Abstract
Acetylation of the ε-amino group of lysine residues (Nε-acetylation) is a reversible post-translational modification with the potential to rival phosphorylation. In addition to histones and many transcription factors such as p53, regulators of DNA repair, replication and recombination are subject to Nε-acetylation. This modification is also important for governing the activities of various enzymes, including histone acetyltransferases, histone deacetylases, bacterial and mammalian acetyl-CoA synthases, kinases, phosphatases, the ubiquitin ligase murine double minute 2 and the chaperonin heat shock protein 90. Furthermore, lysine acetylation occurs in cellular structure proteins such as α-tubulin, actin, cortactin and p120 catenin. Strikingly, the Yersinia outer protein YopJ promotes O-acetylation of crucial serine and threonine residues that are required for activation of the MAPK/ERK kinase and IκB kinase families, which precludes their phosphorylation and blocks signal transduction. Thus, Nε- and O-acetylation are becoming recognized as two prominent mechanisms for regulating protein functions in diverse organisms.
Keywords: multisite modification, lysine acetylation, methylation, ubiquitylation, molecular barcode
Introduction
Among the 20 common amino-acid residues, lysine is unique as its ε-amino group is subject to different modifications (Fig 1A). One of them is acetylation, which is also known as Nε- or lysine acetylation. This modification was identified initially on core histones (Vidali et al, 1968) and then on high mobility group protein 1 (HMG1; Sterner et al, 1979), α-tubulin (L'Hernault & Rosenbaum, 1985; Piperno & Fuller, 1985) and the tumour suppressor p53 (Gu & Roeder, 1997). During the past decade, the prevalence of Nε-acetylation has been extensively characterized for core histones and more than 60 transcription factors (reviewed by Kouzarides, 2000; Sterner & Berger, 2000; Yang, 2004a; Glozak et al, 2005; Nightingale et al, 2006). Recent studies have identified acetyl-lysine residues in many other proteins, including regulators of DNA repair and replication, histone acetyltransferases and deacetylases, metabolic enzymes, kinases, phosphatases and cellular structure proteins. It is noteworthy that this reversible covalent modification is different from Nα-acetylation, which targets the amino terminus of a protein and is mainly co-translational. O-acetylation has been recently identified as a third type of protein acetylation (Mittal et al, 2006; Mukherjee et al, 2006). It was found that Yersinia outer protein J (YopJ) acetylates the side-chains of serine and threonine residues in two families of protein kinases (Fig 1B). In this review, we highlight the importance of Nε-acetylation in various cellular processes, briefly discuss the regulatory potential of O-acetylation, and emphasize how these two types of acetylation interplay with other modifications such as phosphorylation, methylation, ubiquitylation and sumoylation to form dynamic regulatory programmes for the spatiotemporal control of protein function in different organisms.
Figure 1.
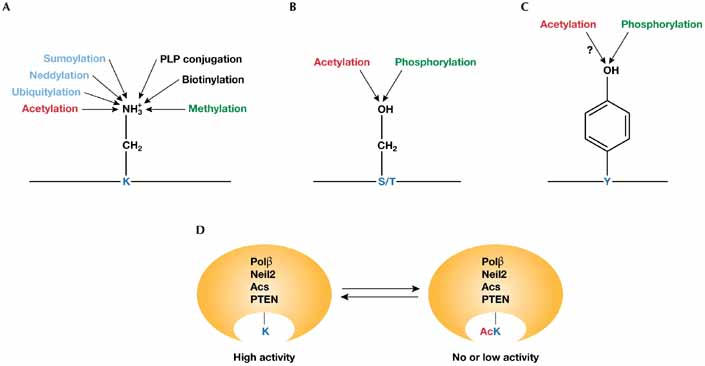
Mechanistic impact of Nε- and O-acetylation. (A) The amino group of a lysine (K) residue is subject to acetylation and other covalent modifications. Acetylation precludes other modifications, and vice versa. Pyridoxal 5′-phosphate (PLP) is a biologically active form of vitamin B6 (Huq et al, 2007). Other, as yet unidentified, lysine modifications, such as propionylation and butyrylation, might exist. (B) The hydroxyl group of a serine (S) or threonine (T) residue is the target of mutually exclusive phosphorylation and acetylation. (C) The hydroxyl group of a tyrosine (Y) residue is the target of competitive phosphorylation and acetylation. (D) Acetylation of a crucial lysine residue in the catalytic centre of DNA polymerase β (Polβ), Neil2, acetyl-CoA synthase (Acs) or the phosphatase and tensin homologue (PTEN) completely or partly inhibits enzymatic activity. AcK, acetyl-lysine.
