Abstract
Mutation of the gene encoding the transcriptional corepressor BCOR results in the X-linked disorder Oculofaciocardiodental Syndrome (OFCD or MCOPS2). Female OFCD patients suffer from severe ocular, craniofacial, cardiac and digital developmental defects and males do not survive through gestation. BCOR can mediate transcriptional repression by the oncoprotein BCL6 and has the ability to reduce transcriptional activation by AF9, a known mixed-lineage leukemia (MLL) fusion partner. The essential role of BCOR in development and its ability to modulate activity of known oncogenic proteins prompted us to determine the expression profile of Bcor during mouse development. Identification of independently transcribed exons in the 5′ untranslated region of Bcor suggests that three independent promoters control the expression of Bcor in mice. Although Bcor is widely expressed in adult mouse tissues, analysis of known spliced isoforms in the coding region of Bcor reveals differential isoform usage. Whole mount in situ hybridization of mouse embryos shows that Bcor is strongly expressed in the extraembryonic tissue during gastrulation and expression significantly increases throughout the embryo after embryonic turning. During organogenesis and fetal stages Bcor is differentially expressed in multiple tissue lineages, with a notable presence in the developing nervous system. Strikingly, we observed that Bcor expression in the eye, brain, neural tube and branchial arches correlates with tissues affected in OFCD patients.
Keywords: Bcor, Oculofaciocardiodental syndrome, MCOPS2, extraembryonic
Results and Discussion
BCOR (BCL6 corepressor) is a transcriptional co-repressor that was identified based on its ability to interact with the POZ domain of the oncoprotein BCL6 (Huynh et al., 2000). Chromosomal translocations in the promoter and 5′ untranslated region of the human BCL6 gene are a common genomic alteration in non-Hodgkin’s B cell lymphomas (Baron et al., 1993; Dalla-Favera et al., 1999; Kerckaert et al., 1993; Miki et al., 1994; Ye et al., 1993). These translocations result in aberrant expression of BCL6 (Chen et al., 1998; Ye et al., 1995). Mice engineered to model one of these translocations develop B cell lymphomas, demonstrating that BCL6 is a bona fide oncogene (Cattoretti et al., 2005). BCOR potentiates BCL6 mediated transcriptional repression of reporter constructs in transiently transfected cells (Huynh et al., 2000). In germinal center B cells, BCOR is found with BCL6 at several known BCL6 target genes, including regulators of cell proliferation and apoptosis (Gearhart et al., 2006). BCOR co-purifies with an 800 kDa complex comprised of Polycomb group transcriptional repressor proteins and SCF ubiquitin E3 ligase components (Gearhart et al., 2006). Epigenetic modification of BCL6 target gene chromatin by the BCOR repression complex is likely to play a role in mediating their repression.
In addition to its role in B-cells, BCOR aids in the control of gene expression in multiple tissues and organ systems during development and into adulthood as mutations in human BCOR result in X-linked Oculofaciocardiodental syndrome (OFCD) (Ng et al., 2004). OFCD is the primary subtype of OMIM #300166 microphthalmia, syndromic 2 (MCOPS2) and is characterized by ocular, dental, cardiac and digital anomalies in heterozygous females (Ng et al., 2004; Schulze et al., 1999). Males with OFCD do not survive due to presumed embryonic lethality (Ng et al., 2004). Since BCOR lies on the X chromosome in both mice and humans, random X-inactivation results in mosaic expression of the mutant allele contributing to varying disease severity in females (Ng et al., 2004; Schulze et al., 1999). Additionally, peripheral leukocytes of female patients show preferential survival of cells in which the mutant allele of BCOR lies on the inactivated X chromosome, indicating a strong requirement for BCOR in hematopoiesis (Hedera and Gorski, 2003; Ng et al., 2004). Another form of MCOPS2, which is distinct from OFCD, occurs in males with a single missense mutation (p.P85L) in the fourth coding exon of BCOR (Horn et al., 2005; Ng et al., 2004). In the described family, this syndrome is inherited in an X-linked recessive pattern and comprises microphthalmia/anophthalmia, mental retardation, and skeletal and other anomalies (Ng et al., 2002). RNAi knock-down of Bcor in zebrafish results in eye, skeletal and nervous system abnormalities consistent with those found in MCOPS2 patients (Ng et al., 2004). BCOR has also been shown to interact with and repress transcriptional activation of the MLL fusion protein AF9 (Srinivasan et al., 2003), a known regulator of Hox gene expression (Collins et al., 2002) and skeletal development, further implicating BCOR as a key developmental regulator. The pleiotropic effects induced by the loss of functional BCOR in humans and zebrafish (Ng et al., 2004) clearly illustrate the essential role of BCOR during embryogenesis and emphasizes the importance of determining the spatial and temporal expression of BCOR during development. Herein, we provide a detailed analysis of Bcor mRNA expression during mouse development and in the adult mouse.
