Abstract
Auxin plays a key role in embryogenesis and seedling development, but the auxin sources for the two processes are not defined. Here, we demonstrate that auxin synthesized by the YUCCA (YUC) flavin monooxygenases is essential for the establishment of the basal body region during embryogenesis and the formation of embryonic and postembryonic organs. Both YUC1 and YUC4 are expressed in discrete groups of cells throughout embryogenesis, and their expression patterns overlap with those of YUC10 and YUC11 during embryogenesis. The quadruple mutants of yuc1 yuc4 yuc10 yuc11 fail to develop a hypocotyl and a root meristem, a phenotype similar to those of mp and tir1 afb1 afb2 afb3 auxin signaling mutants. We further show that YUC genes play an essential role in the formation of rosette leaves by analyzing combinations of yuc mutants and the polar auxin transport mutants pin1 and aux1. Disruption of YUC1, YUC4, or PIN1 alone does not abolish leaf formation, but the triple mutant yuc1 yuc4 pin1 fails to form leaves and flowers. Furthermore, disruption of auxin influx carrier AUX1 in the quadruple mutant yuc1 yuc2 yuc4 yuc6, but not in wild-type background, phenocopies yuc1 yuc4 pin1, demonstrating that auxin influx is required for plant leaf and flower development. Our data demonstrate that auxin synthesized by the YUC flavin monooxygenases is an essential auxin source for Arabidopsis thaliana embryogenesis and postembryonic organ formation.
INTRODUCTION
Auxin has been shown to play an essential role in Arabidopsis thaliana embryogenesis. Genetic screens for mutants with embryo pattern defects have led to the identification of monopteros (mp) (Berleth and Jurgens, 1993; Hardtke and Berleth, 1998) and bodenlos (bdl) (Hamann et al., 1999, 2002), two mutants that fail to develop a hypocotyl and embryonic roots. MP encodes the auxin response factor 5 (ARF5), which belongs to the ARF family, with 23 members in the Arabidopsis genome (Guilfoyle et al., 1998; Okushima et al., 2005). BDL (IAA12) is one of the 29 AUX/IAA transcription factors in Arabidopsis (Kim et al., 1997; Reed, 2001; Liscum and Reed, 2002). It was shown that BDL physically interacts with MP, thus preventing MP from forming homodimers or heterodimers with other ARFs (Weijers et al., 2005a, 2005b). In the presence of auxin, BDL is brought to the SCFTIR1-containing protein degradation machinery mediated by auxin binding directly to the auxin receptor TIR1 (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005; Tan et al., 2007). Auxin-induced degradation of BDL allows MP to form ARF-ARF dimers that can then activate transcription necessary for plant development. The studies on mp and bdl revealed that auxin is necessary for the establishment of the apical-basal axis and the formation of embryonic organs. Recent studies on auxin signaling and polar auxin transport further demonstrate that auxin is required for Arabidopsis embryogenesis. The auxin receptor quadruple mutant tir1 afb1 afb2 afb3 also displays phenotypes similar to mp or bdl (Dharmasiri et al., 2005b). Disruption of auxin efflux during embryogenesis by simultaneously inactivating multiple PIN genes affects the formation of the apical-basal axis during embryogenesis (Friml et al., 2003). Although much is known regarding the roles of auxin signaling and transport in embryogenesis, genes regulating auxin biosynthesis during embryogenesis are not known. De novo auxin biosynthesis is likely to play a critical role in embryogenesis because previous physiological studies have shown that auxin biosynthesis is dynamic during embryogenesis (Fischer-Iglesias et al., 2001; Ribnicky et al., 2002).
Auxin is also known to play a role in leaf development. It has been clearly demonstrated that auxin plays a critical role in phyllotaxis of leaf formation (Reinhardt et al., 2003). Most of the information on the control of leaf formation by auxin comes from analysis of auxin transport mutants and the effects of local application of auxin (Benkova et al., 2003; Reinhardt et al., 2003). However, the sources of auxin and the locations of auxin production during leaf development are not defined.
We previously showed that the YUCCA (YUC) family of flavin monooxygenases are key enzymes in Trp-dependent auxin biosynthesis (Zhao et al., 2001; Cheng et al., 2006). Overexpression of YUC genes leads to long hypocotyls and epinastic cotyledons (Zhao et al., 2001), characteristic auxin overproduction phenotypes observed in iaaM overexpression lines (Romano et al., 1995), superroot1 (Boerjan et al., 1995), and other auxin overproduction mutants (Delarue et al., 1998). There are 11 YUC genes in the Arabidopsis genome (Zhao et al., 2001; Cheng et al., 2006). Inactivation of YUC1 and YUC4 leads to dramatic defects in floral and vascular development (Cheng et al., 2006). Phenotypes of yuc1 yuc4 double mutants are rescued by expressing the bacterial auxin biosynthetic gene iaaM under the control of the YUC1 promoter (Cheng et al., 2006). Deactivation of YUC2 and YUC6 in yuc1 yuc4 further exacerbates the floral and vascular defects of this double mutant, demonstrating that the YUC genes have overlapping functions and are essential for auxin biosynthesis during flower and vascular development (Cheng et al., 2006). Here, we show that YUC1 and YUC4 show distinct and overlapping expression patterns during embryogenesis and seedling development, but the yuc1 yuc4 double mutants do not exhibit any obvious defects in embryogenesis and the formation of leaves. We show that inactivation of YUC10 and YUC11 in the yuc1 yuc4 background leads to phenotypes similar to those of strong mp mutants, indicating that auxin synthesized by the YUC flavin monooxygenases is a required auxin source for embryogenesis. Furthermore, we demonstrate that auxin synthesized by the YUC flavin monooxygenases is also a critical auxin source for leaf development. Together with our previous findings that YUC genes are essential for flower and vascular development, our data show that the YUC genes are necessary for many developmental processes from embryogenesis to seedling development to flower development.