Lysine acetylation in RNA and DNA metabolism
In the past decade or so, many proteins with intrinsic histone acetyltransferase (HAT) or histone deacetylase (HDAC) activity have been identified and characterized. As with histones, these enzymes themselves are targets of Nε-acetylation. Autoacetylation of p300 within its HAT domain leads to activation (Thompson et al, 2004; Karanam et al, 2006), whereas acetylation of two residues upstream of the HAT domain blocks sumoylation (Girdwood et al, 2003). Multiple deacetylases—including sirtuin 1 (SIRT1), HDAC1 and HDAC3—are involved in p300 deacetylation (Bouras et al, 2005; Chuang et al, 2006; Grégoire et al, 2007). Autoacetylation of p300/CBP-associated factor (PCAF; CBP for CREB-binding protein) promotes its nuclear localization (Santos-Rosa et al, 2003). Other HATs, such as monocytic leukaemia zinc finger protein (MOZ) and regulation of retrotransposition 109 (Rtt109), are also autoacetylated (Yang, 2004b; Driscoll et al, 2007; Han J et al, 2007), although the functional impact of this is still unclear. Furthermore, p300 acetylation of HDAC1 inhibits its deacetylase activity (Qiu et al, 2006), and acetyl-lysine is also present in non-catalytic subunits of HDAC and HAT complexes (Buscaino et al, 2003; Morales et al, 2004; Kim et al, 2006). In addition to these enzymes and numerous transcription factors, regulators of RNA processing are subject to this modification (Table 1; Shimazu et al, 2007).
Table 1.
Acetyl-proteins involved in regulating RNA and DNA metabolism
| Transcription & RNA processing |
| DNA-binding transcription factors (>60) |
| Transcription coregulators (>15) |
| General transcription factors: TFIIB, TFIIE, TFIIF |
| RNA processing: CFIm, Poly(A) polymerase |
| DNA repair |
| Polβ, FEN1, Glycosylases (3), AP endonuclease, DNA ligase IIIα, PCNA, Ku70, Werner helicase, ATM kinase, Rtt109 |
| Replication |
| Cellular replication: PCNA, MCM3, FEN1, Ku70, Werner helicase, HBO1–ING4 complex, Rtt109 |
| Viral integration: HIV integrase, SV40 large T |
| Recombination |
| Recombination: Werner helicase, Rtt109 |
| Viral replication: HIV integrase |
AP endonuclease, apurinic/apyrimidinic endonuclease; ATM, ataxia telangiectasia mutated; CFIm, cleavage factor Im; FEN1, Flap endonuclease 1; HBO1, HAT bound to DNA replication origin complex (ORC) 1; HIV, human immunodeficiency virus; ING4, inhibitor of growth 4; MCM3, minichromosome maintenance 3; PCNA, proliferating cell nuclear antigen; Polβ, DNA polymerase β; Rtt109, regulation of retrotransposition 109; SV40 large T, SV40 large T antigen; TFII, transcription factor II.
As clearly established for transcription, histone acetylation affects other chromatin-based processes such as DNA replication, repair and recombination (Peterson & Côté, 2004; Groth et al, 2007). Recent studies have also identified acetyl-lysine in many components of the base excision repair pathway (Table 1; reviewed by Peterson & Côté, 2004), including Flap endonuclease 1 (FEN1; Hasan et al, 2001), DNA polymerase β (Polβ; Hasan et al, 2002), DNA glycosylases (Tini et al, 2002; Bhakat et al, 2004; Mohan et al, 2007), apurinic/apyrimidinic (AP) endonuclease (Bhakat et al, 2003), proliferating cell nuclear antigen (PCNA; Naryzhny & Lee, 2004), DNA ligase III (Bhakat et al, 2004) and Werner helicase (Blander et al, 2002). DNA damage induced by exposure to ultraviolet light, oxidative stress and ionizing radiation regulates acetylation of these and other repair regulators. Acetylation of catalytic lysine residues of Polβ and the DNA glycosylase Neil2 inactivates these enzymes (Fig 1D; Bhakat et al, 2004), whereas acetylation of thymine DNA glycosylase precludes sumoylation (Mohan et al, 2007). Ataxia telangiectasia mutated (ATM), FEN1, Ku70, PCNA and Werner helicase are acetyl-proteins that also have important roles in the double-strand break and nucleotide excision repair pathways (Table 1; Cohen et al, 2004; Peterson & Côté, 2004; Sun et al, 2005). Acetylation of PCNA is cell cycle-dependent and stimulates its binding to Polβ and Polδ (Naryzhny & Lee, 2004), suggesting that the acetylated form of PCNA also participates in DNA replication. Similar to PCNA, Ku70 and Werner helicase regulate both DNA repair and replication. Werner helicase has an additional role in DNA recombination, and human immunodeficiency virus (HIV) integrase is subject to acetylation (Blander et al, 2002; Cereseto et al, 2005). Furthermore, minichromosome maintenance 3-associated protein (MCM3AP) acetylates MCM3 (Takei et al, 2001). Thus, Nε-acetylation is a common mechanism for the regulation of nuclear processes in eukaryotic cells. As discussed below, this modification also has prominent roles in the cytoplasm and various bacteria.