In adult mice and humans, Bcor is widely expressed (Huynh et al., 2000; Nagase et al., 2000). However, these studies are limited in scope, relying on RNA dot blot analysis of human tissues and reverse transcription PCR (RT-PCR) on a limited number of mouse tissues. Expanding upon these results, we have dissected the pattern of Bcor expression by Northern blot analysis, more extensive RT-PCR, and whole mount and section in situ hybridization experiments. To determine the expression pattern of Bcor in adult mouse tissues, we harvested total RNA from 14 different organs and conducted Northern blot analysis on 4 organs and RT-PCR on all 14 organs using a probe that recognizes all mRNA isoforms (Fig. 1F). Three transcripts migrating at approximately 7 kb are found in all four organs analyzed however the stoichiometry of the different transcripts is not identical (Fig. 1A). Bcor mRNA is detected by RT-PCR in all tissues tested (Fig. 1B). The ubiquitous expression of Bcor in adult mouse tissue is consistent with expression of BCOR in human adult tissue (Huynh et al., 2000).
Figure 1.
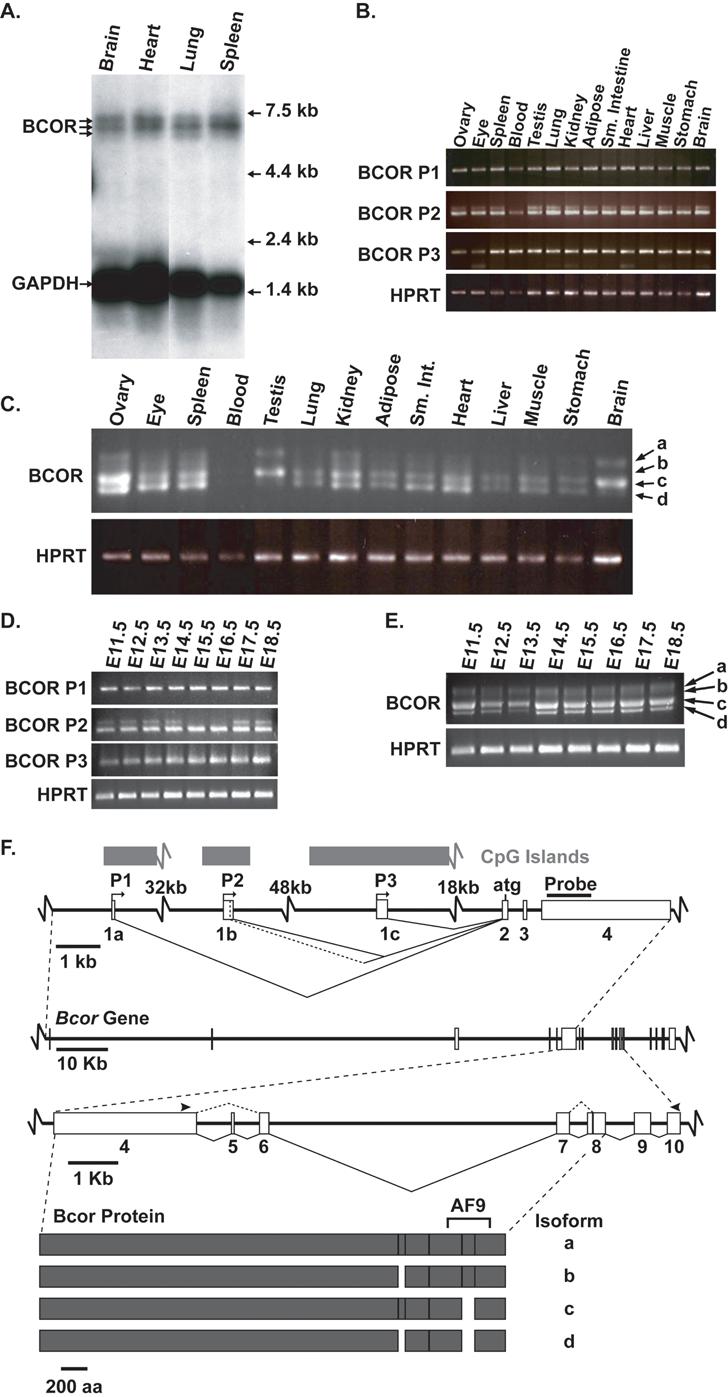
Three independent promoters drive widespread expression of Bcor mRNA in embryonic and adult mouse tissue (A-F). Northern blot analysis of adult mouse organs reveals three major Bcor transcripts. Gapdh was used as a loading control (A). Promoter 1, 2 and 3 (P1, P2 and P3) specific RT-PCR analysis on panel of adult mouse organs (B). RT-PCR product sizes are as follows P1 166 bp, P2 shorter 164 bp, P2 longer 187 bp, P3 159 bp. Bcor isoform a-d specific RT-PCR on mouse organs panel (C). RT-PCR product sizes are as follows a 1642 bp, b 1588 bp, c 1540 bp, and d 1486 bp. P1, P2 and P3 specific RT-PCR analysis of embryonic stages 11.5-18.5 (D). Bcor isoform a-d specific RT-PCR on stages E11.5-E18.5 (E). Hprt was used as a loading control, product 200 bp (B-E). Illustration depicting promoter usage and splicing of the mouse Bcor gene and resulting protein (F). Arrowheads indicate primer binding sites used in C and E. Exon 1b distal splice donor site adds an additional 23 bp. Exon 5 is 54 bp and alternative proximal splice acceptor for exon 8 adds 102 bp. NCBI sequences supporting exons 1a-1c; size given for first exon: exon 1a (AK129398, 67bp), exon 1b (CJ073674, 159 bp; BY207441, 148 bp; CD554764, 99bp; CN677848, 132 bp) exon 1c (AY161171, 51bp; AK086385, 241 bp; BB662958, 242 bp; BC058656, 237 bp; BU056818, 237 bp; AK053309, 736 bp)
The identification of three Bcor transcripts by Northern blot analysis prompted us to consider that the Bcor gene may utilize multiple promoters, alternative splicing and/or polyadenylation sites. Although alternative splicing in the coding region has been reported (Fig. 1F and (Srinivasan et al., 2003)) this can only affect transcript size by up to 156 bp. In silico analysis of spliced expressed sequence tags (EST) databases supported use of alternative promoters and polyadenylation sites