330F), although its amount was lower (roughly two to three fold) than that formed by wild type Flp (Fig. 2A). A trace amount of the product was seen with Flp(W330H), but no cleavage was obtained with Flp(W330A). These results are in agreement with the previous observations by Chen and Rice.20 In the joining reactions, all three Flp mutants including Flp(W330A) gave the hairpin product, albeit at somewhat lower levels than Flp(Y343F) (Fig. 2B). The yields of the joined product were roughly the same for Flp(W330F) and Flp(W330A), the former active as a recombinase and the latter inactive.
The cleavage and joining assays as well as the reactions represented in Fig. 3-6 were performed at substrate concentrations equivalent to about 3.3 nM Flp binding element and 16.5 nM Flp, close to twice its KD.21 We verified that Trp mutants bind to the Flp binding sequence with about the same affinity as wild type Flp (data not shown).
Fig. 3.
Strand cleavage by Flp and Trp-330 mutants of Flp using H2O2 and tyramine as exogenous nucleophiles. Cleavage reactions were carried out on a 5' end-labeled substrate at 30°C using 1% H2O2 (A) or 100 mM tyramine (B) as exogenously supplied cleavage nucleophiles. The products of cleavage, indicated by ‘CP’, were separated from the substrate ‘S’ by denaturing polyacrylamide electrophoresis. In the kinetic assays, samples were quenched by addition of SDS to a final concentration of 0.2% for further processing and analysis. Band intensities were quantitated using a phosphorimager. The values plotted were from four independent assays.
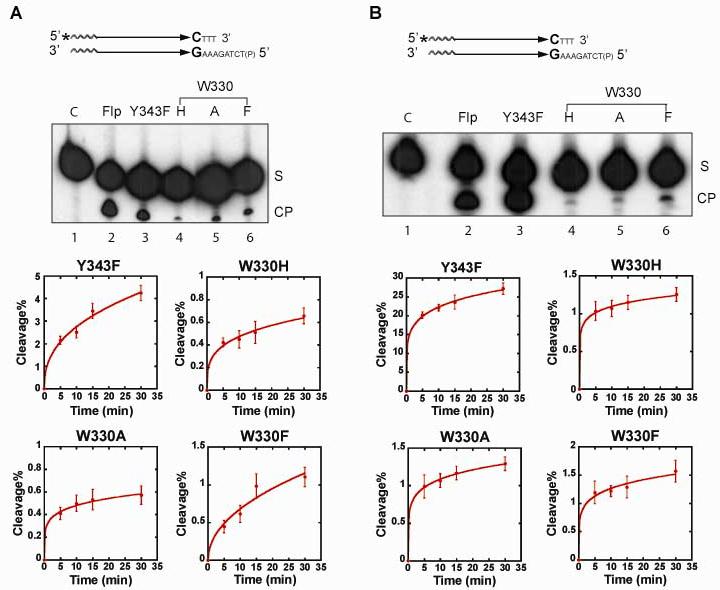
Fig. 6.
Strand cleavage by the Trp-330 Flp mutants in a substrate containing a methylphosphonate substitution at the scissile position. The substrate labeled at the 5′-end is shown schematically at the top. A. Reactions were done for 30 min at 30°C, and analyzed by denaturing polyacrylamide gel electrophoresis. ‘S’ and ‘CP’ stand for substrate and cleavage product, respectively. B. The cleavage yields were estimated by quantitating the ‘S’ and ‘CP’ bands following their electrophoretic separation. The data shown were obtained from four independent reaction sets.
The independent cleavage and joining assays denote that the role of Trp-330 is almost exclusively in the strand cleavage reaction by Flp, with little effect on the strand joining reaction. The deleterious effects of W330H and W330A mutations in the cleavage step are not due to a general perturbation of the Flp active site. The other residues of the catalytic pentad (Arg-191, Lys-223, His-305 and Arg-308) retain their functional configuration. They are essential for the strand joining reaction. They are also indispensable in an RNA cleavage reaction by Flp, which too is unaffected by Trp-330 mutations (see below).
Strand cleavage in half-sites by hydrogen peroxide and tyramine as exogenous nucleophiles
When the scissile phosphodiester bond has been activated/oriented by a bound monomer of Flp, exogenous small nucleophiles such as hydrogen peroxide and tyramine can effect strand cleavage to yield products containing 3'-phosphate and 3'-phosphotyramine ends, respectively (Fig. 3).22,23 These reactions are independent of the native nucleophile,Tyr-343, and can be performed by Flp(Y343F).
To dissociate the hydrophobic role of Trp-330 in positioning Tyr-343 from its potential additional catalytic role(s), we followed half-site cleavage by hydrogen peroxide in the presence of the appropriate Flp variants. The peroxide nucleophile is spontaneously generated in aqueous solution; and unlike Tyr-343, it can derive no orientation advantage from Trp-330. If the catalytic role of Trp-330 is entirely hydrophobic, the Trp-330 Flp mutants are expected to effect normal peroxide mediated strand cleavage. However, cleavage by all of the Trp-330 mutants was much weaker than that by Flp or Flp(Y343F) (Fig. 3A). In a time course assay, there was little difference between the cleavage profiles obtained with Flp(W330A) and Flp(W330H); the cleavage yields from Flp(W330F) were only modestly higher (roughly two fold at 30 min). As with peroxide cleavage, tyramine mediated cleavage was also markedly less efficient for the Trp-330 Flp mutants, with little distinction among Flp(W330A), Flp(W330H) and Flp(W330F) (Fig. 3B).
The above results, in conjunction with the recombination data20 (Fig. 4), are consistent with Trp-330 making a catalytic contribution that is distinct from positioning Tyr-343. This could be either at the level of phosphate activation or leaving group stabilization. The non-hydrophobic effect of Trp-330 is underscored by the uniformly weak peroxide cleavage elicited by all of the Trp-330 mutants, including Flp(W330F), a much more robust recombinase than the weakly active Flp(W330H) or the inactive Flp(W330A) (Fig. 4)20. The inefficiency of Flp(W330F) in tyramine mediated cleavage compared to Tyr-343 mediated cleavage suggests that the catalytic influence of Phe-330 cannot be realized with a freely diffusible tyrosine analog. Rather, as would be consistent with the crystal structure,20 it requires the delivery of Tyr-343 as part of a peptide segment from the neighboring Flp monomer (see also Discussion). The multiple van der Waals contacts made by Trp-330 within the Flp active site are critical in positioning Tyr-343 for cleavage.20 If His-330 fails to partake in one or more of these contacts, its location within the active site may deviate significantly from that of Phe-330. In this case, the hydrogen bonding property of His-330 cannot be functionally utilized in catalysis.