RESULTS
YUC Genes Display Distinct Expression Patterns during Embryogenesis
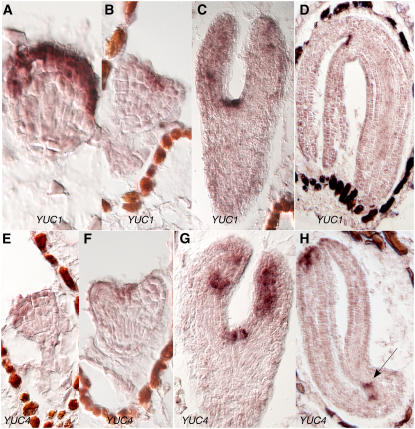
To test whether the YUC genes play a role in synthesizing auxin during embryogenesis, we first analyzed the expression patterns of YUC1 during embryogenesis by RNA in situ hybridization. YUC1 expression became obvious in globular stages of embryo but with expression concentrated in the apical region (Figure 1A). Later, the expression of YUC1 was mainly restricted to the cotyledons and the apical meristem (Figures 1B and 1C). In mature embryos, YUC1 expression was mainly detected at the apex (Figure 1D). Interestingly, the expression domain of YUC1 was relatively broad during early stages of embryogenesis and became more restricted to discrete groups of cells in mature embryos. The in situ data with the YUC1 sense probe are shown in Supplemental Figure 1 online.
Figure 1.
Analysis of the Expression Patterns of YUC1 and YUC4 during Embryogenesis by RNA in Situ Hybridization.
(A) to (D) YUC1 expression patterns. Note that YUC1 is mainly expressed in the first three cell layers in the apical region in the globular-stage embryos (A). At heart stage, the highest expression is at the meristem region (B).
(E) to (H) YUC4 expression patterns. The arrow points to the likely sites for incipient true leaves.
YUC4 was also expressed at various stages of embryogenesis (Figures 1E to 1H). The YUC4 expression patterns were very similar to those of YUC1 throughout embryogenesis, suggesting that YUC1 and YUC4 have overlapping functions during embryogenesis just like their redundant roles in flower and vascular development. Unlike YUC1, YUC4 was also expressed at the apical region of the cotyledons in the mature embryo (Figure 1H). The in situ data with YUC4 sense probe are shown in Supplemental Figure 1 online.
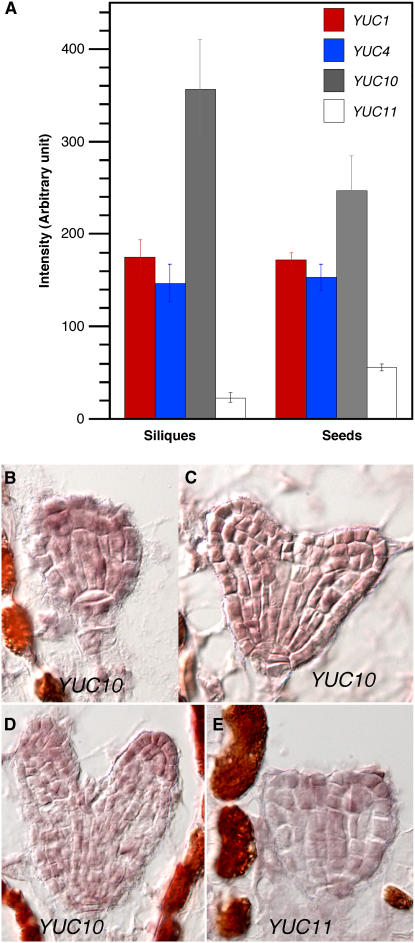
The fact that yuc1 yuc4 double mutants did not display any obvious defects in embryogenesis suggests that other YUC genes may have overlapping functions with YUC1 and YUC4 during embryogenesis. Analysis of the public microarray data (Zimmermann et al., 2004) indicates that YUC10, which is closely related to YUC1 and YUC4 in a phylogenetic tree (Cheng et al., 2006), is highly expressed in siliques (Figure 2A). We further analyzed the expression patterns of YUC10 and its closest homolog YUC11 during embryogenesis by RNA in situ hybridization and found that their expression overlapped with those of YUC1 and YUC4 (Figures 2B to 2E). The data of sense probes are shown in Supplemental Figure 1 online.