at the mouse Bcor genomic locus. Two alternative polyadenylation sites separated by 550 bp are present. CpG islands are also found in close proximity upstream of exons 1a, 1b and 1c, further supporting the presence of three independent promoters (Fig. 1F). To determine whether all three promoters suggested by the EST database are actively used in most tissues, we generated an exon 2 specific reverse primer and three forward primers in putative Bcor exons 1a, 1b and 1c that correspond with sequences specifically driven by putative promoters P1, P2 and P3 (Fig. 1F). RT-PCR and sequencing of the products demonstrates that Bcor uses all three putative independent promoters (Fig. 1B and data not shown). In the panel of adult mouse tissues tested, each promoter appears to be used at similar levels in all tissues tested, with the exception of whole blood, which appears to preferentially use promoter 3 relative to other tissues. Interestingly, amplification from promoter 2 results in two specific amplicons (Fig. 1B) due to the use of an alternative splice donor sites in exon 1b (data not shown). We also examined promoter use during embryonic development (E11.5 - E18.5). Although all promoters are used at these stages the distal splice donor of exon 1b is not used at E15.5 and E16.5 (Fig. 1D).
Splicing bypassing exon 5 and/or alternative splice acceptor usage at exon 8 results in the previously identified Bcor isoforms a-d (Fig. 1F) (Srinivasan et al., 2003). Only isoforms a and b contain sequences required for the interaction with the transcriptional regulator AF9 (Ng et al., 2004; Srinivasan et al., 2003). To determine which isoforms are differentially expressed in the embryo and adult mouse tissues, we conducted RT-PCR using primers that span exon 4 to exon 10 on samples used in Figure 1B and 1D. Bcor isoforms a, c and d can be amplified from the samples tested (Fig. 1C and 1E). Isoform c shows the most ubiquitous expression in the sample set and isoform a shows the most tissue-specific expression. Relative to the other tissues, isoform a is more strongly represented in the brain and testis. Isoform b is only barely detected in ovary, eye, spleen and kidney at this level of amplification but not at embryonic stages.
The clinical presentation of OFCD in female patients and presumed embryonic lethality in males suggested that Bcor might have a unique expression pattern during embryogenesis that might give insight into future studies on Bcor function. To determine the spatial and temporal expression of Bcor during gastrulation and early organogenesis we conducted whole mount and section in situ hybridization on embryonic day 7.5 - 15.5 CD-1 outbred mouse embyros. Using the same probe sequence as was used for northern analysis we generated digoxigenin labeled RNA antisense and sense (- control) probes that would detect all known Bcor transcripts. These probes were used in all subsequent whole mount and section in situ expression images.
