Abstract
The mammalian integration site 6 (INT6) protein has been implicated in breast carcinogenesis and characterized as the eIF3e non-core subunit of the translation initiation factor eIF3, but its role in this complex is not known. Here, we show that INT6 knockdown by RNA interference strongly inhibits nonsense-mediated messenger RNA decay (NMD), which triggers degradation of mRNAs with premature stop codons. In contrast to the eIF3b core subunit, which is required for both NMD and general translation, INT6 is only necessary for the former process. Consistent with such a role, immunoprecipitation experiments showed that INT6 co-purifies with CBP80 and the NMD factor UPF2. In addition, several transcripts known to be upregulated by UPF1 or UPF2 depletion were also found to be sensitive to INT6 suppression. From these observations, we propose that INT6, in association with eIF3, is involved in routing specific mRNAs for degradation.
Introduction
Integration site 6 protein (INT6), first identified as the product of a murine gene in which integration of mouse mammary tumour virus seems to be sufficient to induce breast carcinoma (Marchetti et al, 1995), has been characterized as a subunit (eIF3e) of the eukaryotic translation initiation factor eIF3 (Asano et al, 1997). In addition, this highly conserved protein can interact with subunits of the COP9 signalosome (CSN) and the 26S proteasome (Karniol et al, 1998; Yahalom et al, 2001; Hoareau Alves et al, 2002). Consistently, fission yeast INT6 promotes proteasome assembly by interacting with, and mediating nuclear import of, the Rpn5 (regulatory particle non-ATPase 5) proteasomal subunit (Yen et al, 2003).
In contrast to other eIF3 subunits, INT6 is not essential for global translation. Fission yeast INT6 mutants show only a weak inhibition in the rate of protein synthesis and a slight decrease in the polysome content (Bandyopadhyay et al, 2000; Akiyoshi et al, 2001; Zhou et al, 2005). Budding yeast eIF3 is viewed as a factor comprising a core complex of five stoichiometric subunits required for global translation (Asano et al, 1998; Phan et al, 1998). Mammalian eIF3 is probably organized in a similar way, and non-core subunits, one of which is INT6, might have regulatory roles. For example, Kim et al (2004) have shown in Arabidopsis thaliana that eIF3h was required for translation of messenger RNAs containing particular 5′ leader sequences. In addition, a recent study reported that two distinct eIF3 complexes exist in fission yeast: one is defined by the Csn7Bp/eIF3m subunit and is found associated with an important part of mRNAs, whereas the other complex contains INT6 and is associated with a more restricted mRNA subset (Zhou et al, 2005).
Consistent with the idea that INT6 is a regulatory subunit of eIF3, we searched for a role of human INT6 in nonsense-mediated mRNA decay (NMD), as this mRNA surveillance mechanism is translation dependent (Belgrader et al, 1993; Thermann et al, 1998). Briefly, NMD degrades transcripts with a premature termination codon (PTC) located more than 50 nucleotides upstream of an exon–exon junction (Maquat, 2004; Baker & Parker, 2004; Conti & Izaurralde, 2005). The prevalent model for mammalian NMD suggests a first, or pioneer, round of translation, which recognizes the PTC, and the recruitment of the three Up Frameshift (UPF) factors by the exon junction complex and the translation termination factors. Here, we show that INT6 silencing by RNA interference (RNAi) impairs NMD in a manner similar to UPF1 depletion. This effect relates neither to an alteration in UPF1 or UPF2 protein amounts nor to UPF1 phosphorylation. However, co-immunoprecipitation experiments showed an association of INT6 with UPF2. These data establish INT6 as a non-core eIF3 subunit contributing specifically to NMD.
