A number of crop species of commercial interest have been transformed using either Agrobacterium-mediated, biolistic, or other systems. However, these methods have several limitations. For example, they allow insertion of single or a few genes at random genomic positions, but complex traits cannot be transferred in a coordinated manner. Furthermore, the integrity of the host genome can be disturbed by transgene insertion. These limitations stimulated the development of a chromosome-based vector system suitable for transferring large genes, gene complexes, and/or multiple genes together with regulatory elements for safe, controlled, and persistent expression, avoiding rearrangements that are often linked with insertion events. Additionally, engineered chromosomes could be used to address questions concerning the function of specific chromosomal domains (e.g., centromeric regions). Chromosome engineering has been applied successfully in mammals and yeasts but has lagged in plants. Recent efforts to generate maize minichromosomes by J. Birchler and colleagues from the University of Missouri (Yu et al., 2006, 2007) open new venues for plant biotechnology and chromosome biology.
TOP-DOWN VERSUS BOTTOM-UP APPROACHES
Considerable progress has been made in developing mammalian chromosome-based vector systems either by engineering endogenous chromosomes (top-down approach) or by artificial composition of cloned chromosomal constituents into functional chromosomes (bottom-up approach). The bottom-up strategy relies on cell-mediated chromosome assembly after transfection of a cell line with cloned centromeric sequences and a selectable marker gene, with or without telomeric and genomic DNA (Harrington et al., 1997; Ikeno et al., 1998). However, the process of de novo chromosome assembly within cells is hard to control and has been achieved only in a limited number of mammalian cell lines (Irvine et al., 2005). For plants, the bottom-up strategy has not yet yielded artificial chromosomes.
Our limited understanding of centromere function and maintenance is one of the obstacles on the way to generating artificial chromosomes. In one study, transformation of rice with Mb-sized centromeric repeat arrays from either maize or rice did not result in stable de novo formation of centromeres (Phan et al., 2007). Also, transformation of the fungus Candida albicans with naked homologous centromeric DNA (85 kb) was unable to recruit the centromeric histone variant CENH3 and to form functionally active centromeric chromatin (Baum et al., 2006). Although plant centromeres can be very large (Jin et al., 2004), more recent data suggest that the size of a functional centromere might be only a few hundred kilobases (Nagaki et al., 2004). It has been shown that barley centromeric repeats are neither necessary nor sufficient to establish a centromere (Nasuda et al., 2005). Rapid inactivation of the second centromere of dicentric maize chromosomes with two sequence identical centromeres (Han et al., 2006) supports the idea that the primary DNA sequence alone does not determine centromere identity. Rather, a specific epigenetic mark seems to be responsible for centromere specification (Vig, 1994; Karpen and Allshire, 1997). Although substitution of the histone H3 by CENH3 in centromeric nucleosomes is crucial for kinetochore formation, we do not know what originally triggers this substitution when a new centromere is initiated, or why and how CENH3 gets lost when a centromere becomes inactive.
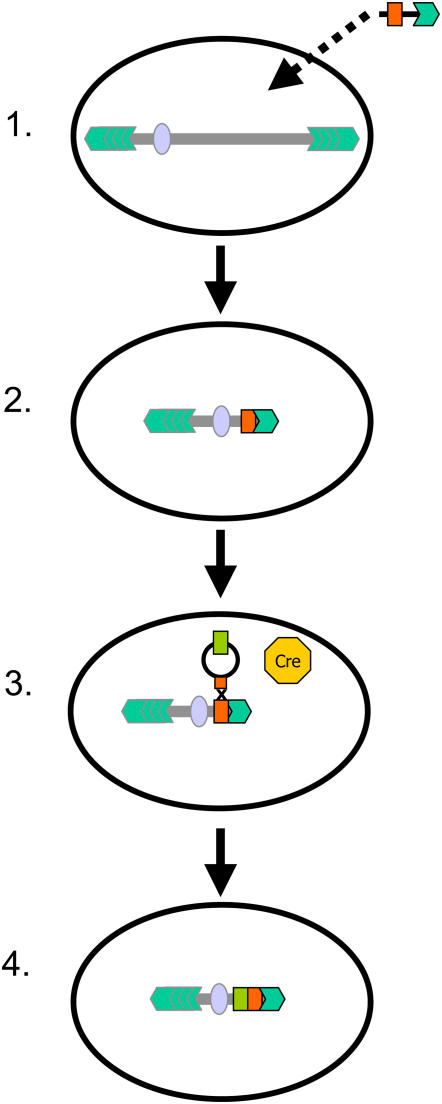
To circumvent the necessity of de novo centromere formation, modification of existing chromosomes to generate a chromosome-based vector can be achieved by at least two different routes. As shown first by Farr et al. (1991), cloned telomeric repeats introduced into cells may truncate the distal portion of a chromosome by the formation of a new telomere at the integration site (Figure 1). This elegant in vivo approach was an important step toward constructing a gene delivery system based on engineered human chromosomes (Lim and Farr, 2004).