Fig. 4.
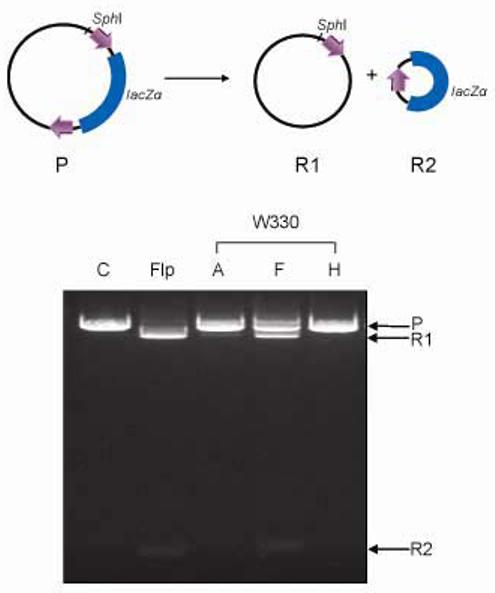
Recombination activities of Flp variants with substitutions at Trp-330. Recombination was assayed using the deletion reaction schematically indicated at the top. The substrate plasmid is the same as that employed in the in vivo assays depicted in Fig. 9 and 12. Reaction mixtures digested with SphI were analyzed by electrophoresis in agarose gels. ‘P’ and ‘R1’ refer to the parent plasmid and the larger deletion circle (or their linear forms in the ethidium bromide stained gel), respectively. ‘R2’ indicates the smaller deletion circle.
The reaction rates for tyramine mediated cleavage suggest an early burst, followed by a slower phase. This is probably due to reactions occurring in distinct oligomeric states of the Flp-bound half-site: Flp-half site monomer, dimer, trimer and tetramer. The monomeric complex may be more accessible to the exogenously supplied tyramine nucleophile compared to the higher-order complexes.
Strand Cleavage by Trp-330 (Flp) mutants in a 5'-phosphorothiolate substrate
As noted before, one of the Flp structures shows Trp-330 in the right position to form a hydrogen bond with the 5'-hydroxyl leaving group.13 It may thus assist the general acid in facilitating leaving group departure. If indeed Trp-330 plays such an accessory role, the Trp-330 mutants should be greatly aided by substituting the 5' bridging oxygen by sulfur. The significantly lower pKa of the 5'-thiol group makes it a much stronger leaving group than the corresponding hydroxyl group. As has been demonstrated for vaccinia topoisomerase, there is a very strong favorable thio-effect on a mutant enzyme that lacks the general acid residue.8
The Trp-330 Flp mutants were all active in cleaving the 5'-thiolate containing substrates (Fig. 5B). Cleavage kinetics of the phosphate and thiolate substrates revealed strong favorable thio-effects for both Flp(W330H) and Flp(W330A) (Fig. 5B). For example, at 120 min, cleavage of the normal ester bond by Flp(W330A) was barely detectable; At the same time, nearly 12% of the thioester bond was cleaved. The thio-effects for Flp(W330F) and Flp were not significantly different from each other. This was not unexpected, given that Flp(W330F) was roughly half as active as Flp in strand cleavage on phosphodiester containing substrates (Fig. 1A and 5B). Recall that the catalytic power generated by the activated Tyr-343 nucleophile is nearly the same in Flp and Flp(W330F).
Fig. 5.
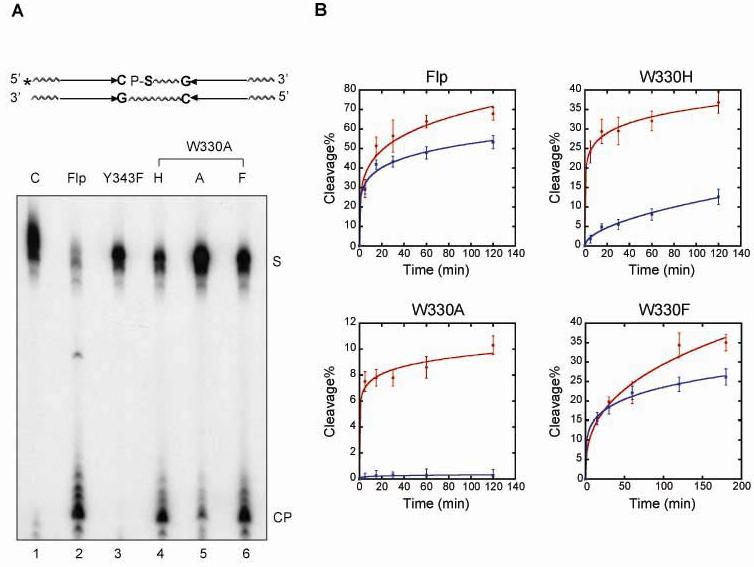
Strand cleavage by the Trp-330 Flp mutants in a substrate containing a 5'-thiolate linkage at the cleavage position. In the 5' end-labeled cleavage substrate, the scissile phosphate on the top strand was linked via a 5'-thiolate bridge to the neighboring nucleotide to the 3' side. A. Reactions from 30 min incubations at 30°C were processed, and fractionated by denaturing polyacrylamide gel electrophoresis. The substrate and product bands are denoted by ‘S’ and ‘CP’, respectively. B. Time course cleavage reactions were carried out using a standard phosphate containing substrate (blue lines) or a 5'-thiolate containing substrate (red lines). Following separation of ‘S’ and ‘CP’ by denaturing polyacrylamide electrophoresis, they were quantitated using a phosphorimager. The estimated cleavage yields for wild type Flp and the mutants were plotted as a function of time. The values were obtained from four separate assays.
Fig. 1.
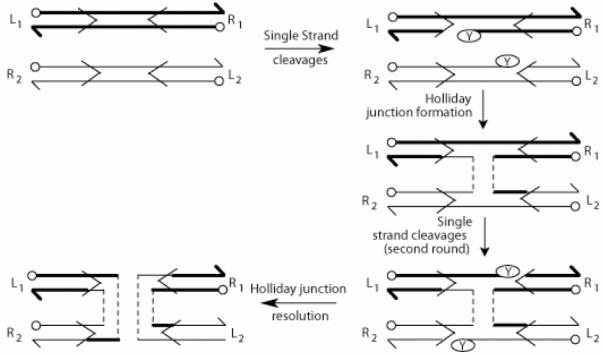
The tyrosine family recombination pathway. The cleavage/exchange of the first and second pair of strands are schematically drawn (details in text). The recombinase binding elements flanking the strand exchange region (spacer) are indicated by the horizontal arrows. The left-right orientation of the recombination partners is designated by L-R. The circular knobs and split arrow heads indicate 5' and 3'ends, respectively.