Figure 2.
Analysis of the Expression Profile of YUC10 and YUC11.
(A) Expression profiles in siliques and seeds. Data were from the public microarray data sets (https://www.genevestigator.ethz.ch).
(B) to (D) RNA in situ hybridization of YUC10.
(E) RNA in situ hybridization of YUC11.
Multiple YUC Genes Contribute to the Formation of the Apical-Basal Axis and Embryonic Organ Formation
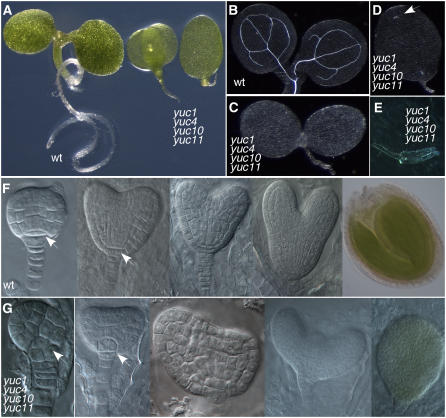
Overexpression of either YUC10 or YUC11 in Arabidopsis led to auxin overproduction phenotypes similar to those of YUC1 overexpression lines (data not shown), suggesting that YUC10 and YUC11 probably also play a role in auxin biosynthesis and have overlapping functions with YUC1 and YUC4 during embryogenesis. The yuc1 yuc4 yuc10 yuc11 quadruple mutants had remarkable developmental defects. Seedlings of the quadruple mutants did not have hypocotyls or roots (Figure 3A). Most of the yuc1 yuc4 yuc10 yuc11 seedlings only had one cotyledon with little vascular tissue (Figures 3B to 3E). The defects of yuc1 yuc4 yuc10 yuc11 occurred as early as the globular stages of the embryo (Figures 3F and 3G). The quadruple mutants lacked a hypophysis (Figures 3F and 3G, arrows), cells that later develop into a root meristem in wild-type plants. The central cells did not elongate and the embryos failed to develop hypocotyls (Figures 3F and 3G). A prominent feature of the yuc1 yuc4 yuc10 yuc11 embryo was that the basal body region was defective and no primary roots were developed (Figure 3A).
Figure 3.
Regulation of Arabidopsis Embryogenesis by YUC Genes.
(A) Developmental defects of yuc1 yuc4 yuc10 yuc11 at seedling stages. The seedling at the left is the wild type, and the two seedlings at the right are yuc1 yuc4 yuc10 yuc11. Note that the mutant lacks a hypocotyl and a root.
(B) to (E) Vascular defects in yuc1 yuc4 yuc10 yuc11.
(B) The wild type.
(C) to (E) yuc1 yuc4 yuc10 yuc11. The arrow points to a short and discontinuous vein. The vascular tissue in (E) was from the seedling of (D).
(F) Various stages of wild-type embryos.
(G) The corresponding stages of embryos of yuc1 yuc4 yuc10 yuc11. The arrows point to where the hypophysis should be.
We analyzed seedlings from a single plant that was yuc1 yuc10 yuc11 homozygous and yuc4 heterozygous to determine whether the mp-like phenotypes are completely penetrant. Among the 369 seedlings analyzed, 73 plants displayed mp-like phenotypes and had the yuc1 yuc4 yuc10 yuc11 genotype. Because we only observed 20% of the progenies that had mp-like phenotypes, we hypothesized that the mp-like phenotypes of yuc1 yuc4 yuc10 yuc11 may not be 100% penetrant or some of the quadruple mutants died during embryogenesis. We then genotyped 96 plants that were from a single yuc1−/− yuc4+/− yuc10−/− yuc11−/− plant and did not display obvious mp-like phenotypes. One of the plants among the 96 plants was identified as yuc1 yuc4 yuc10 yuc11, indicating that occasionally the quadruple mutants could escape from the mp-like phenotypes.
YUC Genes Display Distinct Expression Patterns during Seedling Growth
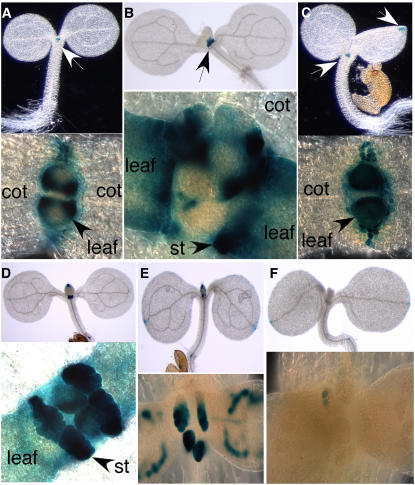
To investigate whether the YUC genes play a role in providing auxin for seedling growth and leaf development, we analyzed the patterns of β-glucuronidase (GUS) staining of YUC1 promoter GUS lines and YUC4 promoter GUS lines. For YUC1-GUS lines, the GUS staining was restricted to the shoot apex and no staining was observed in the cotyledons (Figure 4A, top panel). In the shoot apex, cells flanking the apical meristem showed GUS staining, but the strongest GUS staining was located at the base of young leaves and stipules (Figures 4A and 4B). There was no staining in the apical meristem (Figure 4A, bottom panel). Further analysis of sections of the YUC1-GUS seedlings indicate that YUC1 is not expressed in the apical meristem (data not shown).