Figure 1.
Telomere-Associated Chromosome Fragmentation: A Strategy to Generate Minichromosomes by Truncation of Natural Chromosomes.
A vector containing Arabidopsis-type telomere repeats, a selectable marker, and a homologous recombination site is used to fragment recipient chromosomes (Yu et al., 2006, 2007). The transformation occurs via Agrobacterium-mediated or biolistic transformation of immature embryos prior to selection of cells for antibiotic or herbicide resistance (1 and 2). To add genes onto an engineered minichromosome, targeted transgenes are integrated via site-specific recombinase, as provided by the Cre/lox system, after biolistic transformation (Srivastava and Ow, 2001) of a construct carrying a recombination site that activates a selection marker (3 and 4). The Cre recombinase could be expressed transiently.
A second in vivo approach of engineering endogenous mammalian chromosomes is based on the generation of dicentric chromosomes by amplification of (peri)centromeric satellite DNA and other host sequences, such as rDNA, together with the transgenes after transfection with plasmids (Keresö et al., 1996). Although the generation of a second centromere within transgenic mouse and hamster cell lines seems to be reproducible, the underlying mechanism is not well understood. Nevertheless, the breakage products of such dicentrics can be stabilized by healing the ends via telomere addition. The resulting engineered chromosomes (satellite DNA–based artificial chromosomes) are composed mainly of amplified satellite repeats and ribosomal DNA, interspersed with coamplified transfected foreign DNA (Csonka et al., 2000). A similar process has been associated with the formation of a naturally occurring B chromosome of the plant Plantago lagopus (Dhar et al., 2002). This supernumerary chromosome, which originated from a trisomic A chromosome, underwent a number of structural changes combined with rDNA amplification.
RECENT PROGRESS IN MAIZE
Telomere-mediated chromosome truncation technology recently has been adapted for maize (Figure 2; Yu et al., 2006, 2007). An array of 2.6 kb of Arabidopsis-type telomeric repeats was used for Agrobacterium-mediated or biolistic transformation of immature maize embryos. Because sequence targeting by homologous recombination is inefficient in seed plants (Reiss, 2003), a targeted introgression of cloned telomeres as used in mammals (Lim and Farr, 2004) was not yet possible. Therefore, the sites of chromosomal truncations are random, and in situ hybridization with chromosome-specific marker sequences is required for identifying the origin of the truncated chromosomes.
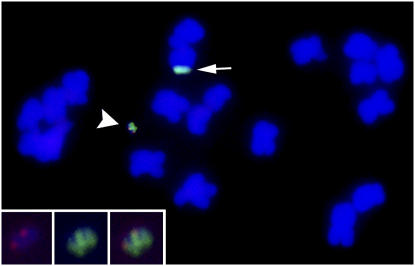
Figure 2.
A Chromosome Spread of Maize Is Shown with a Minichromosome Produced by Truncating a B Chromosome of Maize.
The truncated B chromosome has a specific centromeric DNA sequence that is labeled in green. A normal B chromosome is shown for comparison (arrow). The arrowhead denotes the minichromosome produced by telomere-mediated truncation. The addition of telomeres to the chromosome by genetic transformation fractures the chromosome at that site and leaves the added genes (red) at the tip of the chromosome. The insets (enlarged below) show the added genes in red, the specific centromere region of the minichromosome in green, and the merged image. The chromosomes are stained blue. Modified from Yu et al. (2007). ©2007 National Academy of Sciences, USA.