The kinetics of cleavage (Fig. 5B) suggests an initial rapid phase followed by a slower one. The two phases may represent differences in cleavage rates within a single substrate molecule bound by a Flp dimer versus that within a synapsed pair of molecules bound by a Flp tetramer. Alternatively, the slower part of the curve may indicate cleavage occurring within a Holliday junction intermediate, with isomerization of the junction being the rate limiting step. Note that the bottom strand of the substrate contains a phosphodiester bond at the scissile position. Hence cleavage and exchange can produce a Holliday junction. By contrast, cleavage at the phosphorothiolate position is a dead end reaction, as the 5'-SH group is not active in strand joining. The uncertainty regarding the origin of the two rates does not, however, detract from the distinct catalytic enhancement conferred by thiolate on Flp(W330H) and Flp(W330A).
Combining structural data13 with results on recombination (Fig. 4),20 and strand cleavage in phosphate and 5'-thiolate substrates (Fig. 5), we surmise that Trp-330 plays an accessory role to the general acid in stabilizing the leaving group. One would predict that Flp(W330F) should be comparable to Flp(W330H) and Flp(W330A) in the magnitude of the thiolate response when hydrogen peroxide rather than Tyr-343 acts as the cleavage nucleophile. In the peroxide reaction, unlike the Tyr-343 reaction, Flp(W330F) will have no advantage over the other two mutants in orienting the nucleophile. However, this prediction could not be tested because the thiolate, in contrast to phosphate, was refractory to peroxide cleavage in the presence of wild type Flp or Flp(Y343F). Perhaps the larger size of the sulfur atom sterically precludes exogenous nucleophiles from accessing the reaction center.
Strand cleavage by Flp in a methylphosphonate substrate
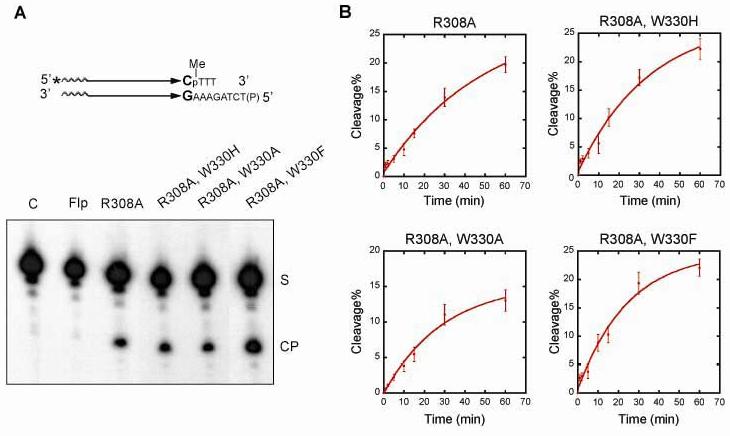
The nearly equidistant positioning of Trp-330, 3.8 Å and 3.6 Å, respectively, from the scissile phosphate and the 5'-hydroxyl group, in the ‘Flpe’-DNA structure24 raises the possibility that Trp-330 might switch hydrogen bonding partners from a non-bridging oxygen atom initially to a bridging oxygen atom subsequently. Although the Flp-DNA and Flpe-DNA structures differ in four amino acid residues, these are located at or close to the protein surface.13, 24 They are distant from the active sites, and do not affect the core packing of the N- and C-terminal domains. The early hydrogen bonding interaction of Trp-330 could assist Arg-191 and Arg-308 in positioning the phosphate and/or balancing the negative charge developing on the non-bridging oxygen atoms in the transition state. To test this idea, we followed the effect of the Trp-330 mutations on the cleavage of a methylphosphonate substrate (Fig. 6).
As elegantly shown by Shuman and colleagues for vaccinia topoisomerase IB,12 the reduction of phosphate charge by methylphosphonate substitution at the scissile position exposes a hydrolytic reaction that proceeds at nearly wild type rate in the absence of Arg-223, which is critical for Tyr-274 mediated strand cleavage in the phosphate substrate. Flp(R308A), which lacks the Arg-223 equivalent of the vaccinia enzyme, can also mediate hydrolysis of a methylphosphonate substrate (Fig. 6A). Unlike the vaccinia topoisomerase reaction, in which hydrolysis occurs on the cleaved tyrosyl intermediate, the Flp reaction proceeds via direct hydrolysis of the methylphosphonate (unpublished data). Mutations of Trp-330 did not show an adverse effect on this reaction (Fig. 6A). Furthermore, Flp(R308A) and the Trp-330 mutant derivatives of Flp(R308A) yielded more or less similar kinetics of hydrolysis (Fig. 6B).
Thus, replacement of a charged oxygen atom on the scissile phosphate by the neutral methyl group eliminates the requirement of Trp-330 during hydrolysis. Since, Tyr-343 does not participate in this reaction, the W330F substitution has no catalytic advantage over the W330H and W330A substitutions in nucleophile positioning. Reducing the phosphate charge to 0 in the ground state (and -1 in the transition state) by the methylphosphonate, aided further by the high concentration (56 M) of water as the nucleophile, is also sufficient to compensate for the negative effects of the Trp-330 mutations on leaving group departure.
The available crystal structures of Flp indicate that Trp-330 is just within van der Waals distance of Arg-308. Trp-330 may thus assist Arg-308 in charge compensation at the scissile phosphate by direct interaction between the two residues. When Arg-308 is no longer required for this function, as in the hydrolysis of methylphosphonate, Trp-330 becomes non-essential as well. It is possible that Arg-308-Trp-330 interaction and hydrogen bonding of Trp-330 with a non-bridging phosphate oxygen serve cooperative functions during the strand cleavage step.
Trp-330 mutants of Flp are unaffected in the type II RNA cleavage activity of Flp
The active site of Flp harbors two cryptic RNA cleavage activities that can be unveiled by incorporating ribonucleotides at appropriate positions in an otherwise deoxyribooligonucleotide substrate.25,26,27. The type I RNase of Flp follows the DNA recombination mechanism (Fig. 7A), and mediates the initial strand cleavage using Tyr-343. The type II RNase activity targets the phosphodiester that is the immediate 3' neighbor of the recombination target (Fig. 7B), and is independent of the Tyr-343 nucleophile. It proceeds via direct attack by the adjacent 2'-hydroxyl group on the scissile phosphate. Previous studies 26,27 and unpublished data demonstrated that the catalytic cluster of Arg-191, Lys-223, His-305 and Arg-308 are critical for both the type I and type II RNA cleavage activities.