Results
To assess whether INT6 is important in NMD, levels of a β-globin mRNA carrying a PTC (NS39; Thermann et al, 1998) were compared with those of the wild-type transcript in HeLa cells treated with INT6 short interfering RNAs (siRNAs). In control cells, levels of the NS39 β-globin mRNA represented only 2–3% of the wild-type levels, owing to its degradation by the NMD process (Fig 1A). As expected, UPF1 silencing caused a tenfold increase in NS39 mRNA levels. A similar effect, between six- and sevenfold, was observed by depleting INT6 (Fig 1A). It should be noted that RNAi against INT6 is slightly less efficient than that against UPF1. That NMD was found to be impaired using two siRNA duplexes targeting different sequences of INT6 mRNA argues against an off-target effect. Accordingly, expression of a Flag-tagged INT6 from a recombinant mRNA lacking the 3′ untranslated region reverted the negative effect on NMD of a third siRNA couple targeting this non-coding part of INT6 mRNA (supplementary Fig S1 online). Next we wanted to determine whether the inhibitory effect of INT6 silencing on NMD was specific of this process or if it was a consequence of an arrest of translation. Therefore, the steady-state translation was evaluated by analysing the amount of exogenous green fluorescent protein (GFP) produced in siRNA-treated cells. We found that GFP mRNA translation was not affected either after INT6 silencing or after UPF1 depletion, as expected (Fig 1B). In agreement with this, we found that INT6 depletion did not significantly affect the amount of other eIF3 subunits (supplementary Fig S2 online).
Figure 1.
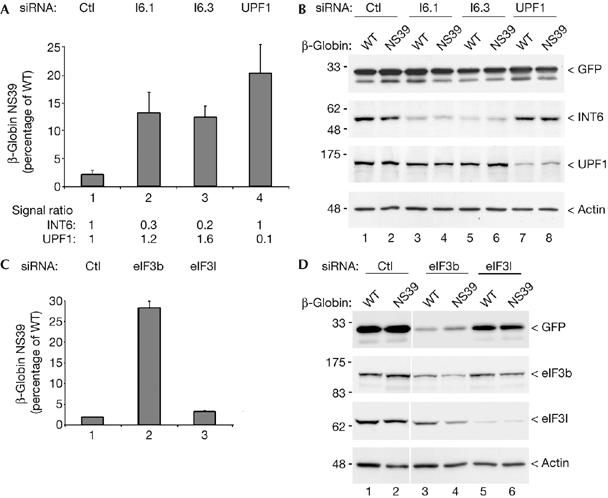
Silencing of INT6 impairs nonsense-mediated messenger RNA decay. (A) Real-time RT–PCR analysis was used to measure the level of the NMD-sensitive NS39 β-globin transcript from HeLa cells treated with control siRNA (lane 1), siRNAs targeting INT6 (lanes 2,3) and the UPF1 essential NMD factor (lane 4). Values were normalized for transfection efficiency to the level of GFP messenger RNA and results are expressed as a percentage of the value obtained for the wild-type (WT) β-globin transcript, which was defined as 100% for each siRNA couple. Data refer to the mean of three independent experiments. INT6 and UPF1 mRNA reduction was monitored by real-time RT–PCR and ratios (set to 1 in control cells) are shown below the graph. (B) The amount of GFP produced in cells treated as above was analysed by immunoblotting and served as a measure of steady-state translation. Efficiency of RNAi was verified by immunoblotting with antibodies to INT6, and UPF1, and equal protein loading was controlled by the detection of actin. (C) The level of the NS39 β-globin NMD substrate was measured as in (A) after RNAi of two other eIF3 subunits. Cells were transfected with control siRNA (lane 1) and siRNAs targeting eIF3b (lane 2) and eIF3l (lane 3). (D) A measure of steady-state translation was performed as in (B) from an aliquot of cells used in (C). Experimental controls were maintained as in (B). Ctl, control; GFP, green fluorescent protein; NMD, nonsense-mediated messenger RNA decay; RNAi, RNA interference; RT–PCR, reverse transcription–PCR; siRNA, short interfering RNA.
The same experiments were carried out with siRNAs targeting the eIF3b core subunit and the eIF3l non-core subunit, which interacts directly with INT6 (Morris-Desbois et al, 2001). Knockdown of eIF3b increases the NS39 mRNA levels (Fig 1C), but because it also strongly inhibits general translation, as shown by immunoblot analyses of GFP and endogenous eIF3l (Fig 1D), the inhibitory effect on NMD should relate to the translational arrest. After depletion of eIF3l, neither NMD nor steady-state translation was significantly affected (Fig 1C,D). Collectively, these data show that INT6 is specifically required for NMD but is not essential for general translation, as anticipated (Bandyopadhyay et al, 2000; Morris & Jalinot, 2005; Zhou et al, 2005).