The presence of telomeric sequences at the double-stranded ends of the transgene insertion sites might be considered as telomere seeding (Yu et al., 2006). The transgenic telomeric sequences might recruit telomerase and telomere binding proteins, thus mediating chromosome healing rather than nonhomologous end joining (Yu et al., 2006). The frequency of chromosomal truncation is low in plants compared with mammalian cells, but testing different constructs and genetic backgrounds might result in an improved truncation efficiency. Because the truncation efficiency of mammalian chromosomes is highest in a hyperrecombinogenic chicken cell line (Buerstedde and Takeda, 1991), plant accessions or mutants with a high somatic recombination frequency might be preferable host organisms.
The lack of meiotic pairing of the derived maize minichromosome with its progenitors suggests that such small chromosomes have a minimal chance of recombination with the normal chromosome and therefore can be used as starting material for plant-engineered chromosomes. The generation of even smaller minichromosomes might be prevented by the requirement of a minimal chromosome size for stable inheritance. While too large chromosomes (with arms longer than half of the average spindle axis) often are not correctly transmitted through mitotic division (Schubert and Oud, 1997), the bottleneck for transmission of too small chromosomes seems to be meiosis. Lower limits of chromosome size for correct transmission seem to vary between species, from ∼50 kb in yeast to <5% of genome size in field bean (Schubert, 2001), while microchromosomes of birds show stable inheritance (Burt, 2002). At present, we do not know whether the lack of meiotic pairing and recombination, the absence of a lateral support for centromeres to maintain centromeric cohesion, or other reasons are responsible for impaired transmission of very small chromosomes.
A VERSUS B CHROMOSOMES
B chromosomes, the often neglected components of the karyotypes of numerous plant and animal species, could become a major player for the generation of engineered chromosomes because of their unique features. B chromosomes are dispensable and do not pair with any of the standard A chromosomes at meiosis by definition. They have irregular modes of inheritance (Jones and Houben, 2003). With respect to the possible use of mini-B chromosomes as a vector, it is important that B chromosomes have little effect on an individual's phenotype. Only in the case of a high number, B chromosomes can reduce vigor (Puertas, 2002). Thus, engineered mini-Bs allow the study of gene dosage effects. Except for the B chromosome–located 45S rRNA gene of Crepis capillaris (Leach et al., 2005) and for transcriptionally active B-specific repetitive sequences (Carchilan et al., 2007; Lamb et al., 2007), there has been no direct molecular evidence for transcription of genes from plant Bs.
The survival rate after telomere-associated truncation was higher for B than for A chromosomes (Yu et al., 2007), most likely because most Bs are genetically inert. Constitutive transgene expression from A and B chromosome–derived minichromosomes suggests that inactivation of genes on B chromosomes, if it occurs, it is at least not a rapid process. It is of interest to compare transgene expression on A and B chromosomes over several generations.
To achieve viability of plants with an A chromosome–derived minichromosome, the truncation event should take place in a polyploid or (for the target chromosome) aneuploid background. Maize A-derived minichromosomes were faithfully transmitted from one generation to the next, whereas the meiotic transmission rate of B-type minichromosomes varied from 12 to 39% via the male parent (Yu et al., 2007), comparable to the transmission rate of mini-B chromosomes generated by breakage-fusion-bridge cycles (Kato et al., 2005). However, due to the intrinsic postmeiotic drive of intact B chromosomes (which includes specific nondisjunction at pollen mitosis and preferential fertilization of the egg by the sperm containing the B chromosomes; Carlson, 1978) a B chromosome–derived vector might potentially reveal an increase of transmission frequency above Mendelian expectation. The truncation of the B eliminates nondisjunction of the mini-B and confers inheritance characteristics equal to any normal A chromosome. However, the nondisjunction process can be restored by the addition of normal B chromosomes to the genotype, which would supply the required trans-acting factor for nondisjunction that is located at the very tip of the chromosome. Early studies indicated that the drive mechanisms for B chromosomes are independent of the background genotype and mitotic nondisjunction is controlled by the B itself (Müntzing, 1970).
Since most of the engineered mammalian chromosomes have been generated and maintained in cell culture, our knowledge on their meiotic transmissibility is rather limited. Studies in transchromosomal animals and nonhuman mammalian tissues suggest a high variability as to the stability of engineered human chromosomes between tissue types and between genetic backgrounds. The meiotic transmission is clearly below that of endogenous chromosomes (Irvine et al., 2005). It remains to be tested whether microinjection or microcell-mediated transfer will broaden the application of engineered chromosomes in cases where the transfer of plant minichromosomes via sexual crossing is not feasible.