Fig. 7.
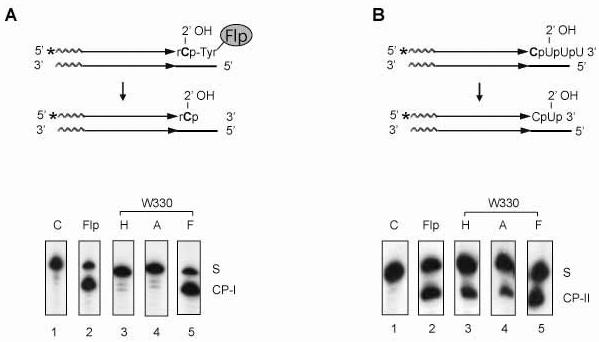
The activities of the Trp-330 mutants of Flp in type I and type II Flp RNA cleavage reactions. A. in the type I RNA cleaving activity of Flp, the vicinal 2'-hydroxyl attacks the phosphate group covalently linked to Flp via Tyr-343. The cleavage product ‘CP-I’, containing a 3'-phopshate end, was separated from the substrate ‘S’ by electrophoresis in a denaturing polyacrylamide gel. B. In the type II RNA cleaving activity of Flp, the vicinal 2'-hydroxyl group directly attacks the target phosphodiester bond. The cleavage product ‘CP-II’ (also with a 3'-phosphate end) was separated from the substrate ‘S’ as under A. The data shown are for 30 min reactions carried out at 30°C.
Based on the results with DNA recombination, one would suspect that Trp-330 is important for the type I RNase activity of Flp. Since Tyr-343 is not involved in the type II reaction, is Trp-330 dispensable for this activity? We therefore assayed the type I and type II activities elicited by the Trp-330 Flp mutants. As suspected, Flp(W330F) was active as the type I RNase, but Flp(W330H) and Flp(W330A) were not (Fig. 7A). By contrast, all three of the mutants exhibited efficient type II RNase activity (Fig. 7B). These data indicate that Trp-330 has no significant effect in orienting the scissile phosphate or promoting the departure of the 5'-hydroxyl group during the type II RNase reaction.
Rescue of Trp330(Flp) mutations by directed incorporation of second site suppressors
The intersubunit interactions that establish the functional configuration of the shared active site of Flp are well displayed in the crystal structure of the Flp recombination complexes13,20 (see Fig. 8). The van der Waals interactions of Trp-330 from the ‘pro-active site’ of one monomer (recipient) with Ser-336,Val-338 and Ala-339 of an adjacent second monomer (donor) are particularly important in correctly positioning Tyr-343 for strand cleavage. All the donated residues, Ser-336, Val-338, Ala-339 and Tyr-343 are housed by the M helix against one face of which Trp-330 is tightly packed, permitting it to contact Tyr-343 as well. In addition, Asn-325 from the recipient makes two hydrogen bonding interactions with the approaching polypeptide chain of the donor, further reinforcing the web of interactions that orient the cleavage nucleophile. Furthermore, His-345 from the Flp donor is nestled within a cleft against helix E and His-309 from the Flp recipient. This interaction almost perfectly stacks- His-309 over Tyr-343, further fine-tuning the alignment of Tyr-343.
Fig. 8.
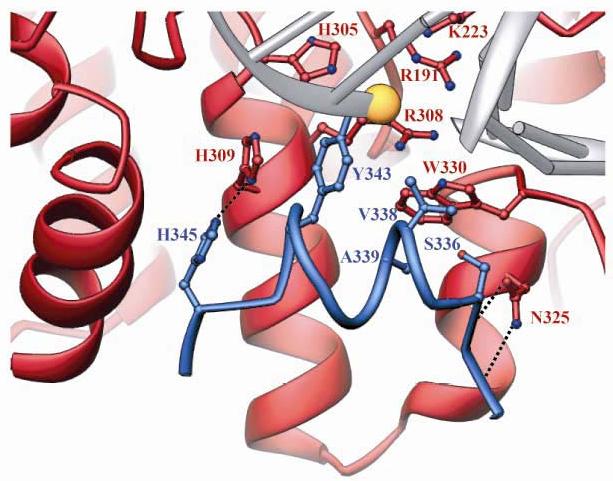
Interactions between helix M of one Flp monomer and its neighbor. In a portion of the Flpe-DNA complex shown, helix M from one protomer (blue) packs into the active site of its neighbor (red). The DNA is cartooned in gray, and the yellow sphere highlights the scissile phosphate, which gets covalently attached to Tyr-343 during strand cleavage. The remainder of the blue protomer is omitted for clarity. Residues that form important van der Walls or H-bond interactions between helix M and its neighbor (Ser-336, Val-338 and Ala-339 in blue; Trp-330 and Asn-325 in red) are shown in ball-and-stick form. Arg-191, Lys-223, His-305, Arg-308 and Trp-330 form the ‘active site pentad’ of the red protomer. His-345 (blue) is positioned in the cleft between helix E (red) to its left and His309 (red) to its right.
Based on the structural features of the Flp active site, it seemed plausible that a mutation at 330 may be suppressed by second site mutation(s) of those residues that cooperatively interact with this position. Flp(W330A) was chosen as the mutant template in the suppression assay, because it showed no detectable recombination activity by in vivo and in vitro assays (Fig. 9AC). Suppression of the catalytic defect of Flp(W330A) was sought by randomizing positions 325, 336 and 339, and was assayed in E. coli by excision of a reporter LacZα cassette flanked by a pair of head-to-tail Flp recombination target (FRT) sites28 (this study). Rescue of Flp(W330A) to nearly 80 percent of the wild type Flp activity, as indicated by recombination in vivo, was possible (Fig. 9A). The suppressor variant contained, in addition to W330A, a single point mutation: A339M.
Fig. 9.