Most capped mRNAs are subjected to NMD soon after their transcription when they are still bound by the nuclear cap binding heterodimer CBP80–CBP20, which drives the pioneer round of translation (Ishigaki et al, 2001; Chiu et al, 2004). Hence, we investigated whether INT6 could be found in complexes including the cap-binding protein (CBP) dimer. Indeed, antibodies to INT6 specifically precipitated CBP80, and this association was still detected when RNase was added before precipitation (Fig 2A), therefore excluding the possibility that it merely results from the presence of both proteins on the same mRNA. In the same experiment, eIF4G-1 was also detected with a weak intensity, but we failed to detect eIF4E (Fig 2A). CBP80 was also detected when precipitation was carried out with an antibody to eIF3 (Fig 2B). INT6 has been shown to be involved in protein degradation (Yen et al, 2003); therefore, we examined whether its silencing affects expression of the UPF1 and UPF2 NMD factors. Immunoblot analysis showed that neither UPF1 nor UPF2 concentration was modified in INT6-silenced cells (Fig 3A). An interaction between these two proteins is required for NMD; therefore this was studied next, and we found that UPF1 was still present in UPF2 immunoprecipitates after INT6 RNAi (Fig 3B). Finally, we investigated whether INT6 depletion inhibits NMD by acting on UPF1 phosphorylation (Pal et al, 2001) mediated by the ataxia telangiectasia mutated (ATM)-like kinase SMG1 (Denning et al, 2001; Yamashita et al, 2001). By using an antibody specific for sites phosphorylated by this kinase family, we found that INT6 RNAi did not modify the extent of UPF1 phosphorylation (Fig 3C). Together, these observations indicate that NMD impairment caused by INT6 knockdown is apparently not due to an effect on the functioning of UPF1 and UPF2 factors.
Figure 2.
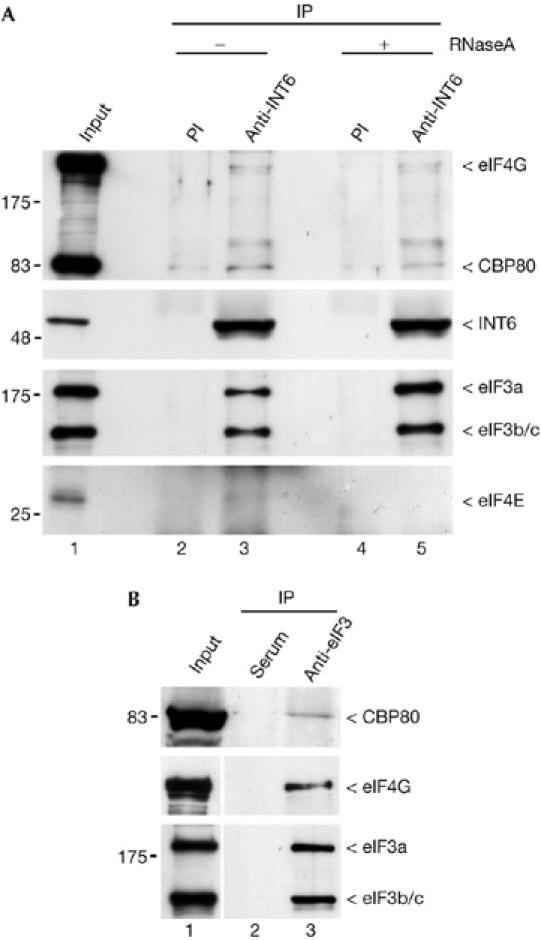
CBP80 co-immunopurifies with INT6. (A) A HeLa whole-cell extract was incubated for 30 min in the absence (lanes 2,3) or presence of 40 μg of RNase A (lanes 4,5) and then immunoprecipitated (IP) with preimmune sera (PI, lanes 2,4) or a mix of C-20 and N-19 antibodies to INT6 (lanes 3,5). Input (1 μl) and pull-down (obtained from 300 μl of lysate) were analysed by immunoblotting using antibodies to CBP80 and eIF4G (top panel). Recovery of INT6 and its co-immunoprecipitation with other eIF3 subunits are shown in the two middle panels. Only the part of the blot corresponding to the largest eIF3 subunits is shown. Similar precipitation reactions were performed with recovered proteins eluted into sample buffer without β-mercaptoethanol and analysed by immunoblotting using an antibody to eIF4E (bottom panel). (B) The extent of CBP80 and eIF4G protein amounts that are pulled down with INT6 was compared with those obtained with the entire eIF3 complex. The same lysate as in (A) was immunoprecipitated with a crude eIF3 antiserum (B, lane 3) or normal goat serum (lane 2). Immunoprecipitates and lysate (lane 1) were analysed by immunoblotting with antibodies to CBP80 (top), eIF4G (middle) and eIF3 (bottom).