Acetylation of acetyl-CoA synthases
An unexpected discovery made several years ago was that acetyl-CoA synthase (Acs) from Salmonella enterica is acetylated at lysine (Lys) 609 within its catalytic centre (Starai et al, 2002). This residue is crucial for synthesis of the acetyl-AMP intermediate from acetate and ATP, so its acetylation inactivates the enzyme (Fig 1D). Lys 609 is evolutionarily conserved from bacteria to humans, suggesting that acetylation might be important for controlling the activity of various Acs enzymes. This idea has received direct support from recent studies of the Acs orthologues in Bacillus subtilis and mammals (Gardner et al, 2006; Hallows et al, 2006; Schwer et al, 2006). Distinct acetylation/deacetylation systems are known to maintain the dynamic acetylation of bacterial Acs—the protein acetyltransferase (Pat) and the CobB sirtuin deacetylase (Starai & Escalante-Semerena, 2004), and the acetoin utilization A (AcuA) and AcuC proteins (Gardner et al, 2006). Related to CobB, mammalian SIRT1 and SIRT3 deacetylate and inactivate Acs1 and Acs2 (Hallows et al, 2006; Schwer et al, 2006). Notably, both Acs2 and SIRT3 localize to mitochondria. AcuA and AcuC are two products of a three-gene operon in B. subtilis and show some sequence similarity to yeast Gcn5 and Hda1, respectively (Gardner et al, 2006), raising the question of whether Gcn5, Hda1 and related proteins regulate acetylation of eukaryotic Acs enzymes.
Acetyl-lysine has been found in many other metabolic enzymes, including catalase, enolase and 6-phosphofructo-2-kinase, although the functional significance remains to be established (Dihazi et al, 2005; Iwabata et al, 2005; Kim et al, 2006). In addition, expression of metabolic enzymes is tightly controlled by acetylation of PPARγ co-activator 1α (PGC1α; Rodgers et al, 2005). During fasting, murine SIRT1 is activated, deacetylates PGC1α and induces gluconeogenic gene expression; conversely, in the presence of sufficient nutrients, GCN5 acetylates PGC1α and inhibits its transcriptional potential (Lagouge et al, 2006; Lerin et al, 2006). Thus, Nε-acetylation is important for regulating amino-acid, glucose and energy metabolism.
Acetylation of cytoskeleton and structural proteins
Acetylation of α-tubulin was mapped to Lys 40 (LeDizet & Piperno, 1987). This residue is conserved from humans to lower eukaryotes, such as Chlamydomonas, and its acetylation is preferentially associated with stable microtubules (Hubbert et al, 2002; Matsuyama et al, 2002; North et al, 2003; Zhang et al, 2003; reviewed by Boyault et al, 2007). A subset of acetylated microtubules was found to be necessary for correct organization of the immune synapse, and tubulin acetylation seemed to affect HIV infection of CD4+ cells (Serrador et al, 2004; Valenzuela-Fernandez et al, 2005). A recent study showed that this modification stimulates kinesin 1 binding and transport (Reed et al, 2006). Other cytoskeleton proteins such as actin and cortactin are also acetylated (Kim et al, 2006; E. Seto, unpublished data). Acetylation of many lysines on cortactin affects actin dynamics and cell motility. Furthermore, p120 catenin is acetylated on at least three lysines, affecting dendrite formation. Catenins are found in complexes with cadherin adhesion molecules and link adherens junctions to actin filaments. Related to this, acetylation of Rho GDP-dissociation inhibitor (RhoGDI) might affect the formation of actin stress fibres (Kim et al, 2006), suggesting that acetylation may act through microtubules and actin fibres to regulate cell motility. Furthermore, acetyl-lysine is present in nuclear lamins (Kim et al, 2006), and acetylation of crystallins occurs in an age-dependent manner (Lin et al, 1998). Therefore, although functional consequences await further analysis, Nε-acetylation occurs in different types of structural protein.