Whole mount in situ hybridization on embryonic day 7.5 and 8.5 shows that Bcor is strongly expressed in the extraembryonic tissue during gastrulation (Fig. 2A, 2C). Anti-sense probe specificity is validated by absence of signal in embryo hybridized with the sense strand control (Fig. 2B, 2D, 2F). Figure 2E and 2G show that expression of Bcor is restricted to the ectoplacental cone and trophectoderm but absent from extraembryonic visceral and parietal endoderm. Consistent with this, trophoblastic stem cells express Bcor (data not shown). Although Bcor is weakly expressed in the neuroectoderm of the embryo proper (Fig. 2E and 2H), the level is significantly lower in comparison to extraembryonic tissues (Fig. 2E and 2G).
Figure 2.
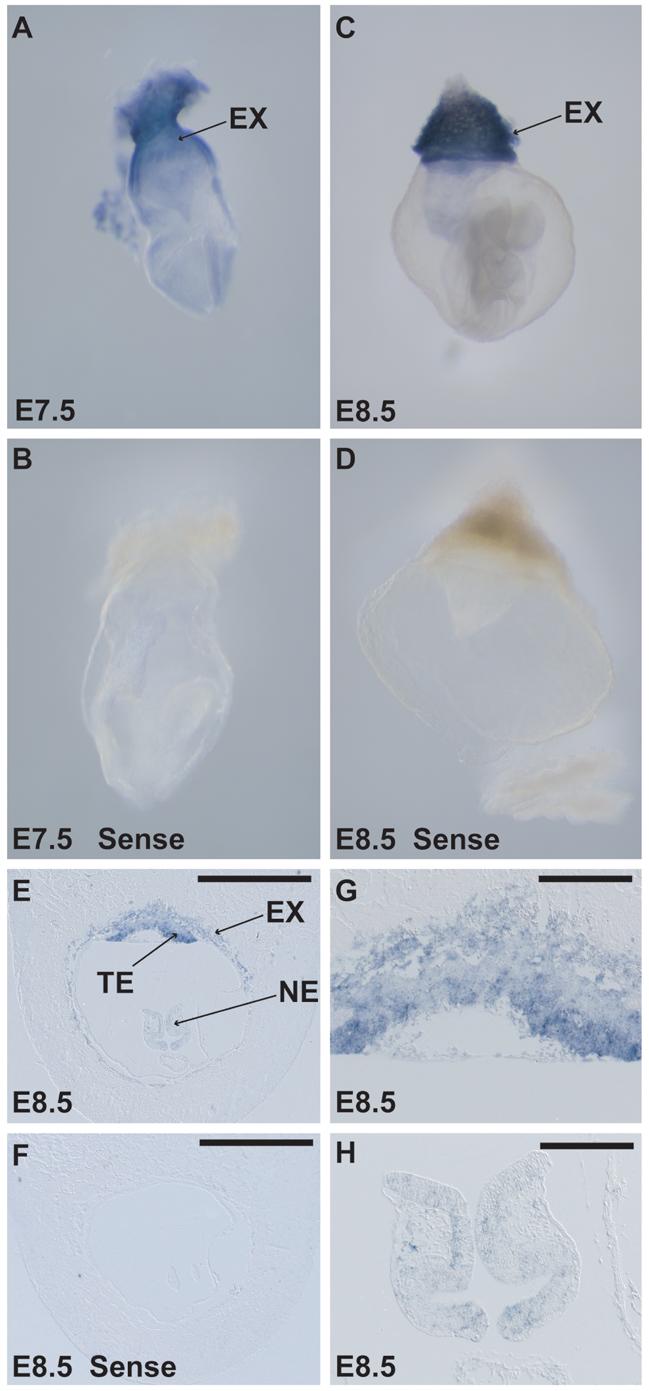
Bcor mRNA is strongly expressed in extraembryonic tissue during early embryogenesis (A-G). Whole mount in situ hybridization of mouse embryos at day 7.5 and 8.5 shows strong expression of Bcor in the extraembryonic tissue (A and C). Sense strand control probes are negative on E7.5 and 8.5 embryos (B and D). Section in situ hybridization of a sagittal section of embryonic day 8.5 shows Bcor expression in the trophectoderm (TE), extraembryonic tissue (EX) and minimally in the neuroectoderm (NE) of the embryo proper (E, G and H). Sense strand control probe is negative on a parallel E8.5 sagittal section (F). (G and H) are insets from panel (E) at higher magnification. Scale bars represent 1 mm in panels (E and F) and 0.1 mm in panels (G and H).