Figure 3.
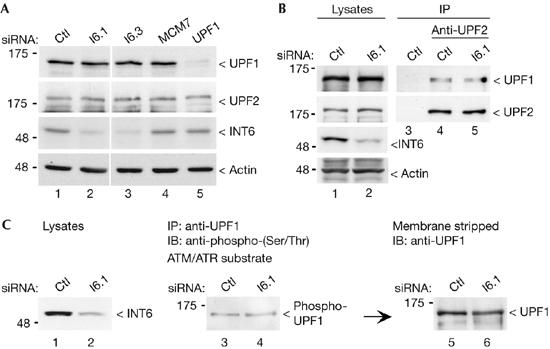
No apparent modification of UPF1 and UPF2 in INT6-silenced cells. (A) INT6 depletion does not change UPF1 or UPF2 protein levels. HeLa cells transfected with control (Ctl) siRNA (lane 1), siRNAs targeting INT6 (lanes 2,3), MCM7 (lane 4) and UPF1 (lane 5) were analysed by immunoblotting with antibodies to UPF1 and UPF2. Efficiency of RNAi and equal protein loading were controlled by the detection of INT6 and actin, respectively. (B) Silencing of INT6 does not affect the association of UPF1 with UPF2. Lysates from cells transfected with control or I6.1 siRNAs, as indicated above the blots, were immunoprecipitated (IP) with an antibody to UPF2 (lanes 4,5), which was omitted in lane 3, and analysed for the presence of UPF1 by immunoblotting (IB; top panel). Lanes 1 and 2 correspond to aliquots of cell lysates. The same blot was probed with the antibody against UPF2 to ensure equal recovery of the protein. Efficiency of RNAi and equal protein loading were controlled as in (A). (C) Phosphorylation of UPF1 was not altered after INT6 RNAi. Lysates from control and INT6-depleted cells (left panel) were immunoprecipitated with the UPF1 antibody and analysed by immunoblotting by using a phospho-(Ser/Thr) ATM/ATR substrate antibody (middle panel). After stripping, the membrane was reprobed with the antibody to UPF1 to verify its precipitation (right panel). MCM, minichromosome maintenance; RNAi, RNA interference; siRNA, short interfering RNA.
To explore further the role of INT6 in NMD, we investigated whether it associates with UPF proteins. UPF1 was weakly detected in the INT6 immunoprecipitates and RNase treatment of cell lysate before precipitation further reduced the amount of UPF1 that was pulled down (Fig 4A, top). By contrast, UPF2 was efficiently precipitated with INT6 and the interaction was unaffected by RNase treatment (Fig 4A, middle). Co-purification of UPF2 was also observed in precipitations using antibodies to whole eIF3 or to the eIF3b core subunit (Fig 4B).
Figure 4.