Nε-acetylation of signalling regulators
Besides metabolic enzymes and structural proteins, acetyl-lysines are present in protein kinases. PCAF and p300/CBP acetylate the tyrosine kinase c-Abl at Lys 730, which inhibits its nuclear localization and promotes myogenic differentiation (di Bari et al, 2006). HDAC3 associates with the Rous sarcoma (Src) tyrosine kinase at the plasma membrane (Longworth & Laimins, 2006), and a Src-related kinase has just been identified as an Nε-acetylated protein (Kim et al, 2006). In response to DNA damage, Tat-interactive protein 60 kDa (TIP60) interacts with, acetylates and activates the ATM kinase (Sun et al, 2005). PCAF acetylates the phosphatase and tensin homologue (PTEN) at two residues within the catalytic cleft, impairing phosphatase activity and promoting phosphatidylinositol 3-kinase signalling (Fig 1D; Okumura et al, 2006). Furthermore, acetylation of insulin receptor substrate 1 stimulates tyrosine phosphorylation and cellular signalling (Kaiser & James, 2004). Acetyl-lysines have also been found in other signalling regulators, including phospholipase C, p120 catenin, RhoGDI, murine double minute 2 (MDM2), Ku70, signal transducer and activator of transcription 3 (Stat3), Sma- and Mad-related protein 7 (Smad7) and heat shock protein 90 (Hsp90). Among these, acetylation of MDM2 impairs its ability to promote p53 ubiquitylation (Wang et al, 2004), Ku70 acetylation promotes apoptosis (Cohen et al, 2004), Stat3 acetylation stimulates dimerization for cytokine signalling (Yuan et al, 2005), Smad7 acetylation increases stability to block transforming growth factor β (TGFβ) signalling (Kume et al, 2007), and Hsp90 acetylation regulates its association with client proteins such as the glucocorticoid receptor (Kovacs et al, 2005; Scroggins et al, 2007). Thus, Nε-acetylation is an additional method of regulation for various signalling pathways.
Interplay of Nε-acetylation with other modifications
Nε-acetylation can act through many different mechanisms (reviewed by Kouzarides, 2000; Sterner & Berger, 2000; Yang, 2004a; Glozak et al, 2005), five of which are noteworthy here. First, acetylation of catalytic lysine residues can abolish or reduce enzymatic activity (Fig 1D). Second, Nε-acetylation can generate specific sites for docking bromodomain proteins such as GCN5, TAFII250 (TBP-associated factor) and Brg1 (Brahma-related gene 1) to promote complex assembly for transcriptional activation (Seet et al, 2006). Third, as shown for p53 and histone H3 (Fig 2), functional consequences of multisite acetylation can be position-dependent. Such an effect has been well documented for Lys 16 acetylation of histone H4 (Shogren-Knaak et al, 2006). Fourth, acetylation competes with other modifications such as methylation, ubiquitylation and sumoylation at the same lysine residue (Fig 1A). This has been documented for histone H3, p53, Smad7, specificity protein 3 (Sp3), sterol regulatory element-binding protein 1α (SREBP1α), p300 and others (reviewed by Brooks & Gu, 2003; Caron et al, 2005; Glozak et al, 2005; Huang et al, 2006; Kim et al, 2006; Stankovic-Valentin et al, 2007; Yang, 2004a; Zheng & Yang, 2005). In this regard, acetylation and methylation of histone H3 at Lys 9 are mutually exclusive and mark genes for activation and repression, respectively (Fig 2B; reviewed by Jenuwein & Allis, 2001; Margueron et al, 2005; Nightingale et al, 2006). Acetylation and methylation of histone H3 at Lys 36 preclude each other, but mark promoters and coding regions of active genes, respectively (Morris et al, 2007), suggesting that same-site modifications might not always yield completely opposite effects. This is also true for two pairs of p53 modifications (Fig 2A). At Lys 320, both acetylation and polyubiquitylation stimulate transcription (Knights et al, 2006; Le Cam et al, 2006). Similarly, at Lys 273, both acetylation and monomethylation lead to transcriptional activation (Chuikov et al, 2004). In the above two cases, the competitive modifications affect distinct sets of genes, revealing a new way for one transcription factor to selectively regulate different gene expression programmes.
Figure 2.
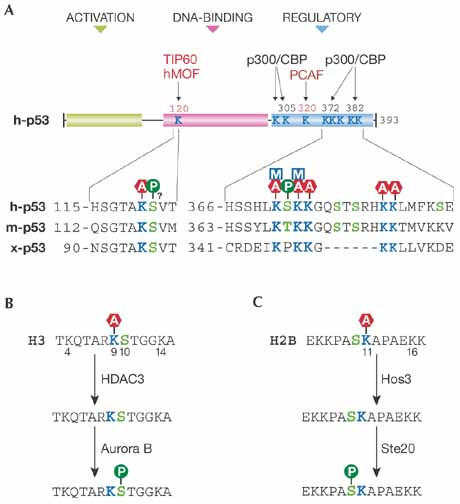
Interplay of acetylation with methylation and phosphorylation in p53 and histones. (A) Domain organization of human (h) p53, with its sequence around lysine (Lys; K) 120 and Lys 370 aligned with the corresponding regions of murine (m) and Xenopus (x) orthologues. Note the potential interplay of acetylation of these two residues with same-site or neighbouring modifications such as methylation and phosphorylation. Also shown are site-specific actions of various acetyltransferases, including TIP60, hMOF, PCAF and p300/CBP (Sykes et al, 2006; Tang et al, 2006; reviewed by Brooks & Gu, 2003). Note that Lys 120 and Lys 373, but not Lys 320, Lys 370 or Lys 372, are conserved in Drosophila p53 (Knights et al, 2006; Tang et al, 2006). Mammalian p53 is also subject to various other modifications (reviewed by Appella & Anderson, 2001; Brooks & Gu, 2003). P in circle, phosphorylation; A in hexagon, acetylation; and M in square, methylation. (B) At the initial stage of mitosis, HDAC3 removes the acetyl group from Lys 9 of histone H3 and facilitates phosphorylation of serine (Ser; S) 10 by Aurora B. (C) In response to oxidative stress, yeast Hos3 deacetylates Lys 11 of histone H2B and promotes phosphorylation of Ser 10 by Ste20 kinase. CBP, CREB-binding protein; HDAC3, histone deacetylase 3; hMOF, human male-absent on the first; Hos3, histone deacetylase one-similar 3; PCAF, p300/CBP-associated factor; Ste20, sterile 20; TIP60, Tat-interactive protein of 60 kDa.