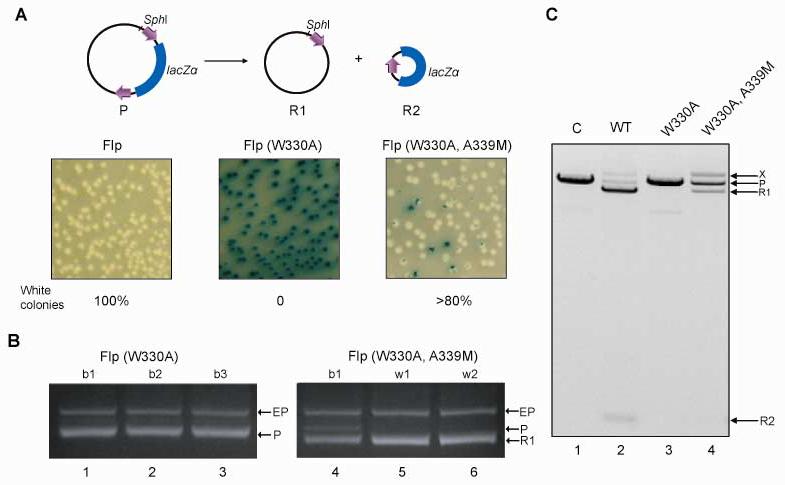
Rescue of Flp(W330A) in recombination by a second site mutation A339M. The excision assay for Flp recombination in E. coli is schematically depicted at the top. A. The recombination event causing deletion of the lacZα cassette is declared by the switch in colony color from blue to white on X-gal plates. The product R2 is lost from the population for lack of a replication origin. B. Resident plasmids from randomly picked blue (b) and white (w) colonies were isolated, digested with SphI, and fractionated by electrophoresis in 1% agarose gels. The expression plasmids, the parent reporter plasmid and the larger of two recombinant products derived from it, all in their linear forms, are indicated by ‘EP’, ‘P’ and ‘R1’, respectively. C. Following in vitro recombination, the extracted DNA was digested with SphI, and subjected to electrophoresis as under A. ‘P’ refers to the linearized substrate, and ‘R1’ to the linear form of the larger deletion product. The smaller deletion circle is denoted by ‘R2’. The band above R1 in the Flp(W330A, A339M) reaction ‘X’ was due to integration of the deletion circle into P via an intermolecular recombination event.
Digestion of plasmids isolated from blue (b) and white (w) colonies with SphI verified the authenticity of the recombination reaction by revealing the deletion plasmid R1 in the white colonies (lanes 5 and 6, Fig. 9B). Even the blue colonies generated in the Flp(W330A, A339M) assay contained a large proportion of R1 with a smaller fraction of the parent reporter P (lane 4, Fig. 9B). By contrast, R1 was undetected in the blue colonies from the Flp(W330A) assay (lanes 1-3, Fig. 9B). The band labeled EP in Fig. 9B refers to the expression plasmids from which the Flp variants were produced.
In agreement with the results of the in vivo assay, the recombination activity of the purified suppressor variant, yielding the deletion products R1 and R2, was readily detected in vitro (lane 4, Fig. 9C). The level of activity, in 30°C reactions for 30 min, was roughly 30-40% of that of wild type Flp (compare lanes 2 and 4, Fig. 9C). This quantitative difference can be accounted for by the much longer incubation period (30 hr at 37°C) during the in vivo assay. The band labeled ‘X’ in Fig. 9C was due to Flp or Flp(W330H, A339M) mediated integration of the excised circle R2 into the parent plasmid P. The reason for the higher yield of the intermolecular reaction by Flp(W330H, A339M) relative to Flp (compare the intensities of ‘X’ in lanes 2 and 4 of Fig. 9B) is not understood. The reduction in the level of R2 in the Flp(W330A, A339M) reaction was consistent with the corresponding increase in that of X (lane 4, Fig. 9C).
The suppression results indicate that the conserved His/Trp-III among the tyrosine family recombinases is not indispensable in catalysis – at least for Flp. Lack of this residue, as in Flp(W330A), would strongly diminish the efficiency of orienting Tyr-343. Likewise, possible catalytic contributions to the cleavage reaction from hydrogen bonding and contact with Arg-308 would be eliminated as well. These cumulative effects can account for the undetectable recombination activity of the mutant protein. In the suppressor variant, the longer side chain of Met-339 is likely to restore, to a considerable extent, the trans interaction between the Flp pro-active site and the M helix (see under Discussion; Fig. 10). As a consequence, helix E-His-309-His345 interactions (Fig. 7), that are also critical for Tyr-343 positioning, may be preserved nearly normally. Flp(W330A, A339M) therefore functions as a reasonably active recombinase.
Fig 10.
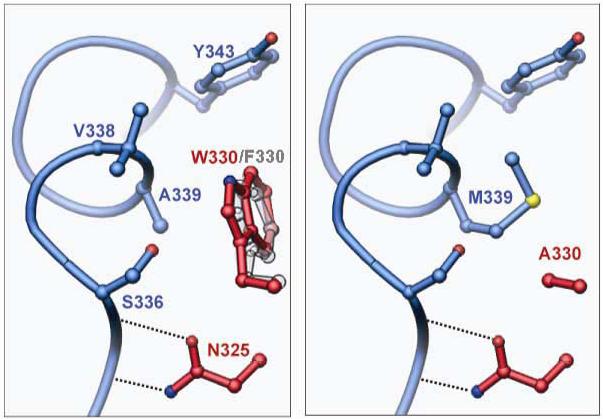
Second site suppression of W330A by A339M. Helix M from the ‘donor’ protomer is shown in blue, and two residues from the ‘recipient’ protomer in red. Left: In wild type Flp or Flp(W330F), Trp-330 or Phe-330 packs against the first turn of helix M, making van der Waals contacts with Ser-336, Val-338 and Ala-339 (see also Fig. 8). Right: Similar view of a model structure for the suppressor variant Flp(W330A, A339M). The program O 40 was used to replace the side chains at 330 and 339, and to find the rotamer of Met-339 that best fills the space vacated by the W330A mutation without causing steric conflicts with other residues.
DISCUSSION
We have addressed the catalytic role of His/Trp-III, a conserved member of the catalytic pentad of the tyrosine family recombinases and type IB topoisomerases, in the chemical steps of Flp recombination. Our three principal findings are as follows. (1) Trp-330 of Flp, corresponding to His/Trp-III, is required for the strand cleavage step, but not for the strand joining step. (2) Whereas the primary role of Trp-330 is in positioning Tyr-343, it has secondary roles in positioning/activating the scissile phosphate and facilitating leaving group departure. (3) It is possible to contrive a new active site arrangement in which the Tyr-343 nucleophile can be positioned in its functional configuration without assistance from Trp-330.
In order to unveil the more subtle catalytic attributes of Trp-330, its primary role in strand cleavage had to be muzzled by employing a combination of non-standard substrate and nucleophile contexts: (a) hydrogen peroxide mediated cleavage in a phosphate substrate, (b) Tyr-343 mediated cleavage in a thiolate substrate and (c) water mediated cleavage in a methylphosphonate substrate. Flp, unlike most other related recombinases, is particularly amenable to these strategies. Among tyrosine recombinases, strand cleavage by hydrogen peroxide or tyrosine mimics has been demonstrated only for Flp. A shared active site configuration, exemplified by Flp, may be a prerequisite for reactions mediated by exogenously supplied small nucleophiles, including perhaps the direct hydrolysis of methyl phosphonate.