UPF2 co-immunopurifies with INT6 and eIF3. (A) A HeLa cell extract was processed as in Fig 2A, and the presence of UPF1 and UPF2 in the immunoprecipitates was analysed by immunoblotting. The same blot was successively probed with antibodies against UPF1 (top), UPF2 (middle) and INT6 (bottom). (B) A lysate as in (A) was immunoprecipitated with a goat serum (lane 1), a crude eIF3 antiserum (lane 2) and an antibody to eIF3b (lane 3). Immunoprecipitates were analysed by immunoblotting with antibodies to UPF2 (top), total eIF3 complex (middle) and eIF3b (bottom). IP, immunoprecipitation; PI, preimmune sera.
These observations are in agreement with a specific role of INT6 in NMD. To substantiate this further, we examined whether INT6 depletion influences the expression of physiologic NMD substrates. Several genes were selected from previous genome-wide studies designed to identify mRNAs sensitive to UPF1 or UPF2 depletion (Mendell et al, 2004; Wittmann et al, 2006), and their transcript levels were quantified in cells treated with siRNAs targeting INT6; UPF1, was used as comparison. Table 1, which summarizes our results and data from previous studies, shows that selected genes can be divided into two groups. The first group includes genes in which the mRNA levels were raised in response to UPF1 or INT6 depletion, and the second group includes genes in which the transcript levels were increased by UPF1 silencing but not by INT6 suppression. This set of data is restricted; however, it establishes that some known NMD substrates require INT6 for degradation, whereas others that are prone to UPF1-mediated degradation do not.
Table 1.
Abundance of INT6-depleted cells in transcripts known to be controlled by the UPF1 essential nonsense-mediated mRNA decay factor
| Gene | *Fold mRNA changes | ||||
|---|---|---|---|---|---|
| UPF1 | I6(1) | I6(3) | Mendell (UPF1) | Wittmann (UPF2) | |
| Group 1 | |||||
| Growth arrest and DNA-damage-inducible α (GADD45A) | 2.7 | 2.3 | 2.9 | 4.7 | 1.2 |
| BCL2-associated athanogene (BAG1) | 2.9 | 1.9 | 2.6 | 2.5 | 2.4 |
| Slit homologue 2 (SLIT2) | 2.3 | 2.1 | 2.0 | 3.7 | 3.4 |
| Activating transcription factor 4 (ATF4) | 1.9 | 1.3 | 1.5 | 4.1 | 0.9 |
| Group 2 | |||||
| Growth arrest and DNA-damage-inducible (GADD45B) | 2.5 | 1.0 | 1.1 | 6.2 | 1.1 |
| Mitogen-activated protein kinase kinase kinase 14 (MAP3K14) | 2.5 | 0.6 | 0.4 | 11.3 | 1.7 |
| Seryl-tRNA synthetase (SARS) | 2.2 | 1.0 | 1.0 | 2.4 | 0.7 |
| Asparagine synthetase (ASNS) | 2.2 | 0.5 | 0.4 | 4.7 | 0.7 |
| Cysteinyl-tRNA synthetase (CARS) | 1.8 | 0.7 | 0.7 | 3.3 | 0.6 |
*‘Fold mRNA changes' is the mean of the ratio of mRNA levels measured by quantitative real-time–PCR in cells treated with UPF1 or INT6 short interference RNA (siRNA) duplexes compared with cells transfected with control siRNA, after normalization with the β-actin signal. Data refer to the mean of three independent experiments. Our results were compared with microarray data obtained from HeLa cells transfected with UPF1 siRNA (Mendell et al, 2004) and in HeLa cells stably depleted for UPF2 (Wittmann et al, 2006).