After completion of embryonic turning at approximately E9.0, Bcor transcript levels in the embryo proper increase dramatically. Whole mount in situ hybridization of E9.0 embryos (Figure 3A and B) reveals the initial increase in Bcor expression throughout the embryo proper, led by striking increases in the ectodermal tissue of the tail bud. By E9.5 Bcor transcripts are differentially expressed showing strong expression in the limb buds and branchial arches (Fig. 3C, 3D, 3E; sense control is 3F), while maintaining less robust expression throughout the rest of the embryo. Strong expression of Bcor also extends from the tail bud (shown in detail in Fig. 3G and 3H) down the ridge of the closing neural tube along the dorsal side of the embryo. Strong expression of Bcor in tissues that will give rise to future craniofacial structures correlates well with craniofacial abnormalities present in female OFCD patients.
Figure 3.
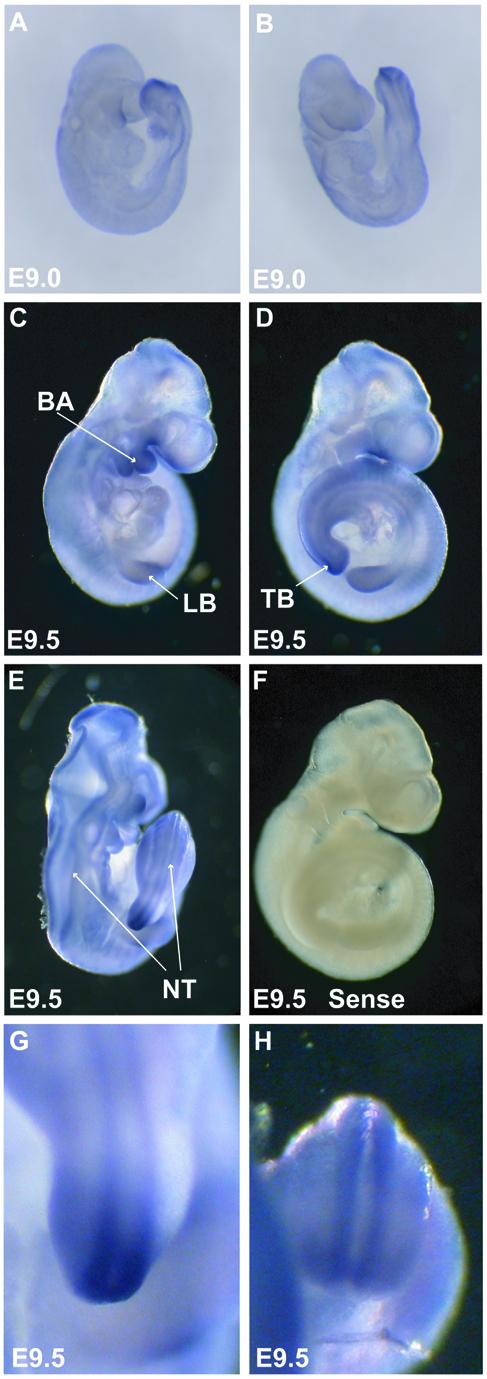
Bcor mRNA expression increases throughout the embryo proper post embryonic turning. Whole mount in situ hybridization shows Bcor expression starting in the tail bud (TB) at embryonic day 9 (A and B) and abundant expression in the tail bud, branchial arches (BA), limb buds (LB) and neural tube (NT) (C, D and E). Sense strand control probes are negative on E9.5 embryos (F). E9.5 tail buds at higher magnification (G and H).
After E9.5, Bcor is differentially expressed in multiple tissue and organ systems during the fetal period of mouse development. Section in situ hybridization of a mid-body cross section at E11.5 reveals strong expression of Bcor in the neural tube and flanking dorsal root ganglion (Fig. 4A and 4D, sense control in 4B). Bcor is also expressed in the notochord and cervical sclerotome and myotome in addition to more subtle and general expression in the forelimb region (Fig. 4A and 4D). Bcor expression can also be found surrounding the esophagus, right and left bronchi and connected lung buds and in the sympathetic chains (Fig. 4A and 4C). A cross section through the head of an E13.5 fetal mouse displays significant expression in the eye, neural tube, the olfactory epithelium and the teeth primordium (Fig. 4E, 4G and 4H, sense control in 4F). Bcor expression in the eye is interestingly found in the retina, lens tissues and in the boundary of the eyelid (Fig. 4E and 4G).