Figure 4.
Expression Patterns of YUC1 and YUC4 during Seedling Development and Effects of yuc1 yuc4 on Auxin Reporter DR5-GUS Expression.
(A) GUS staining of a 3-d-old light-grown YUC1-GUS seedling. Staining was restricted to the shoot apex without any staining in cotyledons (cot) (top panel). In the apex, strong GUS staining was located at the basal parts of the true leaves (bottom panel).
(B) GUS staining of a 5-d-old YUC1-GUS seedling. Staining was observed in stipules (st).
(C) GUS staining of a 3-d-old YUC4-GUS seedling. Staining was located at cotyledon tips and the shoot apex (top panel).
(D) GUS staining of a 5-d-old YUC4-GUS seedling. Note the strong staining in the stipules (bottom panel).
(E) Expression of the auxin reporter DR5-GUS. GUS activities were observed at the cotyledon tip and the shoot apex. At the shoot apex, DR5-GUS is expressed in stipules and vascular tissue (bottom panel).
(F) GUS staining of DR5-GUS in the yuc1 yuc4 background. Inactivation of YUC1 and YUC4 leads to a dramatic decrease of DR5-GUS expression in the apex without affecting the staining at the cotyledon tip.
GUS staining of YUC4-GUS was also mainly restricted to the shoot apex (Figures 4C and 4D, top panels). Like YUC1-GUS, GUS staining was observed in the cells surrounding the apical meristem in YUC4-GUS lines (Figure 4C, bottom panel). Unlike YUC1-GUS, weak staining was observed at the tips of cotyledons (Figures 4C and 4D, top panels). In true leaves, GUS staining was observed initially throughout the young leaf primordia, and later the staining was concentrated at the basal and apical regions of leaves. Strong GUS staining was also observed in stipules (Figures 4C and 4D).
The Expression of the Auxin Reporter DR5-GUS Overlaps with YUC Expression Domains during Seedling Development
In wild-type Arabidopsis seedlings, DR5-GUS mainly expressed at newly developed leaves, the tip of cotyledons, and root tips (Figure 4E). At the apex, DR5-GUS was mainly expressed in young leaves, stipules, and later in vascular tissues (Figure 4E, bottom panel). The DR5-GUS patterns in wild-type plants overlapped significantly with the expression patterns of YUC1 and YUC4 in the shoot apex (Figure 4). Disruption of YUC1 and YUC4 greatly decreased DR5-GUS level in the apex without affecting DR5-GUS at cotyledon tips (Figure 4F) and in roots (data not shown).
PIN1 and YUC Genes Synergistically Control Leaf Development
The YUC expression pattern and DR5-GUS analysis suggest that YUC genes may play a role in leaf development. However, the yuc1 yuc4 double mutants did not show obvious defects in the formation of leaves in terms of the number and position of the leaves, although yuc1 yuc4 leaves had vascular defects. We hypothesized that other YUC genes may compensate for yuc1 yuc4 during leaf initiation and elongation. The yuc1 yuc2 yuc4 yuc6 quadruple mutants have severe defects in vascular and flower defects, but the number of leaves formed in the quadruple mutants appeared to be similar to the wild type. So far, all of the yuc mutant combinations we have constructed either do not have defects in the formation of leaves or have embryonic defects, making it difficult to assess the roles of YUC genes in normal leaf development.
We hypothesized that the roles of YUC genes in leaf development may also be masked by polar auxin transport. To test this hypothesis, we constructed yuc1 yuc4 pin1 triple mutants. Single pin1 mutations or double yuc1 yuc4 mutations only partially disrupt auxin efflux and biosynthesis, respectively, because there are other PINs and YUCs in the Arabidopsis genome with overlapping functions (Galweiler et al., 1998; Cheng et al., 2006). Both yuc1 yuc4 and pin1 had defects in forming normal flowers and vascular tissues (Galweiler et al., 1998; Cheng et al., 2006), but they had leaves and other lateral organs (Figure 5A). The yuc1 yuc4 pin1 triple mutants failed to form true leaves (Figure 5A), a phenotype not observed in either pin1 or yuc1 yuc4 alone, demonstrating the synergistic effects between yuc1 yuc4 and pin1. Electron micrographs showed that the shoot apex of yuc1 yuc4 pin1 was not flat; instead, we observed a dozen or so stipule-like structures with three or four cells (Figures 5B and 5C). The triple mutant yuc1 yuc4 pin1 eventually formed pin-like inflorescences without producing any flowers or cauline leaves (Figure 5D).
Figure 5.
Synergistic Interactions between yuc1 yuc4 and pin1 Mutants during Leaf Development.
(A) The triple mutant yuc1 yuc4 pin1 did not make a leaf. Seedlings shown are 7 d old and grown on 0.5× Murashige and Skoog media. Arrow points to the apex of yuc1 yuc4 pin1. The pin1 mutant was a null mutant caused by a T-DNA insertion. From left to right: wild type, yuc1 yuc4, pin1, and yuc1 yuc4 pin1.
