Transfer RNAs (tRNAs) are involved in many cellular functions distributed throughout the cellular space. In addition to their role in protein translation, these molecules are engaged in tasks as diverse as the regulation of gene expression, amino-acid synthesis, protein degradation, cell-wall synthesis, porphyrin biosynthesis, priming of replication, RNA interference and the transport of macromolecules. The nucleus and the mitochondria of eukaryotic cells, as well as the chloroplasts in plants, contain their own genome that encodes a range of proteins and nucleic acids. Early evidence suggested that some of the RNA content of mitochondria could be transcribed from non-mitochondrial DNA (Suyama, 1967). It is now well established that a variable number of tRNA species present in the mitochondria are indeed nucleus-encoded (Fig 1). The phenomenon of mitochondrial tRNA import has been reported in plants, marsupials, the yeast Saccharomyces cerevisiae, and the protozoa Leishmania, Trypanosoma and Tetrahymena. When certain tRNA species are not encoded by the mitochondrial genome, there is a predictable requirement for the import of nucleus-encoded tRNAs into the mitochondria (Schneider & Marechal-Drouard, 2000). This is the case, for example, for the mitochondria of Leishmania, which are completely devoid of tRNA-encoding genes. However, there are a few examples of tRNA import into mitochondria that already have a complete set of mitochondria-encoded tRNAs. Other tRNA transport systems have also been identified (Fig 1), which include their retrograde transport from the cytoplasm to the nucleus (Takano et al, 2005; Shaheen & Hopper, 2005), the export of mitochondria-encoded tRNA to the cytoplasm (Maniataki & Mourelatos, 2005), and the packaging of tRNAs into retroviruses (Waters & Mullin, 1977). The roles of some of these transport systems have yet to be defined.
Figure 1.
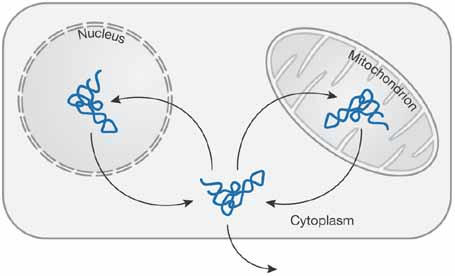
The travels of transfer RNA. tRNAs are either nucleus- or mitochondria-encoded. Several tRNA transport systems have been described: from the nucleus or the mitochondria to the cytoplasm; from the cytoplasm to the nucleus or the mitochondria; and export out of the cell.
In yeast, the cytoplasmic, nucleus-encoded tRNA(Lys,CUU) is imported into the mitochondria, although the mitochondrial genome encodes the full set of tRNAs, including an authentic tRNALys (Tarassov & Entelis, 1992). Furthermore, the yeast mitochondrial lysyl-tRNA synthetase (mLysRS) cannot aminoacylate the nucleus-encoded tRNA(Lys,CUU). This is because the mitochondrial, prokaryotic-type enzyme requires an adenine at position 73 to aminoacylate its cognate tRNA, whereas the eukaryotic enzyme accepts any of the four nucleotides at this position. Therefore, the physiological significance of this import is unclear. When the import of a tRNA species is not a prerequisite for mitochondrial protein synthesis, one interesting possibility is that import of that tRNA is a means to transport a protein into the mitochondrial compartment in a co-import mechanism. This suggests that the co-imported protein is not able to use the protein-import system. Such a mechanism has been described for the nuclear export of human elongation factor EF1A, which is efficiently excluded from the nucleus by a co-transport process involving exportin 5 and tRNA as a carrier molecule (Bohnsack et al, 2002). This explanation might hold true in some circumstances for mitochondrial import, but is not a universal answer to the problem. Indeed, the nucleus-encoded tRNA(Lys,CUU) referred to above is actually co-imported through the protein-import pathway.