We find that the strong hydrophobic contribution of Trp-330 in strand cleavage is not due to the shared architecture of the Flp active site, with Tyr-343 being donated from one monomer to its neighbor. The equivalent residue Trp-315 in the Cre recombinase, which assembles its active site entirely within a monomer, behaves similarly. The effects of phenylalanine, histidine and alanine substitutions at position 315 in Cre roughly parallel those observed for the corresponding substitutions at position 330 in Flp (Fig. S1, Supplementary Material).
In the sections below, we discuss the key experimental outcomes from this work in the context of the structures of Flp recombination complexes.13,20,24
Potential hydrogen bonding contributions of Trp-330 of Flp towards strand cleavage
The recombinationally active Flp(W330F), the inactive Flp(W330A) and the weakly active Flp(W330H) are uniformly inefficient in hydrogen peroxide mediated strand cleavage (within a factor of two or so; Fig. 3A). However, their overall active site configurations are not perturbed, as evidenced by their activities in strand joining (Fig. 2B) and type II RNA cleavage (Fig. 7B). These results suggest an accessory role for Trp-330 during orientation of the scissile phosphate or departure of the leaving group or both. The structures of Flp-DNA complexes are consistent with the hydrogen bonding potential of Trp-330 being utilized towards either or both of these steps. The lack of hydrogen bonding from position 330 in Flp(W330F) is then expected to cause a finite drop in its catalytic power. However, the handicap is redressed to a large extent by the strong hydrophobic character at this position and the resulting salutary effect on Tyr-343 orientation. As a result, Flp(W330F) is reasonably efficient in recombination. The extensive inter-subunit van der Waals contacts required for the correct alignment of position 33013,20 are likely disturbed when its occupant is histidine. Such misalignment would prevent His-330 from providing catalytically relevant hydrogen bonding interactions.
When constraints on leaving group departure are relaxed in a 5'-thiolate substrate, the strong impediment to cleavage by Flp(W330H) or Flp(W330A) can be substantially ameliorated. Similarly, when the negative charge in the transition state is partially neutralized in a methylphosphonate substrate, hydrolysis of this bond by the Trp-330 mutants can proceed efficiently. Consistent with Flp structural data, these results may be accommodated by a dynamic role for Trp-330 during the progression of catalysis: initially hydrogen bonding to a non-bridging oxygen atom on the scissile phosphate group and subsequently moving in position to hydrogen bond with the 5'-hydroxyl group. The first interaction would help the catalytic arginines (Arg-191/Arg-308) in positioning the scissile phosphate and/or opposing the negative charge within the transition state; the second would assist the general acid in leaving group stabilization. As noted earlier, van der Waals interaction of Trp-330 with Arg-308 could assist the latter in effecting charge compensation at the scissile phosphate. In principle, hydrogen bonding of Trp-330 to phosphate and direct interaction between Trp-330 and Arg-308 may act in concert to promote strand cleavage.
The two Flp-DNA structures that suggest the 5'-hydroxyl group and the scissile phosphate as potential hydrogen bonding partners of Trp-330 differ from each other by one bp in the length of their FRT spacer sequences.13,24 This is the result of a ‘slippage ligation’ event that has occurred in one of them. However, the overall arrangement of the active sites and the relative locations of key catalytic residues are nearly identical between the two. There is an intrinsic flexibility to the Flp active site, which permits recombination not only on substrates with the optimal 8 bp spacer but also on those with 7 or 9 bp spacers.29
The strand joining reaction and type II RNA cleavage by Flp are independent of Trp-330
The finding that the strand joining reaction by Flp proceeds efficiently without Trp-330 is consistent with the architecture of the Flp active site. Once Tyr-343, delivered in trans, is linked to the 3'-end of the scissile phosphate, it is likely well positioned in the cleaved intermediate without assistance from Trp-330. Furthermore, the 5-hydroxyl group, which is the nucleophile for strand joining, is aligned by base pairing between the invading cleaved strand and the uncleaved complementary strand.30-32
The close mechanistic similarity between DNA recombination and type I RNA cleavage by Flp accounts for the requirement of Trp-330 in both these reactions. Conversely, the mechanistic distinction and Tyr-343 independence of type II RNA cleavage affords the dispensability of Trp-330. The orientation of the two sequentially employed nucleophiles in this reaction, a vicinal 2'-hydroxyl group and a water molecule, respectively, will not be assisted by the hydrophobicity of Trp-330. Lack of Trp-330 does not significantly hinder steps of scissile phosphate activation and leaving group stabilization during the type II RNase activity.
It has been suggested that the simple pancreatic RNase-like active site (exemplified by the type II activity) in Flp was the progenitor of the more complex recombination active site, likely optimized through an intermediate topoisomerase-like active site.127,33 Perhaps the incorporation of a tryptophan residue into the catalytic scheme of recombination may have occurred concomitant with or subsequent to the emergence of a tyrosine nucleophile for strand cutting, signifying a topoisomerase-like active site.
Second site suppression of the W330A mutation in Flp: Potential configuration of the restored active site
Flp(W330A), with no detectable recombination function in vivo and in vitro, can be restored to a respectable level of activity by a single additional mutation, A339M. A modeled structure incorporating Ala-330 and Met-339, respectively, is compared to the structures of wild type Flp and Flp(W330F) in Fig. 10. The Ala-330-Ser-336-Val-338-Met-339 van der Waals interactions in the suppressor variant, aided by the longer side chain at position 339, may stabilize the dimer interface sufficiently to maintain helix E, His-309 and His-345 close to their normal relative configurations. This, in turn, would permit Tyr-343 to be orientated even in the absence of Trp-330. The loss of catalytic power in this altered active site is quite modest. Consistent with the proposed scheme for suppression, we noticed that leucine-339 also has suppressor activity, and is less efficient than Met-339 (Fig. S2, Supplementary Material). By contrast, isoleucine is quite weak in its suppression of W330A.
Catalytic contribution of His/Trp-III: Novel ensembles of catalytic residues for tyrosine family recombination
A dual catalytic role for the conserved His-Trp-III, hydrophobic as well as hydrogen bonding, is perhaps generally true among type IB topoisomerases and tyrosine recombinases. The relative strengths of these two contributions may vary depending upon individual systems. Thus, amino acid conservation at this position may not necessarily translate into strict mechanistic conservation as well.