(B) to (D) Electron micrographs of yuc1 yuc4 pin1.
(B) Seven-day-old yuc1 yuc4 pin1. Note that no true leaves were visible at the shoot apex.
(C) A close-up of the shoot apex of a yuc1 yuc4 pin1 seedling.
(D) Pin-like structure in 2-week-old yuc1 yuc4 pin1.
(E) Genetic interactions between yuc1 yuc4 and pin1-5, a weak allele of pin1. From left to right: wild type, yuc1, yuc4, yuc1 yuc4 double mutants, pin1-5, two plants of pin1 null, two plants of yuc1 pin1-5 double mutants, two plants of yuc4 pin1-5 double mutants, and yuc1 yuc4 pin1-5 triple mutants. Note that pin1-5 never forms pin-like inflorescences, but yuc1 pin1-5 or yuc4 pin1-5 generated pin-like inflorescences. The yuc1 yuc4 pin1-5 triple mutants had stronger phenotypes than pin1 null. Arrows point to pin-like inflorescences.
Analysis of the interactions between yuc1 yuc4 double mutants and the weak mutant pin1-5 (Bennett et al., 1995; Sohlberg et al., 2006) further supports that PIN1 and YUC1/YUC4 have synergistic interactions. The pin1-5 allele never forms pin-like inflorescences and is fertile (Figure 5E). When pin1-5 was combined with yuc1, the yuc1 pin1-5 double mutants produced pin-like inflorescences (Figure 5E). The yuc4 pin1-5 double mutants displayed stronger phenotypes than yuc1 pin1-5 double mutants (Figure 5E). The yuc1 yuc4 pin1-5 triple mutants produced pin-like inflorescences with a stature smaller than either yuc1 pin1-5 or yuc4 pin1-5 (Figure 5E). Our data indicate that auxin synthesized by the YUC flavin monooxygenases is a critical auxin source required for leaf and flower development.
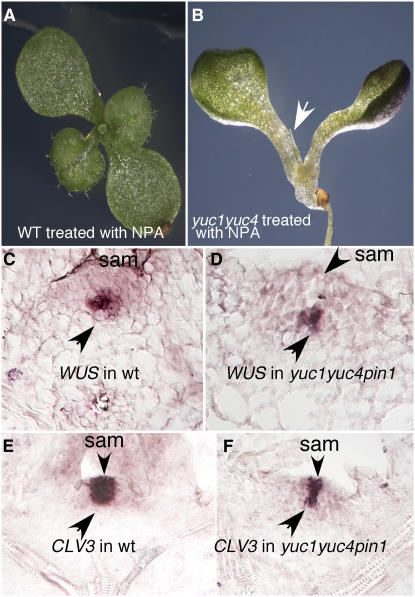
yuc1 yuc4 Double Mutants Treated with the Auxin Transport Inhibitor Naphthylphthalamic Acid Phenocopy yuc1 yuc4 pin1 Triple Mutants
Treatment of wild-type Arabidopsis plants with the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) leads to the formation of pin-like inflorescences (Okada et al., 1991) but does not inhibit leaf formation (Figure 6A). When yuc1 yuc4 was grown on NPA-containing media, no true leaves were formed (Figure 6B). The observation that yuc1 yuc4 pin1 phenotypes can be phenocopied by treating yuc1 yuc4 with NPA further supports the conclusion that auxin synthesized by the YUCs contributes to leaf development (Figures 6A and 6B). NPA treatment also led to a further decrease of DR5-GUS expression in the apex of yuc1 yuc4 (see Supplemental Figure 2 online).
Figure 6.
Analysis of Expression Patterns of Apical Meristem Markers in yuc1 yuc4 pin1 and the Effects of NPA on yuc1 yuc4 Development.
(A) Wild-type plants grown on 25 μM NPA for 12 d.
(B) yuc1 yuc4 seedlings grown on NPA for 12 d. Arrow points to a pin-like structure.
(C) Expression patterns of WUS in wild-type shoot apical meristem (sam). WUS was expressed in a small group of cells several layers away from the epidermis.
(D) WUS expression in yuc1 yuc4 pin1. Note the similar patterns in the wild type and the mutant.
(E) and (F) CLV3 expression in the shoot apical meristem in the wild type (E) and yuc1 yuc4 pin1 (F). The expression patterns were analyzed by RNA in situ hybridization using 8-d-old light-grown seedlings.
The Apical Meristem in yuc1 yuc4 pin1 Triple Mutants Is Not Collapsed
We analyzed whether the failure to form true leaves in yuc1 yuc4 pin1 was caused by a collapse of the apical meristem. We chose to analyze the meristem markers CLV3 and WUS by RNA in situ hybridization (Fletcher et al., 1999; Lenhard et al., 2001; Lohmann et al., 2001). In the wild type, WUS mRNA was detected in a small group of cells in the center of the shoot apical meristem (Figure 6C), while CLV3 expression was restricted to a small zone of cells at the meristem apex, including cells in the L1 and L2 layers (Figure 6E). In yuc1 yuc4 pin1 triple mutants, both CLV3 and WUS displayed expression patterns similar to those of the wild type, suggesting that yuc1 yuc4 pin1 still had an apical meristem (Figures 6D and 6F). Therefore, the yuc1 yuc4 pin1 triple mutants mainly failed to differentiate the cells flanking the meristem into leaves and flowers.