Fifth, acetylation crosstalks with neighbouring modifications such as phosphorylation and methylation (Fig 2). Deacetylation of histone H3 at Lys 9 and histone H2B at Lys 11 cooperates with serine (Ser) 10 phosphorylation (Fig 2B,C; Ahn et al, 2006; Li et al, 2006). By analogy, acetylation of p53 at Lys 120, Lys 370 and Lys 372 might cooperate with phosphorylation of adjacent serine residues (Fig 2A). For cooperative actions on Lys 9 and Ser 10 of histone H3, HDAC3 associates with Aurora B kinase (Li et al, 2006). Reminiscent of this, HDACs form complexes with phosphatases (Canettieri et al, 2003; Zhang et al, 2005). Similarly, ubiquitin protease 8 (Ubp8) associates with Gcn5 (Daniel et al, 2004; Henry et al, 2003). Furthermore, HDAC6 has a zinc-finger domain for ubiquitin interaction, allowing the recognition and transport of ubiquitylated proteins, and controlling polyubiquitin-chain turnover (Seigneurin-Berny et al, 2001; Hook et al, 2002; Boyault et al, 2006). The deacetylase and ubiquitin-binding activities of HDAC6 are both required for correct management of misfolded proteins (Kawaguchi et al, 2003; Iwata et al, 2005). Furthermore, an O-linked N-acetylglucosamine (O-GlcNAc) transferase has a HAT domain (Toleman et al, 2004), which might promote crosstalk between acetylation and glycosylation. Therefore, in the context of multisite modification, Nε-acetylation might exert its effect through an interplay with phosphorylation, methylation, ubiquitylation and other modifications to form concerted regulatory programmes.
These programmes have characteristics of dynamic molecular barcodes (reviewed by Yang, 2005). In terms of multisite modification, p53 and core histones have clusters of acetylation and other modification sites in their regulatory regions. An important question is whether principles drawn from modifications of p53 and core histones can be extended to other proteins such as Ku70, p300 and DNA methyltransferase 1 (DNMT1), which are known to have many acetylation sites in their regulatory loops (Cohen et al, 2004; Thompson et al, 2004; Kim et al, 2006). If so, studies of p53 and core histones shall form informative conceptual frameworks for understanding the regulation of many other Nε-acetylated proteins.
Competition of O-acetylation with phosphorylation
YopJ is a virulence factor of Yersinia pestis, the cause of the plague centuries ago; two other Yersinia species cause septicaemic and gastrointestinal disorders. YopJ compromises the host's defence systems by blocking the mitogen-activated protein kinase (MAPK) and nuclear factor κB (NFκB) signalling pathways (reviewed by Worby & Dixon, 2006). Two groups recently reported that YopJ catalyses O-acetylation to inhibit these pathways (Mittal et al, 2006; Mukherjee et al, 2006). It acetylates MAPK/ERK kinase (MEK) 6 (or MKK6) at the activation-loop residues Ser 207 and threonine (Thr) 211, the phosphorylation of which is required for kinase activation (Mukherjee et al, 2006). This indicates that O-acetylation precludes phosphorylation and inhibits MAPK signalling (Fig 3). Two other members of the MEK family are similarly acetylated. Furthermore, YopJ acetylates IκB kinase (IKK) α and β at two serine residues equivalent to Ser 207 and Thr 211 of MEK6, and the acetylation blocks NFκB activation (Fig 3; Mittal et al, 2006; Mukherjee et al, 2006). These findings raise two exciting issues: whether MEKs, IKKs and other mammalian proteins are subject to O-acetylation under physiological conditions (Table 1); and whether this modification occurs in other bacteria. If so, this would take signalling research and bacterial pathology to new and totally uncharted territory.
Figure 3.