The ease with which Trp-330 of Flp can be eliminated, while preserving recombination capacity, is consistent with the notion that His/Trp-III is perhaps a late draftee to the active site of tyrosine recombinases and type IB topoisomerases. However, once enlisted, His/Trp-III could add to the catalytic efficiency by better utilization of the tyrosine nucleophile for strand cleavage. In addition, it could serve an accessory role in refining the orientation of the scissile phosphate and/or facilitating the departure of the 5'-hydroxyl group. Consistent with these ideas, in ResT, a member of the telomere resolvase family that generates hairpin ends in linear bacterial chromosomes and plasmids by the type IB topoisomerase mechanism, the histidine residue corresponding to His/Trp-III is dispensable with only a 30 per cent drop in enzyme activity.34
The structure based randomization strategy employed in this study may prove useful in suppressing amino acid substitutions at other active site positions of Flp as well. Novel catalytic constellations for performing the phosphoryl transfer steps of tyrosine family recombination may thus be unveiled.
Materials and Methods
Plasmids and strains
The plasmids for expressing Flp or Flp variants and the reporter plasmid for assaying recombination have been previously described.28 All in vivo recombination assays were performed in the E. coli strain DH10B from Invitrogen (F, mcrA, (mrr-hsdRMS-mcrBC), φ80dlacZM15, lacX74, deoR, recA1, endA1, ara139, D(ara, leu)7697, galU, galK, λ−, rpsL, nupG). The screen is based on the excision and loss of a lacZα reporter cassette and the resulting change in colony color from blue to white on X-gal indicator plates.28
Synthetic oligonucleotides
Synthetic oligonucleotides were purchased from IDT (Coralville, IA)) or Trilink Technologies (San Diego, CA). Several of the substrates used in assays in vitro were assembled by hybridizing the appropriate oligonucleotides as described previously.35 The exact sequences of the oligonucleotides employed in this study are available upon request.
Substrate containing specific 5'-thiolate linkage
A chemical ligation between an oligonucleotide containing a 3'-thiophosphate and one containing a 5'-iodide was employed to incorporate a 5'-thiolate linkage at a desired position.36 The thiophosphate and iodide containing oligonucleotides were purchased from Trilink Biotechnologies (San Diego, CA). The method is the same as that was recently used to construct suicide cleavage substrates in the Cre-loxP recombination system.37
In vitro Flp reactions
In vitro recombination reactions were performed according to Konieczka et al.38 using Flp or Flp variants purified to roughly 60-70% homogeneity. Strand cleavage using H2O2 and tyramine as nucleophiles was carried out as described previously.22; 23 Strand cleavage reactions by the native nucleophile (Tyr-343) and strand joining reactions were conducted under the recombination conditions employed by Konieczka et al.38 A subset of the cleavage reactions was assayed by running the SDS (0.2% final concentration)-quenched samples in 12% SDS-polyacrylamide gels (acrylamide to bis-acrylamide, 1:29). Others were treated with proteinase K prior to DNA extraction and electrophoresis in 10-15% denaturing polyacrylamide gels (acrylamide to bisacrylamide, 1:19). The type I and type II Flp RNA cleavage activities were assayed as detailed previously.26,27 All the in vitro reactions contained approximately 4 to 6 pmoles of Flp or a Flp variant per pmole of Flp binding element.
In vitro Cre reactions
Cre and variants of Cre, tagged at their C-termini with (His)6 tags, were purified to 70-80% homogeneity using Ni affinity chromatography. Recombination assays (shown in Fig.S2, Supplementary Material) were carried out under Flp reaction conditions.
Amino acid randomization in Flp and screening of randomized libraries
Selected amino acid positions within Flp were randomized by PCR using synthetic oligonulceotide primers containing appropriate degeneracies.39 A subset of the clones from the randomized library was sequenced within the region of interest to assess the efficiency of randomization. The library was tested in the in vivo Flp recombination screen.28 Expression plasmids from white colonies (indicating recombination) were separated from the reporter, and were re-screened for their recombination phenotypes.
Modeling amino acid substitutions and presentation of Flp structures
The program O40 was used to perform side chain replacements and to find the rotamer at a position that is accommodated best without steric conflicts. Figures 7 and 9 were prepared from PDB 1D 1M6X 24 using the program Ribbons.41
Supplementary Material
Acknowledgements
This work was supported by NIH grant GM35654. Partial support was provided by the Robert F. Welch Foundation. We thank Qian Wu for performing some of the early experiments. We gratefully acknowledge P.A. Rice (University of Chicago) for providing the structural representations shown in Fig. 8 and 10.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jayaram M, Tribble G, Grainge I. Site-specific recombination by the Flp protein of Saccharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington, DC: 2002. pp. 192–218. [Google Scholar]
- 2.Rice PA. Theme and variation in tyrosine recombinases: structure of a Flp-DNA complex. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 219–229. [Google Scholar]
- 3.Van Duyne GD. A structural view of tyrosine recombinase site-specific recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 93–117. [Google Scholar]
- 4.Azaro MA, Landy A. λ integrase and λ int family. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 118–148. [Google Scholar]
- 5.Barre F-X, Sherratt DJ. Xer site-specific recombination: promoting chromosome segregation. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 149–161. [Google Scholar]
- 6.Esposito D, Scocca JJ. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucl. Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunes-Duby SE, Kwon HJ, Tirumalai RS, Ellenberger T, Landy A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucl. Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krogh BO, Shuman S. Catalytic mechanism of DNA topoisomerase IB. Mol. Cell. 2000;5:1035–1041. doi: 10.1016/s1097-2765(00)80268-3. [DOI] [PubMed] [Google Scholar]
- 9.Krogh BO, Shuman S. Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J. Biol. Chem. 2002;277:5711–5714. doi: 10.1074/jbc.C100681200. [DOI] [PubMed] [Google Scholar]
- 10.Nagarajan R, Kwon K, Nawrot B, Stec WJ, Stivers JT. Catalytic phosphoryl interactions of topoisomerase IB. Biochem. 2005;44:11476–11485. doi: 10.1021/bi050796k. [DOI] [PubMed] [Google Scholar]