Figure 4.
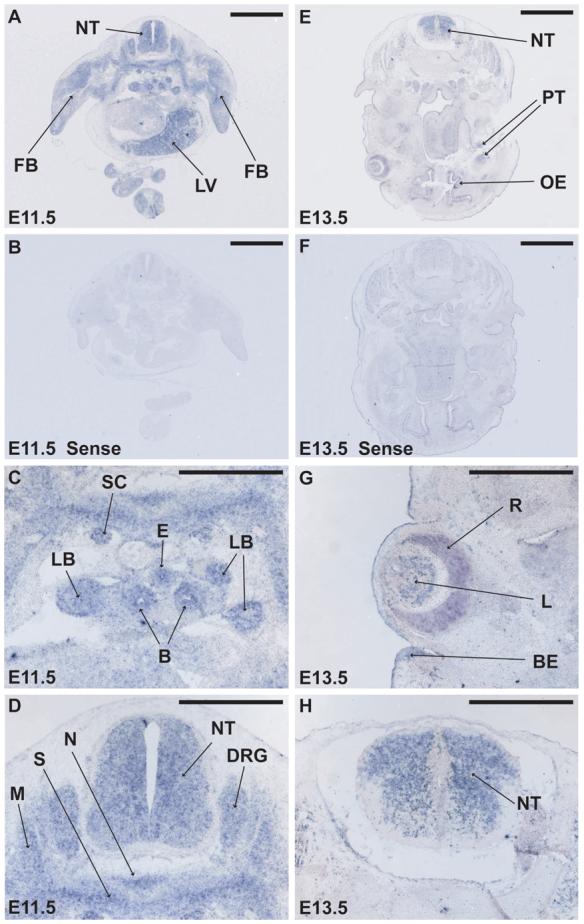
Early fetal stage mouse embryos show widespread but differential expression of Bcor mRNA (A-H). Section in situ hybridization on horizontal sections of an E11.5 embryo shows Bcor expression in multiple tissues including the right and left forelimb buds (FB), liver (LV), left and right lung buds (LB), sympathetic chains (SC), esophagus (E), bronchi (B), myotome (M), sclerotome (S), notochord (N), neural tube (NT), and dorsal root ganglion (DRG) (A, B, C and D). C and D are enlargements from A. Near horizontal section of E13.5 embryo shows expression in the tooth primordium (PT), olfactory epithelium (OE), lens (L) retina (R), boundary of the eyelid (BE), and neural tube (NT), (D, E and F). Sense strand control probes are negative on E11.5 and E13.5 equivalent horizontal sections (B and F). G and H are enlargements form E. Scale bars represent 1 mm in panels (A, B, E and F) and 0.5 mm in panels (C, D, G and H).
At E14.5, a sagittal section near the midline of a fetus reveals expression of Bcor in the trigeminal (V) ganglion, the neopallial cortex, the corpus striatum and the olfactory epithelium (Fig. 5A, 5C and 5D, sense control in 5B). In more lateral sagittal sections through an E15.5 fetus, Bcor expression is retained in the eye but is localized to the retina and the dorsal side of the lens (Fig. 5E and 5G, sense control in 5F). Bcor is also expressed in the lung and gut epithelium and muscle tissue in the fetus e.g. in the temporalis and masseter muscles (Fig. 5E and 5H).
Figure 5.