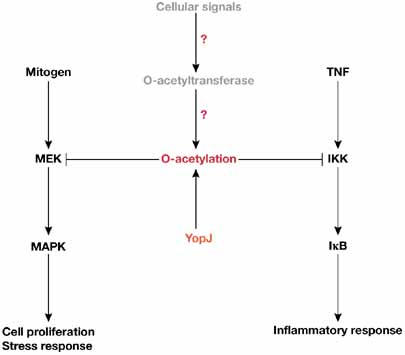
Negative regulation of the MAPK/ERK kinase and IκB kinase families by Yersinia outer protein J. IκB, inhibitor of NFκB; IKK, IκB kinase; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; TNF, tumour necrosis factor; YopJ, Yersinia outer protein J.
Conclusions and perspectives
Studies of Nε-acetylation have gradually expanded from histones and transcription factors to other proteins, including DNA repair regulators, metabolic enzymes, structural proteins and signalling modulators. Most of these acetylation events were identified by the biased-candidate approach. Three proteomic studies have recently taken a more systematic approach and identified a diverse array of Nε-acetylated proteins (Iwabata et al, 2005; Kim et al, 2006; Xie et al, 2007). Although acetylation of these proteins awaits further substantiation by different experimental means, it is reasonable to conclude that additional Nε-acetylated proteins have yet to be identified and characterized. This idea is also consistent with the following considerations. First, given the dynamic nature of Nε-acetylation and the low abundance of various cellular proteins, such as signalling regulators, acetyl-lysine in many proteins might have evaded identification. Second, it occurs in different bacteria, so this modification could be a rather ancient regulatory mechanism and various prokaryotic proteins might be subject to such regulation. Third, different substrates might exist for many uncharacterized plant proteins that are related to p300/CBP and class II HDACs (Han SK et al, 2007). Fourth, there are atypical Nε-acetyltransferases, including bacterial Pat and AcuA (Gardner et al, 2006), yeast establishment of cohesion 1 (Eco1; Ivanov et al, 2002) and Rtt109 (Driscoll et al, 2007; Han J et al, 2007), and a mammalian glycosylation enzyme with a lysine acetyltransferase domain (Toleman et al, 2004). Such atypical enzymes might act on novel substrates. Finally, TIP60 and HDAC3 target signal transduction pathways through unusual mechanisms (Zhang et al, 2002; Ceol & Horvitz, 2004). One possibility is that TIP60 and HDAC3 directly modify the signalling regulators involved. Thus, it will be important to complete the catalogue of Nε-acetylated proteins—that is, the Nε-acetyl proteome—and to characterize the functional impact on metabolism, cytoskeleton dynamics, signalling and other cellular processes. In this regard, it is noteworthy that Nε-acetylation often does not act alone but interplays with other covalent modifications to form dynamic regulatory programmes for orchestrated regulation of protein functions in vivo (Fig 2). If there is a protein modification code, Nε-acetylation must be an important element in the coding system.
Besides this modification, cellular proteins might be subject to O-acetylation of serine, threonine and perhaps tyrosine residues (Fig 1B,C). An important issue to address is whether there are intrinsic enzymatic systems to govern protein O-acetylation in eukaryotic cells. If so, this modification would compete with phosphorylation for the same residues and, as protein phosphorylation is crucial to cellular signalling, this would revolutionize our fundamental view of cellular regulation. It is notable that acetyl salicylic acid (Aspirin) promotes O-acetylation of cyclooxygenase enzymes. In addition, Nε-deacetylase inhibitors and activators are being actively evaluated in clinical trials as new therapeutic agents for cancer, heart diseases and neurodegenerative disorders. Thus, studies of protein Nε- and O-acetylation not only are for fundamental research and intellectual curiosity, but also will provide vital information on how to use related enzymes and substrates as new molecular targets for developing preventive, diagnostic and therapeutic strategies against various diseases.

Xiang-Jiao Yang

Serge Grégoire
Acknowledgments
This work was supported by funds from National Cancer Institute of Canada and Canadian Institutes for Health Research to X.J.Y.; S.G. received a Canderel studentship from McGill Cancer Center.