Discussion
Our findings show that INT6 is required for degradation of a model NMD-sensitive transcript and also for several cellular mRNAs regulated by this process. Although it has been shown that eIF3 participates in the pioneer round of translation (Chiu et al, 2004), the functional contribution of its different subunits to the NMD process has only been addressed in one study, by Welch & Jacobson (1999). They reported that mutation of PRT1, the eIF3b yeast homologue, causes increased suppressor of Ty 10 (SPT10) mRNA levels, in a manner similar to UPF1 depletion. In agreement with this study, we found that silencing of eIF3b, a core subunit of the initiation factor, hinders both the pioneer round and steady-state translation. Concerning INT6, our results show that it is not required for general translation, but its silencing inhibits NMD. This was not observed after depletion of eIF3l, a protein also viewed as an eIF3 subunit. eIF3l is found in the cytoplasm in association with INT6 and eIF3 (Morris-Desbois et al, 2001), and in the nucleus, it is nucleolar and acts as a cofactor of RNA PolI (Seither et al, 2001; Yuan et al, 2002). By contrast, nuclear INT6 is present in the nucleoplasm and is partly associated with chromatin. As previously reported in other species (Yahalom et al, 2001), cell fractionation experiments showed that all eIF3 subunits are not equally distributed between the nucleus and the cytoplasm (C. Morris, data not shown). Hence, the composition of eIF3 is likely to be dynamic and one possibility is that a specific subcomplex is implicated in the nuclear pioneer round of translation. Co-precipitation of UPF2 with some eIF3 subunits, including INT6, is interesting and might represent a direct link between NMD core factors and eIF3. INT6 within eIF3 has recently been shown to interact in vitro with eIF4G-1 (LeFebvre et al, 2006). Our results show that INT6 interacts with both CBP80 and UPF2, and also less intensely with eIF4G-1, even when RNA is degraded. These proteins include the middle domain of eukaryotic initiation factor 4G (MIF4G) motifs. Collectively, these observations raise the possibility that INT6 binds specifically to MIF4G domains and this would allow efficient recruitment of eIF3 complexes containing INT6 to mRNAs loaded with UPF2, thereby promoting the pioneer round of translation. INT6 was not found associated with eIF4E; therefore, this eIF3 subunit would not intervene in steady-state translation.
In conclusion, our findings support a model in which INT6 acts in CBP80-mediated translation and, in combination with UPF2, directs mRNAs towards degradation rather than active translation. They establish that human INT6, in the eIF3 complex, is involved in the quality control of specific mRNAs, the abundance of which is regulated by NMD.
Methods
Cell culture, transfection of plasmids and short interference RNAs. HeLa cells were cultured in Dulbecco's modified Eagle's medium under standard conditions. Cells were co-transfected with an NMD reporter plasmid (pBluescript-β-globin WT or pBluescript-β-globin NS39; Thermann et al, 1998), a reference plasmid (pSG–Flag–GFP) together with different siRNA duplexes listed in Table 2. Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) was used according to the supplier's protocol. Cells were split 24 h after transfection and cultured for another 48 h. HeLa cells treated with siRNAs alone were collected 72 h after transfection. One portion of cells was used as a source of RNA, whereas the other was used as a source of protein.
Table 2.
Synthetic short interference RNA used in this study
| siRNA | 19-nt mRNA targeted sequence (sense strand) |
|---|---|
| I6.1 | 5′-CAGGGAUGGUAGGAUGCUC-3′ |
| I6.3 | 5′-GUCUUUCCGCUUCUUGAAU-3′ |
| I6.4 | 5′-AGGGUGACUUACAUUUUGG-3′ |
| UPF1 | 5′-GAUGCAGUUCCGCUCCAUU-3′ |
| eIF3b | 5′-CGCCGACGGCUACAAGCUU-3′ |
| eIF3l | 5′-GUCGUGUCUCCAGUGAUGU-3′ |
| MCM7 | 5′-GAGCUGCUGCCUCAGUACA-3′ |
| Control | 5′-CGGGCAGAGCCUGCUGGGC-3′ |
nt, nucleotide; siRNA, short interfering RNA.
Nonsense-mediated messenger RNA decay analysis and real-time reverse transcription–PCR. For quantification of exogenous β-globin transcripts or endogenous NMD substrates, total RNA was isolated using the RNeasy Mini kit (Qiagen, Hilden, Germany), and real-time reverse transcription–PCR (RT–PCR) analysis was performed using the QuantiTect SYBR green RT–PCR kit (Qiagen) and a LightCycler apparatus (Roche, Basel, Switzerland) according to cycling conditions specified in the handbook of the kit. Primer sequences were designed using the Primer3 software and are listed in Table 3.
Table 3.
