Abstract
The maize (Zea mays) floury1 (fl1) mutant was first reported almost 100 years ago, but its molecular identity has remained unknown. We report the cloning of Fl1, which encodes a novel zein protein body membrane protein with three predicted transmembrane domains and a C-terminal plant-specific domain of unknown function (DUF593). In wild-type endosperm, the FL1 protein accumulates at a high level during the period of zein synthesis and protein body development and declines to a low level at kernel maturity. Immunogold labeling showed that FL1 resides in the endoplasmic reticulum surrounding the protein body. Zein protein bodies in fl1 mutants are of normal size, shape, and abundance. However, mutant protein bodies ectopically accumulate 22-kD α-zeins in the γ-zein–rich periphery and center of the core, rather than their normal discrete location in a ring at outer edge of the core. The 19-kD α-zein is uniformly distributed throughout the core in wild-type protein bodies, and this distribution is unaffected in fl1 mutants. Pairwise yeast two-hybrid experiments showed that FL1 DUF593 interacts with the 22-kD α-zein. Results of these studies suggest that FL1 participates in protein body formation by facilitating the localization of 22-kD α-zein and that this is essential for the formation of vitreous endosperm.
INTRODUCTION
The development of a vitreous endosperm at kernel maturity is essential for many important maize (Zea mays) grain characteristics, and there is much evidence to support the hypothesis that zein protein bodies play an important role in this trait. For example, the vitreous portion of the kernel contains much more zein than the soft, starchy interior (Dombrink-Kurtzman and Bietz, 1993), and environmental conditions that reduce zein synthesis, such as nitrogen deprivation, result in kernels that are soft and starchy (Tsai et al., 1978). Perhaps the most compelling evidence for this relationship comes from the molecular characterization of several maize mutants that have a starchy, opaque kernel and are caused by either quantitative or qualitative alterations in zein proteins. For example, the opaque2 (o2) mutant accumulates low levels of α-zeins and has protein bodies that are much smaller than the wild type. O2 encodes a transcription factor that regulates several endosperm-expressed genes, in particular those encoding 22-kD α-zeins (Schmidt et al., 1990). Several other opaque mutants have structural defects in different types of zein proteins. Defective endosperm B30 (De-B30) and floury2 (fl2) have small, misshapen protein bodies (Lending and Larkins, 1992; Kim et al., 2004), resulting from single amino acid substitutions that cause an uncleaved signal peptide in a 19-kD (Kim et al., 2004) and a 22-kD α-zein (Coleman et al., 1997), respectively. The Mucronate (Mc) mutant, which also has misshapen protein bodies, results from an abnormal 16-kD γ-zein. In this case, a 38-bp deletion creates a frame-shift mutation and an abnormal sequence for the last 63 amino acids of the protein (Kim et al., 2006). The defective 16-kD γ-zein in Mc appears to have a reduced interaction with 22-kD α-zeins (Kim et al., 2006), which could disrupt the organization of zeins within the protein body. De-B30, fl2, and Mc result in increased expression of genes associated with the unfolded protein response (UPR) (Hunter et al., 2002), consistent with the expression of a mal-folded, endoplasmic reticulum (ER)–localized protein. o2 and o1 (Nelson et al., 1965) also show an increased UPR, but to a lesser extent than in De-B30, fl2, and Mc (Hunter et al., 2002). The molecular nature of the o1 mutation is unknown, but its phenotype is interesting because it does not reveal obvious differences in zein accumulation. This suggests that factors other than zein structure, possibly the UPR itself, may contribute to the generation of the opaque kernel phenotype.
The discovery that kernels of the o2 mutant have almost twice the Lys content of wild-type maize as a result of the reduced accumulation of zeins and increased synthesis of Lys-containing proteins (Mertz et al., 1964) fueled interest in other opaque mutants, including o1, fl1, and fl2. Fl2, which also has a reduced level of zeins, was also shown to have a substantially increased Lys level (Nelson et al., 1965). Although o1 and fl1 do not manifest an increase in Lys (Nelson et al., 1965), their study has merit because they can provide insight into the factors influencing the development of a vitreous endosperm.
Here, we describe the cloning of the Fl1 gene, for which the mutant was first described almost a century ago (Hayes and East, 1915). Fl1 encodes a transmembrane protein that is located in the protein body ER membrane and appears to be involved with targeting the 22-kD α-zein to a location at the interface between the γ-zein–rich periphery and the core of the protein body.