THE NEXT STEP: MAKING USE OF ENGINEERED PLANT MINICHROMOSOMES
How are genes of interest introduced onto engineered minichromosomes? Targeted transgene integration into unique chromosomal loci might be achieved using gene constructs in combination with a site-specific recombinase cassette as provided by the Cre/lox system. The proof of principle has been demonstrated by Yu et al. (2007). The engineered maize chromosome R2 provides a defined recipient locus for site-specific integration of transgenes, enabling genetic manipulation via site-specific recombination. After crossing a transgenic plant supplying the Cre recombinase and a lox recombination site at the terminus of chromosome 3 with a plant carrying the minichromosome R2 with a defined recipient site, composition of two complementary parts at the recipient locus led to activation of the DsRed reporter gene in some tissues, indicating reciprocal translocation between lox sites. Although the recombination efficiency is not as high as in many nonplant organisms, this approach offers a possible way of adding transgenes to a minichromosome. Ayabe et al. (2005) used an engineered human chromosome to introduce the native human HPRT gene together with ∼40 kb upstream and ∼10 kb downstream regions that include all regulatory elements needed for correct expression in Hprt-deficient cells and demonstrated functional complementation. To harness targeted gene loading onto the engineered chromosome, an elegant combination of transformation-associated recombination cloning of the gene-containing genomic fragment into a YAC vector (Kouprina et al., 1998) and Cre/loxP-based site-specific recombination of the circular YAC-DNA into the recipient loxP site of the engineered human minichromosome has been developed (Ayabe et al., 2005) and might be adapted for plants as well.
In summary, the future for engineered plant chromosomes as a fascinating new tool for basic research on chromosomes and biotechnology is promising. The initial demonstration of their construction and behavior provides the foundation for this technology in plants, onto which further developments can be built. However, an increase of truncation efficiency, the introduction of multiple site-specific recombination systems, and a demonstration of full meiotic transmissibility of site-specific recombination products are required before engineered plant chromosomes will enter commercial application.
Acknowledgments
We thank James Birchler and colleagues (University of Missouri) for providing the image of the maize minichromosome and for comments on the manuscript.
References
- Ayabe, F., Katoh, M., Inoue, T., Kouprina, N., Larionov, V., and Oshimura, M. (2005). A novel expression system for genomic DNA loci using a human artificial chromosome vector with transformation-associated recombination cloning. J. Hum. Genet. 50 592–599. [DOI] [PubMed] [Google Scholar]
- Baum, M., Sanyal, K., Mishra, P.K., Thaler, N., and Carbon, J. (2006). Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc. Natl. Acad. Sci. USA 103 14877–14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde, J.M., and Takeda, S. (1991). Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67 179–188. [DOI] [PubMed] [Google Scholar]
- Burt, D.W. (2002). Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96 97–112. [DOI] [PubMed] [Google Scholar]
- Carchilan, M., Delgado, M., Ribeiro, T., Costa-Nunes, P., Caperta, A., Morais-Cecílio, L., Jones, R.N., Viegas, W., and Houben, A. (2007). Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell 19 1738–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, W.R. (1978). B-chromosome of corn. Annu. Rev. Genet. 12 5–23. [DOI] [PubMed] [Google Scholar]
- Csonka, E., Cserpan, I., Fodor, K., Hollo, G., Katona, R., Keresö, J., Praznovszky, T., Szakal, B., Telenius, A., deJong, G., Udvardy, A., and Hadlaczky, G. (2000). Novel generation of human satellite DNA-based artificial chromosomes in mammalian cells. J. Cell Sci. 113 3207–3216. [DOI] [PubMed] [Google Scholar]
- Dhar, M.K., Friebe, B., Koul, A.K., and Gill, B.S. (2002). Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma 111 332–340. [DOI] [PubMed] [Google Scholar]