The essential requirement of tRNA import for protein synthesis to occur in the mitochondria of some organisms stimulated the initial research in this field (Marechal-Drouard et al, 1988; Hancock & Hajduk, 1990; Lye et al, 1993; Schneider et al, 1994). However, no unifying mechanism has emerged from the various in vitro and in vivo studies performed so far. In S. cerevisiae, cytosolic factors are involved in this process. Co-import of a tRNA(Lys,CUU) with the mitochondrial precursor of LysRS has been established (Tarassov et al, 1995), but recruitment of this tRNA to the mitochondrial compartment also requires the glycolytic enzyme, enolase (Entelis et al, 2006). In this system, it is likely that the protein-import channels are used to translocate tRNA(Lys,CUU). By contrast, import of tRNA in Leishmania mitochondria does not require cytosolic factors and the existence of a receptor-mediated import pathway has been uncovered (Mahapatra et al, 1994; Mahapatra & Adhya, 1996). A large multisubunit complex, isolated from the inner mitochondrial membrane of this organism, promotes tRNA import when incorporated into phospholipid vesicles (Bhattacharyya et al, 2003). Transport is ATP-dependent. Previous studies identified two components of this RNA import complex—RIC1 and RIC8A—as tRNA import receptors. RIC1 is homologous to the α-subunit of F1 ATP synthase (Goswami et al, 2006), and RIC8A is homologous to subunit 6b of the respiratory complex III (Chatterjee et al, 2006). Interestingly, RIC1 and RIC8A are responsible for the transport of type I and type II RNAs, respectively: RIC1 specifically recognizes import determinants localized in the DHU stem loop of tRNAs, whereas RIC8A recognizes determinants in the variable-TΨC stem loop structure.
In this issue of EMBO reports, Mukherjee and colleagues provide a comprehensive description of the isolation and preliminary characterization of the RIC that mediates mitochondrial tRNA import in the kinetoplastid protozoon Leishmania tropica (Mukherjee et al, 2007). The authors affinity-purified the RIC through a column containing a tRNA-import signal oligonucleotide, and, by using mass spectrometry, the various components were identified. A total of 11 subunits were detected, eight of which are nucleus-encoded. Probably the most surprising feature of the RIC is that five of its components are shared with respiratory complexes, and therefore are likely to be bifunctional: RIC6 and RIC8A are the iron sulphur protein and subunit 6b of the respiratory complex III, respectively; RIC5 and RIC9 are components of complex IV; and RIC1 is also known as the F1 ATP synthase subunit of complex V. Consequently, RIC provides a spectacular example of a complex that is a reservoir of multifunctional proteins and strengthens the view that the association of a polypeptide with a variable set of partners expands the functions of a given protein. Systematic knockdown of the RIC components by RNA interference was used to determine their role in mitochondrial tRNA import. The six subunits—RIC1, 4A, 6, 8A, 8B and 9—are essential in vivo. They are also necessary and sufficient to support tRNA transport in vitro in a reconstitution assay. Indeed, the various components were expressed in Escherichia coli and an active RIC could be reconstituted into liposomes. This complex displayed the regulatory properties of a native RIC, suggesting that the identified components are necessary and sufficient for tRNA import. This is a significant advance towards the understanding of the import process, and these new data will certainly stimulate further investigations in the field. Now that the components of the molecular machinery responsible for mitochondrial tRNA import are known, further study will undoubtedly lead to the elucidation of the function of all the components of the RIC and provide a detailed description of the process.
Additional studies will be necessary to understand fully the mitochondrial tRNA-import mechanism. Intriguingly, only a fraction of a cytosolic tRNA is imported into the mitochondria, so what mechanism governs the balance between the mitochondrial and cytosolic fractions of a given tRNA? Another challenge is to understand the role of ATP hydrolysis and the proton gradient in tRNA translocation. A working model has been proposed (Bhattacharyya & Adhya, 2004), which suggests that binding of tRNAs to the RIC triggers a series of events that leads to tRNA translocation: activation of an ATPase, ATP hydrolysis in the mitochondrial matrix, export of protons and, finally, proton-dependent tRNA import through an RNA channel (Fig 2). How universal this mechanism is remains to be established. The extent of mitochondrial tRNA import differs between species, suggesting that import machineries have been independently selected as a result of the loss of organellar tRNA genes during evolution. It is noteworthy that in plant mitochondria, a distinct import machinery—the voltage-dependent anion channel—has been identified (Salinas et al, 2006). Finally, several human diseases are associated with mutations in mitochondrial tRNA genes. Human mitochondria do not naturally import tRNA, but the possibility of rescuing mutations through the import of nucleus-encoded tRNAs has been established (Kolesnikova et al, 2004; Mahata et al, 2006). Therefore, a fuller understanding of the tRNA-import process as provided by Mukherjee and colleagues might also have an impact on the rescue of mitochondrial dysfunctions.