Quantitative real-time PCR primers used in this study
| Gene-specific primers | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|
| β-Globin | TTGGGGATCTGTCCACTCC | CACACCAGCCACCACTTTC |
| GFP | ACGTAAACGGCCACAAGTTC | AAGTCGTGCTGCTTCATGTG |
| INT6 | TTCTTCAATCACCCCAAAGG | TAGAACCTGCCGACGTTTTC |
| UPF1 | AGGTACCGACAGTCCCTGTG | CAATGACACCACACCCTCAG |
| GADD45A | ACGAGGACGACGACAGAGAT | GCAGGATCCTTCCATTGAGA |
| ATF4 | GAGCGTCCATTTTGTGGAAC | GAAGCCAACTCCCATTAGAGG |
| SLIT2 | CCCCACAAATCTTCCAGAGA | AGCGTAGTCCTTGGAAAGCA |
| BAG1 | TGCCGGGTCATGTTAATTGGG | AGAACCAGTGTGAGAGTAGGAAA |
| ASNS | CGACCAAAAGAAGCCTTCAG | CCACTTGGGCATCCAGTAAT |
| CARS | ATGACATGGAGGGCAAAGAG | TTAGGACCCAAGGGTGACTG |
| MAP3K14 | TCAGTGCAGAACCAGGTCAG | GGGGACTGAGAACCACTTCA |
| GADD45B | TCGGATTTTGCAATTTCTCC | GACTCGTACACCCCCACTGT |
| SARS | CTGGCCTGTCTACCTGCTTC | CTGGCAGCATGATTCAAAGA |
| β-Actin | GGACTTCGAGCAAGAGATGG | AGCACTGTGTTGGCGTACAG |
ASNS, asparagine synthetase; ATF4, activating transcription factor 4; BAG1, BCL2-associated athanogene; CARS, cysteinyl-tRNA synthetase; GADD45, growth arrest and DNA-damage-inducible; GFP, green fluorescent protein; INT6, integration site 6; MAP3K14, mitogen-activated protein kinase kinase kinase 14; SARS, seryl-tRNA synthetase; SLIT2, slit homologue 2; UPF1, Up Frameshift 1.
Immunoprecipitation and immunoblotting. For immunoprecipitation, 1 × 107 cells were lysed in 300 μl of buffer N (50 mM Tris–HCl, pH 7.4; 300 mM NaCl; 0.05% NP-40; 0.5 mM TCEP (Sigma, St Louis, MO, USA); 0.5 mM Pefabloc (Roche) and 50 U of RNase inhibitor (Promega, Madison, WI, USA)) by four 10 s sonications using a Bioruptor system (Diagenode, Liège, Belgium) and centrifuged at 10,000g for 10 min at 4°C. Lysates were precleared by incubation with 40 μl of protein A–agarose beads for 1 h and the supernatants were incubated overnight at 4°C with primary antibodies. An 80 μl volume of protein A–agarose beads was then added for 1 h, washed three times for 10 min and resuspended in 20 μl of 2 × SDS sample buffer. After separation by SDS–polyacrylamide gel electrophoresis, proteins were transferred to a PVDF membrane and immunoblotting was performed using protein A/G coupled to peroxidase. The following antibodies have been previously described: INT6 C-20 and N-19 (Morris-Desbois et al, 1999); eIF3l (Morris-Desbois et al, 2001); CBP80 (Izaurralde et al, 1994); UPF1 (Applequist et al, 1997); UPF2 (Wittmann et al, 2006); eIF3 (Asano et al, 1997); and eIF4E (Morley & McKendrick, 1997). The following antibodies were purchased: Flag (clone M2; Sigma), β-actin (Sigma), eIF3b (A-20; Santa Cruz Biotechnology, Santa Cruz, CA, USA), eIF4G (H-300; Santa Cruz) and anti-phospho-(Ser/Thr) ATM/ATR (ataxia telangiectasia and Rad3-related) substrate antibody (#2851; Cell Signaling Technology, Danvers, MA, USA).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Fig S1
supplementary Fig S2
Acknowledgments
We thank A. Kulozik, N. Gehring, S. Mazoyer, E. Izaurralde, S. Morley and J. Hershey for kindly providing the plasmids and antibodies. We are also grateful to A. Roisin and S. Buchsbaum for cells and plasmid. This work was supported by grants from the Interdisciplinary Center of Clinical Research (IZKF) and the University's ELAN fund (H.-M.J. and J.W.) and from the ‘Fondation pour la Recherche Médicale' (P.J.).