- Farr, C., Fantes, J., Goodfellow, P., and Cooke, H. (1991). Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 88 7006–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, F., Lamb, J.C., and Birchler, J.A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 103 3238–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, J.J., Van Bokkelen, G., Mays, R.W., Gustashaw, K., and Willard, H.F. (1997). Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15 345–355. [DOI] [PubMed] [Google Scholar]
- Ikeno, M., Grimes, B., Okazaki, T., Nakano, M., Saitoh, K., Hoshino, H., McGill, N.I., Cooke, H., and Masumoto, H. (1998). Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16 431–439. [DOI] [PubMed] [Google Scholar]
- Irvine, D.V., Shaw, M.L., Choo, K.H., and Saffery, R. (2005). Engineering chromosomes for delivery of therapeutic genes. Trends Biotechnol. 23 575–583. [DOI] [PubMed] [Google Scholar]
- Jin, W., Melo, J.R., Nagaki, K., Talbert, P., Henikoff, S., Dawe, R.K., and Jiang, J. (2004). Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, N., and Houben, A. (2003). B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 8 417–423. [DOI] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13 489–496. [DOI] [PubMed] [Google Scholar]
- Kato, A., Zheng, Y.Z., Auger, D.L., Phelps-Durr, T., Bauer, M.J., Lamb, J.C., and Birchler, J.A. (2005). Minichromosomes derived from the B chromosome of maize. Cytogenet. Genome Res. 109 156–165. [DOI] [PubMed] [Google Scholar]
- Keresö, J., et al. (1996). De novo chromosome formations by large-scale amplification of the centromeric region of mouse chromosomes. Chromosome Res. 4 226–239. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., Annab, L., Graves, J., Afshari, C., Barrett, J.C., Resnick, M.A., and Larionov, V. (1998). Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3′ end target sequence. Proc. Natl. Acad. Sci. USA 95 4469–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.C., Riddle, N.C., Cheng, Y.M., Theuri, J., and Birchler, J.A. (2007). Localization and transcription of a retrotransposon-derived element on the maize B chromosome. Chromosome Res. 15 383–398. [DOI] [PubMed] [Google Scholar]
- Leach, C.R., Houben, A., Field, B., Pistrick, K., Demidov, D., and Timmis, J.N. (2005). Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris. Genetics 171 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, H.N., and Farr, C.J. (2004). Chromosome-based vectors for Mammalian cells: An overview. Methods Mol. Biol. 240 167–186. [DOI] [PubMed] [Google Scholar]
- Müntzing, A. (1970). Chromosomal variation in the Lindström strain of wheat carrying accessory chromosomes in rye. Hereditas 66 279–286. [Google Scholar]
- Nagaki, K., Cheng, Z.K., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J.M. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Nasuda, S., Hudakova, S., Schubert, I., Houben, A., and Endo, T.R. (2005). Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, B.H., Jin, W., Topp, C.N., Zhong, C.X., Jiang, J., Dawe, R.K., and Parrott, W.A. (2007). Transformation of rice with long DNA-segments consisting of random genomic DNA or centromere-specific DNA. Transgenic Res. 16 341–351. [DOI] [PubMed] [Google Scholar]
- Puertas, M.J. (2002). Nature and evolution of B chromosomes in plants: A non-coding but information-rich part of plant genomes. Cytogenet. Genome Res. 96 198–205. [DOI] [PubMed] [Google Scholar]
- Reiss, B. (2003). Homologous recombination and gene targeting in plant cells. Int. Rev. Cytol. 228 85–139. [DOI] [PubMed] [Google Scholar]
- Schubert, I. (2001). Alteration of chromosome numbers by generation of minichromosomes - Is there a lower limit of chromosome size for stable segregation? Cytogenet. Cell Genet. 93 175–181. [DOI] [PubMed] [Google Scholar]
- Schubert, I., and Oud, J.L. (1997). There is an upper limit of chromosome size for normal development of an organism. Cell 88 515–520. [DOI] [PubMed] [Google Scholar]
- Srivastava, V., and Ow, D.W. (2001). Biolistic mediated site-specific integration in rice. Mol. Breed. 8 345–350. [Google Scholar]
- Vig, B.K. (1994). Do specific nucleotide bases constitute the centromere? Mutat. Res. 309 1–10. [DOI] [PubMed] [Google Scholar]
- Yu, W., Han, F., Gao, Z., Vega, J.M., and Birchler, J. (2007). Construction and behavior of engineered mini-chromosomes in maize. Proc. Natl. Acad. Sci. USA 104 8924–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., Lamb, J.C., Han, F., and Birchler, J.A. (2006). Telomere-mediated chromosomal truncation in maize. Proc. Natl. Acad. Sci. USA 103 17331–17336. [DOI] [PMC free article] [PubMed] [Google Scholar]