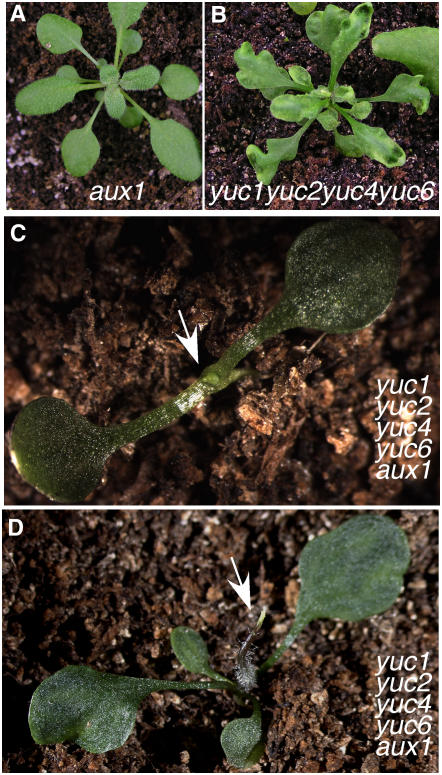
Inactivating AUX1 in yuc1 yuc2 yuc4 yuc6 Mutants Phenocopies yuc1 yuc4 pin1 Triple Mutants
Polar auxin transport has been explained by the Chemiosmotic Hypothesis of auxin transport in which directional auxin transport is mediated by specific auxin influx and efflux carriers (Rubery and Sheldrake, 1974; Raven, 1975). Both passive diffusion and carrier-mediated influx are proposed for auxin influx, whereas auxin efflux totally depends on efflux carriers. The AUX1 family proteins have been shown to be influx carriers (Bennett et al., 1996; Swarup et al., 2001; Yang et al., 2006). Previous studies have shown that AUX1 plays an important role in root development (Swarup et al., 2001), but the contributions of AUX1 to leaf and flower development are not as defined. We hypothesized that AUX1 and the YUCs may mask each other's functions during leaf development.
To test this hypothesis, we introduced aux1 mutation into various yuc mutant combinations. The quintuple mutants of yuc1 yuc2 yuc4 yuc6 aux1 had fewer leaves than either yuc1 yuc2 yuc4 yuc6 quadruple mutants or aux1 single mutants (Figures 7A to 7D). The overall developmental defects of the quintuple mutants yuc1 yuc2 yuc4 yuc6 aux1 were very similar to those of yuc1 yuc4 pin1 (Figure 5). The fact that aux1 and pin1 displayed similar phenotypes in the yuc mutant background clearly demonstrates that both auxin influx and efflux are very important to leaf development. Our data also indicate that auxin synthesized by the YUCs is a necessary auxin source for leaf development.
Figure 7.
A Role for AUX1 in Leaf and Flower Development.
(A) Adult aux1.
(B) Adult yuc1 yuc2 yuc4 yuc6.
(C) and (D) yuc1 yuc2 yuc4 yuc6 aux1.
Note that both aux1 and yuc1 yuc2 yuc4 yuc6 developed leaves, whereas the yuc1 yuc4 yuc6 aux1 quintuple mutants developed fewer leaves and showed phenotypes similar to yuc1 yuc4 pin1.
DISCUSSION
YUC Genes Are Essential for Embryogenesis
We have shown that simultaneous disruption of four YUC genes (YUC1, YUC4, YUC10, and YUC11) led to seedlings without a hypocotyl and a root meristem. The phenotypes of yuc1 yuc4 yuc10 yuc11 quadruple mutants resembled the strong alleles of mp (Berleth and Jurgens, 1993), bdl (Hamann et al., 2002), and tir1 afb1 afb2 afb3 auxin receptor mutants (Dharmasiri et al., 2005b), which are defective in auxin signaling. Similarities of embryo defects between yuc1 yuc4 yuc10 yuc11 and pin1 pin3 pin4 pin7 mutants (Friml et al., 2003) are also remarkable, indicating that auxin synthesized by YUC flavin monooxygenases is a critical auxin source for embryogenesis.