References
- Ahn SH, Diaz RL, Grunstein M, Allis CD (2006) Histone H2B deacetylation at lysine 11 is required for yeast apoptosis induced by phosphorylation of H2B at serine 10. Mol Cell 24: 211–220 [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268: 2764–2772 [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S (2003) Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J 22: 6299–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Hazra TK, Mitra S (2004) Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res 32: 3033–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M (2002) DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem 277: 50934–50940 [DOI] [PubMed] [Google Scholar]
- Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG (2005) SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem 280: 10264–10276 [DOI] [PubMed] [Google Scholar]
- Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, Matthias P, Muller CW, Khochbin S (2006) HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J 25: 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault C, Sadoul K, Pabion M, Khochbin S (2007) HDAC6, at the crossroads between cytoskeleton and cell signalling by acetylation and ubiquitination. Oncogene (in press) [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2003) Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol 15: 164–171 [DOI] [PubMed] [Google Scholar]
- Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, Wilm M, Akhtar A (2003) MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell 11: 1265–1277 [DOI] [PubMed] [Google Scholar]
- Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD, Yates JR III, Montminy M (2003) Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat Struct Biol 10: 175–181 [DOI] [PubMed] [Google Scholar]
- Caron C, Boyault C, Khochbin S (2005) Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays 27: 408–415 [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR (2004) A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signalling. Dev Cell 6: 563–576 [DOI] [PubMed] [Google Scholar]
- Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M (2005) Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J 24: 3070–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HC, Chang CW, Chang GD, Yao TP, Chen H (2006) Histone deacetylase 3 binds to and regulates the GCMa transcription factor. Nucleic Acids Res 34: 1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S et al. (2004) Regulation of p53 activity through lysine methylation. Nature 432: 353–360 [DOI] [PubMed] [Google Scholar]
- Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA (2004) Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell 13: 627–638 [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, Grant PA (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871 [DOI] [PubMed] [Google Scholar]
- di Bari MG, Ciuffini L, Mingardi M, Testi R, Soddu S, Barila D (2006) c-Abl acetylation by histone acetyltransferases regulates its nuclear-cytoplasmic localization. EMBO Rep 7: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihazi H, Kessler R, Muller GA, Eschrich K (2005) Lysine 3 acetylation regulates the phosphorylation of yeast 6-phosphofructo-2-kinase under hypo-osmotic stress. Biol Chem 386: 895–900 [DOI] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG, Grundy FJ, Henkin TM, Escalante-Semerena JC (2006) Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD+ involvement in Bacillus subtilis. J Bacteriol 188: 5460–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT (2003) p300 transcriptional repression is mediated by SUMO modification. Mol Cell 11: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E (2005) Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23 [DOI] [PubMed] [Google Scholar]
- Grégoire S, Xiao L, Nie J, Xu M, Wong J, Seto E, Yang XJ (2007) Histone deacetylase 3 interacts with and deacetylates MEF2 transcription factors. Mol Cell Biol 27: 1280–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G (2007) Chromatin challenges during DNA replication and repair. Cell 128: 721–733 [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90: 595–606 [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA 103: 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655 [DOI] [PubMed] [Google Scholar]
- Han SK, Song JD, Noh YS, Noh B (2007) Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J 49: 103–114 [DOI] [PubMed] [Google Scholar]
- Hasan S, Stucki M, Hassa PO, Imhof R, Gehrig P, Hunziker P, Hubscher U, Hottiger MO (2001) Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol Cell 7: 1221–1231 [DOI] [PubMed] [Google Scholar]
- Hasan S, El-Andaloussi N, Hardeland U, Hassa PO, Burki C, Imhof R, Schar P, Hottiger MO (2002) Acetylation regulates the DNA end-trimming activity of DNA polymerase β. Mol Cell 10: 1213–1222 [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SS, Orian A, Cowley SM, Eisenman RN (2002) Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc Natl Acad Sci USA 99: 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632 [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- Huq MD, Tsai NP, Lin YP, Higgins L, Wei LN (2007) Vitamin B6 conjugation to nuclear corepressor RIP140 and its role in gene regulation. Nat Chem Biol 3: 161–165 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K (2002) Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol 12: 323–328 [DOI] [PubMed] [Google Scholar]
- Iwabata H, Yoshida M, Komatsu Y (2005) Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics 5: 4653–4664 [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 280: 40282–40292 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kaiser C, James SR (2004) Acetylation of insulin receptor substrate-1 is permissive for tyrosine phosphorylation. BMC Biol 2: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA (2006) Kinetic and mass spectrometric analysis of p300 histone acetyltransferase domain autoacetylation. J Biol Chem 281: 40292–40301 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Kim SC et al. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618 [DOI] [PubMed] [Google Scholar]
- Knights CD, Catania J, Di Giovanni S, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J 19: 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP (2005) HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 18: 601–607 [DOI] [PubMed] [Google Scholar]
- Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki SI, Isshiki K, Isono M, Uzu T, Guarente L, Kashiwagi A, Koya D (2007) SIRT1 inhibits TGFβ-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J Biol Chem 282: 151–158 [DOI] [PubMed] [Google Scholar]
- L'Hernault SW, Rosenbaum JL (1985) Chlamydomonas α-tubulin is post-translationally modified by acetylation on the ε-amino group of a lysine. Biochemistry 24: 473–478 [DOI] [PubMed] [Google Scholar]
- Lagouge M et al. (2006) Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127: 1109–1122 [DOI] [PubMed] [Google Scholar]
- Le Cam L et al. (2006) E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G (1987) Identification of an acetylation site of Chlamydomonas α-tubulin. Proc Natl Acad Sci USA 84: 5720–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P (2006) GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3: 429–438 [DOI] [PubMed] [Google Scholar]