References
- Akiyoshi Y, Clayton J, Phan L, Yamamoto M, Hinnebusch AG, Watanabe Y, Asano K (2001) Fission yeast homolog of murine Int-6 protein, encoded by mouse mammary tumor virus integration site, is associated with the conserved core subunits of eukaryotic translation initiation factor 3. J Biol Chem 276: 10056–10062 [DOI] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Raman C, Jack HM (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Merrick WC, Hershey JW (1997) The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem 272: 23477–23480 [DOI] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG (1998) Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem 273: 18573–18585 [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R (2004) Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 16: 293–299 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Matsumoto T, Maitra U (2000) Fission yeast Int6 is not essential for global translation initiation, but deletion of int6(+) causes hypersensitivity to caffeine and affects spore formation. Mol Biol Cell 11: 4005–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Cheng J, Maquat LE (1993) Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA 90: 482–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE (2004) The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev 18: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Izaurralde E (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 17: 316–325 [DOI] [PubMed] [Google Scholar]
- Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP (2001) Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem 276: 22709–22714 [DOI] [PubMed] [Google Scholar]
- Hoareau Alves K, Bochard V, Rety S, Jalinot P (2002) Association of the mammalian proto-oncoprotein Int-6 with the three protein complexes eIF3, COP9 signalosome and 26S proteasome. FEBS Lett 527: 15–21 [DOI] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668 [DOI] [PubMed] [Google Scholar]
- Karniol B, Yahalom A, Kwok S, Tsuge T, Matsui M, Deng XW, Chamovitz DA (1998) The Arabidopsis homologue of an eIF3 complex subunit associates with the COP9 complex. FEBS Lett 439: 173–179 [DOI] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG (2004) Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16: 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE (2006) Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R (1995) Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol 69: 1932–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Morley SJ, McKendrick L (1997) Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T3 cells. J Biol Chem 272: 17887–17893 [DOI] [PubMed] [Google Scholar]
- Morris C, Jalinot P (2005) Silencing of human Int-6 impairs mitosis progression and inhibits cyclin B–Cdk1 activation. Oncogene 24: 1203–1211 [DOI] [PubMed] [Google Scholar]
- Morris-Desbois C, Bochard V, Reynaud C, Jalinot P (1999) Interaction between the Ret finger protein and the Int-6 gene product and co-localisation into nuclear bodies. J Cell Sci 112: 3331–3342 [DOI] [PubMed] [Google Scholar]
- Morris-Desbois C, Rety S, Ferro M, Garin J, Jalinot P (2001) The human protein HSPC021 interacts with Int-6 and is associated with eukaryotic translation initiation factor 3. J Biol Chem 276: 45988–45995 [DOI] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, Maquat LE (2001) Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol 18: 4935–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seither P, Iben S, Thiry M, Grummt I (2001) PAF67, a novel protein that is associated with the initiation-competent form of RNA polymerase I. Biol Chem 382: 1163–1170 [DOI] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Jacobson A (1999) An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J 18: 6134–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jack HM (2006) hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol 26: 1272–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom A, Kim TH, Winter E, Karniol B, von Arnim AG, Chamovitz DA (2001) Arabidopsis eIF3e (INT-6) associates with both eIF3c and the COP9 signalosome subunit CSN7. J Biol Chem 276: 334–340 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Gordon C, Chang EC (2003) Schizosaccharomyces pombe Int6 and Ras homologs regulate cell division and mitotic fidelity via the proteasome. Cell 112: 207–217 [DOI] [PubMed] [Google Scholar]
- Yuan X, Zhao J, Zentgraf H, Hoffmann-Rohrer U, Grummt I (2002) Multiple interactions between RNA polymerase I, TIF-IA and TAF(I) subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep 3: 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA (2005) PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Fig S1
supplementary Fig S2