YUC1 and YUC4 were mainly expressed in the apical region of globular or heart stages of embryos with little expression in the hypophysis, yet the main defects in the yuc1 yuc4 yuc10 yuc11 quadruple mutants were in hypocotyl and root meristem. We envision two mechanisms for how auxin synthesized in the apical region can regulate the development of hypophysis and root meristem. Our results suggest that auxin synthesized by the YUCs may be transported to the hypophysis to regulate the root specification. This hypothesis is consistent with the observations that there is a DR5-GFP maximum at the hypophysis and there is a PIN1 polarity switch during embryogenesis (Friml et al., 2003). However, it is also known that MP and BDL are not expressed in the hypophysis (Weijers et al., 2006), yet both mp and bdl mutants fail to form a root meristem (Hardtke and Berleth, 1998; Hamann et al., 2002). An alternative mechanism for controlling root meristem development by YUC genes during embryogenesis is to degrade BDL and to activate MP by YUC-mediated auxin synthesis in the apical region. Once BDL is degraded and MP is activated to initiate transcription, an unidentified MP-dependant signal is sent to the hypophysis to specify its development. We should point out that these two mechanisms are not mutually exclusive. Nevertheless, the elucidation of the roles of YUC genes in embryogenesis and the YUC expression patterns provides a foundation for further analyzing the mechanisms by which auxin controls embryogenesis.
Genetic Interactions between YUCs and Polar Auxin Transport
Previous work shows that both pin1 mutants and yuc1 yuc4 double mutants produce fewer flowers than the wild type and that the flowers in pin1 or yuc1 yuc4 are defective and completely sterile (Galweiler et al., 1998; Tobena-Santamaria et al., 2002; Cheng et al., 2006), but it is not clear whether the pin mutants and yuc mutants have epistatic interactions. The fact that yuc1 yuc4 pin1 triple mutants displayed phenotypes (Figure 5) that are not seen in either pin1 or yuc1 yuc4 alone demonstrates that polar auxin transport and YUC-mediated auxin biosynthesis have strong genetic interactions. This conclusion is further supported by the findings that aux1 and yuc mutants also synergistically enhance each other (Figure 7). Arabidopsis organogenesis probably requires that the founder cells for an organ primordium must have a threshold of auxin. The auxin threshold hypothesis is also consistent with our previous observations that the double, triple, and quadruple yuc mutants displayed increasingly severe defects in vascular development and overall plant architecture (Cheng et al., 2006).
Because both auxin transport and auxin biosynthesis are redundantly regulated and the pin1 and yuc1 yuc4 mutants only partially disrupt auxin transport and biosynthesis, respectively, it is more difficult to interpret the synergistic interactions between yuc and transport mutants. The simplest interpretation for the synergistic interactions is that in the yuc1 yuc4 double mutant background, auxin levels are low and there is not sufficient auxin to be transported to the founder cells of leaf primordia. Another explanation is that auxin in the founder cells of leaf primordia comes from auxin transported from surrounding cells and from de novo synthesis by the YUCs in the founder cells as well. The latter explanation is consistent with the observations that YUC genes are expressed at the incipient sites of new leaves and emerging leaf primordia. The two interpretations are not mutually exclusive because it is likely both auxin levels and the locations of auxin production are important for the establishment of an auxin gradient for leaf formation. Regardless of which mechanisms are involved, our data have clearly shown that auxin produced by the YUC genes is an essential auxin source for leaf and flower development.
Auxin Influx by AUX1 Plays a Key Role in Leaf and Flower Development
The role of AUX1 in root development and root gravitropic responses is well established (Bennett et al., 1996; Swarup et al., 2001). However, the role of auxin influx mediated by AUX1 in leaf and flower development received much less attention compared with PIN-mediated auxin efflux, partly because disruption of AUX1 causes no obvious developmental defects in the shoot, although AUX1 is highly expressed in the shoot (https://www.genevestigator.ethz.ch). It is argued that auxin influx is probably not regulated because most of the IAA molecules outside of the cell are protonated and potentially can cross the plasma membrane by diffusion. The fact that aux1 developed pin-like inflorescences in yuc1 yuc2 yuc4 yuc6 quadruple mutants, but not in wild-type background, demonstrates that the role of AUX1 in plant development is masked by YUC-mediated auxin biosynthesis. Our data have clearly shown that AUX1-mediated auxin influx becomes limiting in the yuc1 yuc2 yuc4 yuc6 background in terms of providing auxin to the founder cells of leaf primordia.
METHODS
Plant Materials
We obtained the T-DNA insertion mutants from either the ABRC or from Institut National de la Recherche Agronomique (INRA) (Samson et al., 2004). The mutants were genotyped according to the published protocols (Alonso et al., 2003; Samson et al., 2004). Genotyping primers for yuc1, yuc2, yuc4, and yuc6 were described previously (Cheng et al., 2006). The insertion mutant for YUC10 (At1g48910) was the FLAG_599G05 line from INRA (Samson et al., 2004). Our DNA sequencing data showed that the T-DNA was inserted 653 bp downstream of the start codon of YUC10. Primers for genotyping yuc10 are 5′-CCTGAATCTCGCCATCGCGAATC-3′, 5′-CCAAAGAGCTTTTCGTCAACTACC-3′, and the T-DNA specific primer FLAG_LB4 5′-CGTGTGCCAGGTGCCCACGGAATAGT-3′.
The T-DNA line for YUC11 (At1g21430) was the SALK_073485 line from ABRC. The T-DNA was inserted in the second exon of the gene, 822 bp downstream of the start codon. Genotyping primers for yuc11 were as follows: LP, 5′-TGTCAACTCCCTCACATGCCA-3′; RP, 5′-CAGATCTCCATCATCGACCTGTGT-3′; and JMLB1 as the T-DNA–specific primer.