Figure 2.
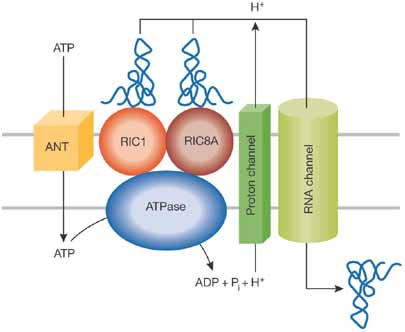
The machinery of nucleus-encoded tRNA import through the inner mitochondrial membrane in Leishmania. Adapted with the permission of Landes Bioscience from Bhattacharyya & Adhya, 2004. ANT, a transporter similar to the adenine nucleotide translocator; RIC, RNA import complex.

References
- Bhattacharyya SN, Adhya S (2004) The complexity of mitochondrial tRNA import. RNA Biol 1: 84–88 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Chatterjee S, Goswami S, Tripathi G, Dey SN, Adhya S (2003) ‘Ping-pong' interactions between mitochondrial tRNA import receptors within a multiprotein complex. Mol Cell Biol 23: 5217–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Görlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Home P, Mukherjee S, Mahata B, Goswami S, Dhar G, Adhya S (2006) An RNA-binding respiratory component mediates import of type II tRNAs into Leishmania mitochondria. J Biol Chem 281: 25270–25277 [DOI] [PubMed] [Google Scholar]
- Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I (2006) A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev 20: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S, Dhar G, Mukherjee S, Mahata B, Chatterjee S, Home P, Adhya S (2006) A bifunctional tRNA import receptor from Leishmania mitochondria. Proc Natl Acad Sci USA 103: 8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K, Hajduk SL (1990) The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem 265: 19208–19215 [PubMed] [Google Scholar]
- Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I (2004) Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet 13: 2519–2534 [DOI] [PubMed] [Google Scholar]
- Lye LF, Chen DHT, Suyama Y (1993) Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol Biochem Parasitol 58: 233–245 [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Adhya S (1996) Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J Biol Chem 271: 20432–20437 [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Ghosh T, Adhya S (1994) Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res 22: 3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahata B, Mukherjee S, Mishra S, Bandyopadhyay A, Adhya S (2006) Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science 314: 471–474 [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z (2005) Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the argonaute 2 protein. RNA 11: 849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal-Drouard L, Weil JH, Guillemaut P (1988) Import of several tRNAs from the cytoplasm into the mitochondria in bean Phaseolus vulgaris. Nucleic Acids Res 16: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Basu S, Home P, Dhar G, Adhya S (2007) Necessary and sufficient factors for import of tRNA into the kinetoplast mitochondrion. EMBO Rep 8: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas T, Duchene AM, Delage L, Nilsson S, Glaser E, Zaepfel M, Marechal-Drouard L (2006) The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc Natl Acad Sci USA 103: 18362–18367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Marechal-Drouard L (2000) Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol 10: 509–513 [DOI] [PubMed] [Google Scholar]
- Schneider A, Martin J, Agabian N (1994) A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol Cell Biol 14: 2317–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK (2005) Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama Y (1967) The origins of mitochondrial ribonucleic acids in Tetrahymena pyriformis. Biochemistry 6: 2829–2839 [DOI] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T (2005) tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Tarassov IA, Entelis NS (1992) Mitochondrially-imported cytoplasmic tRNALys(CUU) of Saccharomyces cerevisiae—in vivo and in vitro targetting systems. Nucleic Acids Res 20: 1277–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Entelis N, Martin RP (1995) Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J 14: 3461–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LC, Mullin BC (1977) Transfer RNA in RNA tumor viruses. Prog Nucleic Acid Res Mol Biol 20: 131–160 [DOI] [PubMed] [Google Scholar]