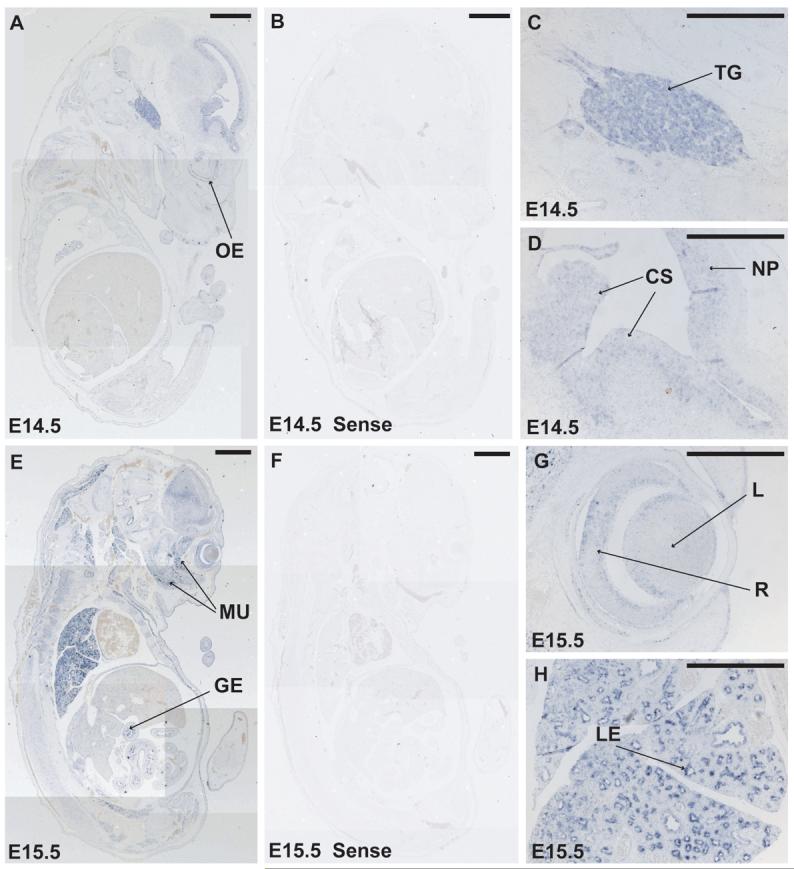
Later fetal stage mouse embryos continue to express Bcor mRNA in a widespread yet tissue specific manner (A-H). Section in situ hybridization on a sagittal section of an E14.5 embyro (A-D) shows Bcor expression in trigeminal ganglion (TG), neopallial cortex (NP), the corpus striatum (CS) and the olfactory epithelium OE). At E15.5 Bcor continues to be expressed in the lens (L) and retina (R) of the eye and in the lung epithelium (LE), gut epithelium (GE), and temporalis and masseter muscle (MU) (E-H). Sense strand control probes are negative on E14.5 and E15.5 equivalent sagittal sections (B and E). Scale bars represent 1 mm in panels (A, B, E and F) and 0.5 mm in panels (C, D, G and H).
Here we have characterized the unique expression pattern of the transcriptional corepressor Bcor. We have shown that Bcor is widely expressed in adult tissue, is expressed in ES cells and displays a widespread but specific expression during embryonic development. There is very strong expression of Bcor in extraembryonic tissue. Expression of Bcor in the developing eye, tooth primordium, limb buds, branchial arches and multiple nervous system tissues also distinctly correlates with many of the tissues adversely affected in OFCD and LM patients (Ng et al., 2004; Schulze et al., 1999). This analysis provides important information for the analysis of future mouse models of OFCD.
Experimental Procedures
Mice
CD-1 outbred mice from Jackson Laboratory were kept on a normal light dark cycle and maintained by Research Animal Resources at the University of Minnesota.
Probe Generation
An amplicon containing nucleotides 280 to 1294 of the Bcor open reading frame was generated by PCR from a bacterial artificial chromosome containing the mouse Bcor gene using the following primers: Forward: 5′-TACGGGTACCATCGATGGCCTGGCAGCACT-3′ and Reverse: 5′-CATGTCTAGATTGGACAGCCTGGATGTAGAG-3′. The PCR amplicon was gel purified (Qiagen, PCR clean up kit) and directly cloned into the Kpn and Xba restriction sites of a pBluescript KS+ vector (Strategene). Radiolabeled DNA probes for Northern blot analysis were generated using a Promega Prime-A-Gene kit and 32P labeled dCTP. Digoxigenin labeled RNA antisense and sense in situ hybridization probes were in vitro transcribed from the T7/T3 promoter containing pKS+ plasmid with Bcor insert using an Ambion Maxiscript kit and Digoxigenin labeleld dUTP from Roche.
Northern Blot Analysis
Total RNA was harvested from embryonic mice and adult mouse organs using TriZol (Invitrogen). Messenger RNA was then extracted using a Miltenyi mRNA isolation kit. Northern blot analysis using one ug per lane was carried out using standard procedures (Ausubel, 1988).
RT-PCR / qRT-PCR
Total RNA was isolated from mouse embryos and tissues using TriZol and cDNA was generated using SuperScript III cDNA sythesis kit (Invitrogen). Bcor promoter RT-PCR was performed using the following primers: Bcor exon 1a forward: 5′-TGCTGGGTTTGAACGGAATG-3′, Bcor exon 1b forward: 5′-GAGAGATAAATGGCCTCTGACAGC-3′, Bcor exon 1c forward: 5′-GAAAACTTCAAAGCGCCGGATC-3′ and Bcor exon 2 reverse: 5′-GACCCTCTCGCTGTTCATCCAG-3′. Bcor isoform a-d RT-PCR was performed using the following primers: Bcor isoform a-d forward: 5′-CTAGCCGGGCAGCCCGCTGCAGGAG-3′ and Bcor isoform a-d reverse: 5′-GTGGGCGGTGCCCCTCCAAACATGGA-3′. HPRT expression was used as a loading control for Bcor promoter and isoform a-d RT-PCR analysis. These HPRT primers are as follows: HPRT forward: 5′-AGCTACTGTAATGATCAGTCAACG-3′ and HPRT reverse: 5′-AGAGGTCCTTTTCACCAGCA-3′.