The pin1 mutant contains a T-DNA insertion 1945 bp downstream of the ATG site. The mutant was genotyped with the following primers: 5′-ACAACCAGTACGTGGAGAGGG-3′ and 5′-TCATAGACCCAAGAGAATGTAGTAG-3′. The T-DNA insertion in aux1 was located at 1893 bp from the ATG site. The aux1 mutant is genotyped using the following primers: 5′-CGATCATCTGGACAAGAGAACATG-3′ and 5′-TCCTCCACCGACTCTTCATTTC-3′.
DR5-GUS and YUC promoter GUS lines were described previously (Cheng et al., 2006).
Methods
In situ RNA hybridization and scanning electron microscopy analysis were performed according to the methods described by Dinneny et al. (2004). Vascular visualization was performed using protocols from Cheng et al. (2006). We performed GUS staining as previously described (Cheng et al., 2006).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_175321 (YUC10) and NP_173564 (YUC11).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RNA in Situ Hybridization Using Sense Probes of YUC Genes.
Supplemental Figure 2. Effects of NPA Treatment on DR5-GUS Expression in yuc1 yuc4 Double Mutants.
Supplementary Material
Acknowledgments
We thank J. Chory, M. Chrispeels, R. Schmidt, and M. Yanofsky for advice and comments on the manuscript. We also thank J. Long and G. Ditta for technical assistance on RNA in situ hybridization and analysis of embryos, E. York for scanning electron microscopy analysis, and E. Sundberg for the pin1-5 mutant. We thank F. Pierri, K. Saroca, L. Guerra, S. Lee, A. Fang, Y. Liu, M. Chang, Jin O, Y. Sun, and M. Lewis for DNA preps and genotyping various mutants. This work is supported by a grant from the National Institutes of Health (R01GM68631) to Y.Z.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yunde Zhao (yzhao@biomail.ucsd.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273 948–950. [DOI] [PubMed] [Google Scholar]
- Bennett, S.R.M., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8 505–520. [Google Scholar]
- Berleth, T., and Jurgens, G. (1993). The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development 118 575–587. [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inze, D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue, M., Prinsen, E., Onckelen, H.V., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14 603–611. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005. a). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J.S., Jurgens, G., and Estelle, M. (2005. b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131 1101–1110. [DOI] [PubMed] [Google Scholar]
- Fischer-Iglesias, C., Sundberg, B., Neuhaus, G., and Jones, A.M. (2001). Auxin distribution and transport during embryonic pattern formation in wheat. Plant J. 26 115–129. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jurgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T.J., Ulmasov, T., and Hagen, G. (1998). The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 54 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Baurle, I., Kientz, M., and Jurgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jurgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49 387–400. [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. [DOI] [PubMed] [Google Scholar]
- Okada, K., Ueda, J., Komaki, M.K., Bell, C.J., and Shimura, Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven, J.A. (1975). Transport of indoleacetic-acid in plant cells in relation to pH and electrical potential gradients, and its significance for polar IAA transport. New Phytol. 74 163–172. [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6 420–425. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Ribnicky, D.M., Cohen, J.D., Hu, W.S., and Cooke, T.J. (2002). An auxin surge following fertilization in carrots: A mechanism for regulating plant totipotency. Planta 214 505–509. [DOI] [PubMed] [Google Scholar]
- Romano, C.P., Robson, P.R., Smith, H., Estelle, M., and Klee, H. (1995). Transgene-mediated auxin overproduction in Arabidopsis: Hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27 1071–1083. [DOI] [PubMed] [Google Scholar]
- Rubery, P.H., and Sheldrake, A.R. (1974). Carrier-mediated auxin transport. Planta 118 101–121. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Duchene, S., De Oliveira, Y., Caboche, M., Lecharny, A., and Aubourg, S. (2004). FLAGdb++: A database for the functional analysis of the Arabidopsis genome. Nucleic Acids Res. 32 D347–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg, J.J., Myrenas, M., Kuusk, S., Lagercrantz, U., Kowalczyk, M., Sandberg, G., and Sundberg, E. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 47 112–123. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., and Bennett, M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.I., Sharon, M., Zheng, C., Robinson, C.V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Tobena-Santamaria, R., Bliek, M., Ljung, K., Sandberg, G., Mol, J.N., Souer, E., and Koes, R. (2002). FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 16 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Benkova, E., Jager, K.E., Schlereth, A., Hamann, T., Kientz, M., Wilmoth, J.C., Reed, J.W., and Jurgens, G. (2005. b). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Sauer, M., Meurette, O., Friml, J., Ljung, K., Sandberg, G., Hooykaas, P., and Offringa, R. (2005. a). Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17 2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers, D., Schlereth, A., Ehrismann, J.S., Schwank, G., Kientz, M., and Jurgens, G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10 265–270. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Hammes, U.Z., Taylor, C.G., Schachtman, D.P., and Nielsen, E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16 1123–1127. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Christensen, S.K., Fankhauser, C., Cashman, J.R., Cohen, J.D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.