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA (2006) A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev 20: 2566–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PP, Barry RC, Smith DL, Smith JB (1998) In vivo acetylation identified at lysine 70 of human lens αA-crystallin. Protein Sci 7: 1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Laimins LA (2006) Histone deacetylase 3 localizes to the plasma membrane and is a substrate of Src. Oncogene 25: 4495–4500 [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D (2005) The key to development: interpreting the histone code? Curr Opin Genet Dev 15: 163–176 [DOI] [PubMed] [Google Scholar]
- Matsuyama A et al. (2002) In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J 21: 6820–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Peak-Chew SY, McMahon HT (2006) Acetylation of MEK2 and IκB kinase (IKK) activation loop residues by YopJ inhibits signalling. Proc Natl Acad Sci USA 103: 18574–18579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan RD, Rao A, Gagliardi J, Tini M (2007) SUMO-1 dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment. Mol Cell Biol 27: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB (2004) Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J 23: 2258–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD (2007) Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J Biol Chem 282: 7632–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312: 1211–1214 [DOI] [PubMed] [Google Scholar]
- Naryzhny SN, Lee H (2004) The post-translational modifications of proliferating cell nuclear antigen (PCNA): acetylation, not phosphorylation, plays an important role in the regulation of its function. J Biol Chem 279: 20194–20199 [DOI] [PubMed] [Google Scholar]
- Nightingale KP, O'Neill LP, Turner BM (2006) Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev 16: 125–136 [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444 [DOI] [PubMed] [Google Scholar]
- Okumura K, Mendoza M, Bachoo RM, Depinho RA, Cavenee WK, Furnari FB (2006) PCAF modulates PTEN activity. J Biol Chem 281: 26562–26568 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Côté J (2004) Cellular machineries for chromosomal DNA repair. Genes Dev 18: 602–616 [DOI] [PubMed] [Google Scholar]
- Piperno G, Fuller MT (1985) Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol 101: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y et al. (2006) HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell 22: 669–679 [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ (2006) Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol 16: 2166–2172 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M (2003) Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res 31: 4285–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E (2006) From the cover: reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA 103: 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scroggins BT et al. (2007) An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell 25: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T (2006) Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol 7: 473–483 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signalling pathways. Mol Cell Biol 21: 8035–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador JM, Cabrero JR, Sancho D, Mittelbrunn M, Urzainqui A, Sanchez-Madrid F (2004) HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity 20: 417–428 [DOI] [PubMed] [Google Scholar]
- Shimazu T, Horinouchi S, Yoshida M (2007) Multiple histone deacetylases and CBP regulates pre-mRNA 3′-end processing. J Biol Chem 282: 4470–4478 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D (2007) An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved ψKxEP motif in the tumor suppressor HIC1 (Hypermethylated in Cancer 1) regulates transcriptional repression activity. Mol Cell Biol 27: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Escalante-Semerena JC (2004) Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340: 1005–1012 [DOI] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298: 2390–2392 [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R, Vidali G, Allfrey VG (1979) Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMG-1. J Biol Chem 254: 11577–11583 [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102: 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Swietlik M, Tanoue A, Tsujimoto G, Kouzarides T, Laskey R (2001) MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep 2: 119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W (2006) Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 24: 827–839 [DOI] [PubMed] [Google Scholar]
- Thompson PR et al. (2004) Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol 11: 308–315 [DOI] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P (2002) Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell 9: 265–277 [DOI] [PubMed] [Google Scholar]
- Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE (2004) Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem 279: 53665–53673 [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A et al. (2005) Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell 16: 5445–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali G, Gershey E, Allfrey VG (1968) Chemical studies of histone acetylation. The distribution of ε-N-acetyllysine in calf thymus histones. J Biol Chem 243: 6361–6366 [PubMed] [Google Scholar]
- Wang X, Taplick J, Geva N, Oren M (2004) Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett 561: 195–201 [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE (2006) Bacteria seize control by acetylating host proteins. Science 312: 1150–1151 [DOI] [PubMed] [Google Scholar]
- Xie H, Bandhakavi S, Roe MR, Griffin TJ (2007) Preparative peptide isoelectric focusing as a tool for improving the identification of lysine-acetylated peptides from complex mixtures. J Proteome Res doi:10.1021/pr060691j [DOI] [PubMed] [Google Scholar]
- Yang XJ (2004a) Lysine acetylation and the bromodomain: a new partnership for signalling. BioEssays 26: 1076–1087 [DOI] [PubMed] [Google Scholar]
- Yang XJ (2004b) The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res 32: 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ (2005) Multisite protein modification and intramolecular signalling. Oncogene 24: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307: 269–273 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG (2002) The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell 9: 611–623 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E (2005) Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev 19: 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P (2003) HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J 22: 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Yang YC (2005) Sumoylation and acetylation play opposite roles in the transactivation of PLAG1 and PLAGL2. J Biol Chem 280: 40773–40781 [DOI] [PubMed] [Google Scholar]