Whole mount and Section In Situ Hybridization
Whole mount in situ hybridization was performed as described (Nagy, 2003). Section in situ hybridization was performed as described (http://genetics.med.harvard.edu/∼cepko/protocol/insituprotocol.pdf).
Acknowledgements
This work was supported by NCI (R01 CA071540) and NIH (T32DE07288). We thank members of the Bardwell laboratory and Anna Petryk for helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ausubel FM.Current protocols in molecular biology 1988v.Greene Pub.Associates; Wiley-Interscience; New York: (loose-leaf) [Google Scholar]
- Baron BW, Nucifora G, McCabe N, Espinosa R, 3rd, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci U S A. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Chen W, Iida S, Louie DC, Dalla-Favera R, Chaganti RS. Heterologous promoters fused to BCL6 by chromosomal translocations affecting band 3q27 cause its deregulated expression during B-cell differentiation. Blood. 1998;91:603–607. [PubMed] [Google Scholar]
- Collins EC, Appert A, Ariza-McNaughton L, Pannell R, Yamada Y, Rabbitts TH. Mouse Af9 is a controller of embryo patterning, like Mll, whose human homologue fuses with Af9 after chromosomal translocation in leukemia. Mol Cell Biol. 2002;22:7313–7324. doi: 10.1128/MCB.22.20.7313-7324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla-Favera R, Migliazza A, Chang CC, Niu H, Pasqualucci L, Butler M, Shen Q, Cattoretti G.Molecular pathogenesis of B cell malignancy: the role of BCL-6 Curr Top Microbiol Immunol 1999246257–263. discussion 263-255 [DOI] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedera P, Gorski JL. Oculo-facio-cardio-dental syndrome: skewed X chromosome inactivation in mother and daughter suggest X-linked dominant Inheritance. Am J Med Genet A. 2003;123:261–266. doi: 10.1002/ajmg.a.20444. [DOI] [PubMed] [Google Scholar]
- Horn D, Chyrek M, Kleier S, Luttgen S, Bolz H, Hinkel GK, Korenke GC, Riess A, Schell-Apacik C, Tinschert S, et al. Novel mutations in BCOR in three patients with oculo-facio-cardio-dental syndrome, but none in Lenz microphthalmia syndrome. Eur J Hum Genet. 2005;13:563–569. doi: 10.1038/sj.ejhg.5201391. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- Miki T, Kawamata N, Arai A, Ohashi K, Nakamura Y, Kato A, Hirosawa S, Aoki N. Molecular cloning of the breakpoint for 3q27 translocation in B-cell lymphomas and leukemias. Blood. 1994;83:217–222. [PubMed] [Google Scholar]
- Nagase T, Kikuno R, Nakayama M, Hirosawa M, Ohara O. Prediction of the coding sequences of unidentified human genes. XVIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:273–281. doi: 10.1093/dnares/7.4.271. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo: a laboratory manual. 3rd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2003. [Google Scholar]
- Ng D, Hadley DW, Tifft CJ, Biesecker LG. Genetic heterogeneity of syndromic X-linked recessive microphthalmia-anophthalmia: is Lenz microphthalmia a single disorder? Am J Med Genet. 2002;110:308–314. doi: 10.1002/ajmg.10484. [DOI] [PubMed] [Google Scholar]
- Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- Schulze BR, Horn D, Kobelt A, Tariverdian G, Stellzig A. Rare dental abnormalities seen in oculo-facio-cardio-dental (OFCD) syndrome: three new cases and review of nine patients. Am J Med Genet. 1999;82:429–435. doi: 10.1002/(sici)1096-8628(19990219)82:5<429::aid-ajmg13>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Srinivasan RS, de Erkenez AC, Hemenway CS. The mixed lineage leukemia fusion partner AF9 binds specific isoforms of the BCL-6 corepressor. Oncogene. 2003;22:3395–3406. doi: 10.1038/sj.onc.1206361. [DOI] [PubMed] [Google Scholar]
- Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. Embo J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large- cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]