- 11.Tian L, Claeboe CD, Hecht SM, Shuman S. Guarding the genome: electrostatic repulsion of water by DNA suppresses a potent nuclease activity of topoisomerase IB. Mol. Cell. 2003;12:199–208. doi: 10.1016/s1097-2765(03)00263-6. [DOI] [PubMed] [Google Scholar]
- 12.Tian L, Claeboe CD, Hecht SM, Shuman S. Mechanistic plasticity of DNA topoisomerase IB: phosphate electrostatics dictate the need for a catalytic arginine. Structure. 2005;13:513–520. doi: 10.1016/j.str.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Narendra U, Iype LE, Cox MM, Rice PA. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 14.Guo F, Gopaul DN, Van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 15.Gelato KA, Martin SS, Baldwin EP. Reversed DNA strand cleavage specificity in initiation of Cre-LoxP recombination induced by the His289Ala active-site substitution. J. Mol. Biol. 2005;354:233–245. doi: 10.1016/j.jmb.2005.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons RL, Prasad PV, Harshey RM, Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol. Cell. Biol. 1988;8:3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies DR, Mushtaq A, Interthal H, Champoux JJ, Hol WG. The structure of the transition state of the heterodimeric topoisomerase I of Leishmania donovani as a vanadate complex with nicked DNA. J. Mol. Biol. 2006;357:1202–1210. doi: 10.1016/j.jmb.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 19.Stivers JT, Jagadeesh GJ, Nawrot B, Stec WJ, Shuman S. Stereochemical outcome and kinetic effects of Rp- and Sp-phosphorothioate substitutions at the cleavage site of vaccinia type I DNA topoisomerase. Biochem. 2000;39:5561–5572. doi: 10.1021/bi992429c. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Rice PA. The role of the conserved Trp330 in Flp-mediated recombination: functional and structural analysis. J. Biol. Chem. 2003;278:24800–24807. doi: 10.1074/jbc.M300853200. [DOI] [PubMed] [Google Scholar]
- 21.Ringrose L, Lounnas V, Ehrlich L, Buchholz F, Wade R, Stewart AF. Comparative kinetic analysis of FLP and Cre recombinases: mathematical models for DNA binding and recombination. J. Mol. Biol. 1998;284:363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 22.Kimball AS, Lee J, Jayaram M, Tullius TD. Sequence-specific cleavage of DNA via nucleophilic attack of hydrogen peroxide, assisted by Flp recombinase. Biochem. 1993;32:4698–4701. doi: 10.1021/bi00069a002. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Jayaram M. Mechanism of site-specific recombination. Logic of assembling recombinase catalytic site from fractional active sites. J. Biol. Chem. 1993;268:17564–17570. [PubMed] [Google Scholar]
- 24.Conway AB, Chen Y, Rice PA. Structural plasticity of the Flp-Holliday junction complex. J. Mol. Biol. 2003;326:425–434. doi: 10.1016/s0022-2836(02)01370-0. [DOI] [PubMed] [Google Scholar]
- 25.Sau AK, Tribble GD, Grainge I, Frohlich RF, Knudsen BR, Jayaram M. Biochemical and kinetic analysis of the RNase active sites of the integrase/tyrosine family site-specific DNA recombinases. J. Biol. Chem. 2001;276:46612–46623. doi: 10.1074/jbc.M106492200. [DOI] [PubMed] [Google Scholar]
- 26.Xu CJ, Ahn YT, Pathania S, Jayaram M. Flp ribonuclease activities. Mechanistic similarities and contrasts to site-specific DNA recombination. J. Biol. Chem. 1998;273:30591–30598. doi: 10.1074/jbc.273.46.30591. [DOI] [PubMed] [Google Scholar]
- 27.Xu CJ, Grainge I, Lee J, Harshey RM, Jayaram M. Unveiling two distinct ribonuclease activities and a topoisomerase activity in a site-specific DNA recombinase. Mol. Cell. 1998;1:729–739. doi: 10.1016/s1097-2765(00)80072-6. [DOI] [PubMed] [Google Scholar]
- 28.Voziyanov Y, Stewart AF, Jayaram M. A dual reporter screening system identifies the amino acid at position 82 in Flp site-specific recombinase as a determinant for target specificity. Nucl. Acids Res. 2002;30:1656–1663. doi: 10.1093/nar/30.7.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senecoff JF, Cox MM. Directionality in FLP protein-promoted site-specific recombination is mediated by DNA-DNA pairing. J. Biol. Chem. 1986;261:7380–7386. [PubMed] [Google Scholar]
- 30.Lee J, Jayaram M. Role of partner homology in DNA recombination. J. Biol. Chem. 1995;270:4042–4052. doi: 10.1074/jbc.270.8.4042. [DOI] [PubMed] [Google Scholar]
- 31.Nunes-Duby SE, Azaro MA, Landy A. Swapping DNA strands and sensing homology without branch migration in λ site-specific recombination. Curr. Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhu XD, Pan G, Luetke K, Sadowski PD. Homology requirements for ligation and strand exchange by the FLP recombinase. J. Biol. Chem. 1995;270:11646–11653. doi: 10.1074/jbc.270.19.11646. [DOI] [PubMed] [Google Scholar]
- 33.Jayaram M, Mehta S, Uzri D, Voziyanov Y, Velmurugan S. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. In: Moldave K, editor. Prog. Nucl. Acid Res. Mol. Biol. Vol. 77. Academic Press; 2004. pp. 127–172. [DOI] [PubMed] [Google Scholar]
- 34.Deneke J, Burgin AB, Wilson SL, Chaconas G. Catalytic residues of the telomere resolvase ResT: a pattern similar to, but distinct from, tyrosine recombinases and type IB topoisomerases. J. Biol. Chem. 2004;279:53699–53706. doi: 10.1074/jbc.M409001200. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Whang I, Jayaram M. Assembly and orientation of Flp recombinase active sites on two-, three- and four-armed DNA substrates: implications for a recombination mechanism. J. Mol. Biol. 1996;257:532–549. doi: 10.1006/jmbi.1996.0183. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, Kool ET. A novel 5′-iodonucleoside allows efficient nonenzymatic ligation of single stranded and duplex DNAs. Tetrahedron Lett. 1997;38:5595–5598. doi: 10.1016/S0040-4039(97)01266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh K, Lau CK, Gupta K, Van Duyne GD. Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat. Chem. Biol. 2005;1:275–82. doi: 10.1038/nchembio733. [DOI] [PubMed] [Google Scholar]
- 38.Konieczka JH, Paek A, Jayaram M, Voziyanov Y. Recombination of hybrid target sites by binary combinations of Flp variants: mutations that foster interprotomer collaboration and enlarge substrate tolerance. J. Mol. Biol. 2004;339:365–78. doi: 10.1016/j.jmb.2004.03.060. [DOI] [PubMed] [Google Scholar]
- 39.Bolusani s., Ma C-H, Paek A, Konieczka JH, Jayaram M, Voziyanov Y. Evolution of variants of yeast site-specific recombinase Flp that utilize native genomic sequences as recombination target sites. Nucl. Acids Res. 2006;34:5259–5269. doi: 10.1093/nar/gkl548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 41.Carson M. Ribbons 2.0. J. Appl. Crystallogr. 1991;24:958–961. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