RESULTS
Identification and Molecular Characterization of the mto222 Mutant
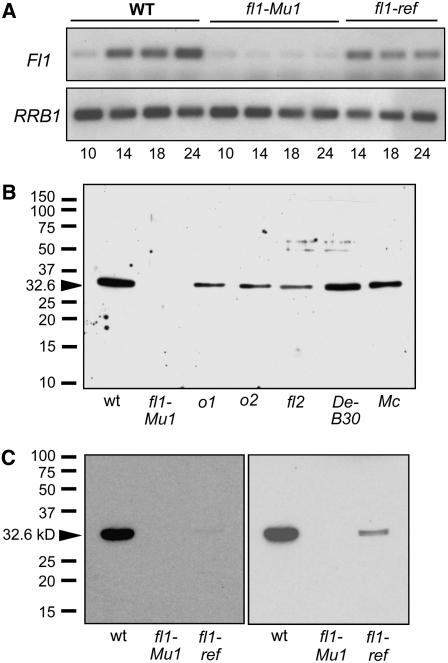
The mto222 mutant was identified during a screen of Pioneer Hi-Bred's Trait Utility System for Corn (TUSC) mutant collection (Benson et al., 1995), while searching for Mutator (Mu)-tagged opaque (mto) mutants. mto222 was introgressed to the BC6 generation in the W64A background at the University of Arizona and B73 at Pioneer Hi-Bred. Mutant kernels of both inbreds have a starchy endosperm at maturity and an opaque appearance when viewed on a light box (Figure 1A). To identify a Mu insertion linked to the mutant phenotype, selective amplification of insertion-flanking fragments (Muszynski et al., 2006) was performed using DNA from 10 independent homozygous mutant and 10 wild-type B73 plants. A perfect cosegregation with the mutant phenotype was observed for a PCR product of 945 bp. This DNA fragment was sequenced and used to design a gene-specific primer in the Mu flanking DNA (see Methods). With this primer and a Mu-TIR primer, it was possible to confirm that the Mu insertion was linked in 38 additional independent mutant plants (22 in the W64A background and 16 in the B73 background). The Mu-flanking sequence matched a 1473-bp cDNA in the Pioneer maize sequence database (GenBank accession number EF536720). This cDNA contained a 906-bp open reading frame (ORF). The Mu insertion site is 4 bp upstream of the start codon of this ORF and 173 bp downstream of the 5′ end of the cDNA (Figure 1C). The cDNA sequence was used to identify corresponding maize genome survey sequences in the Genome Survey Sequences (GSS) section of GenBank, which were subsequently assembled into an extended GSS contig of 5.9 kb. Comparison of the GSS sequence with the cDNA sequence indicated that the gene contains no introns and that the cDNA likely contains the full-length coding sequence for a 302–amino acid protein of 32.6 kD (Figure 1D).
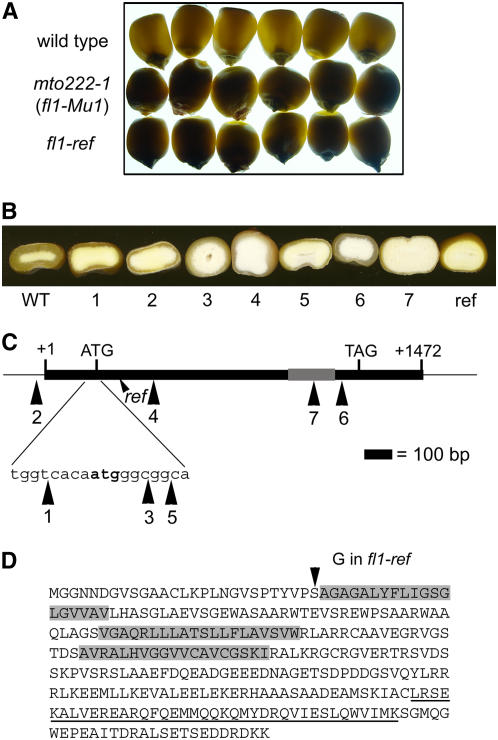
Figure 1.
Phenotype and Gene Structure of the fl1-Mu and fl1-ref Mutations.
(A) Light transmission by mature kernels. Kernels were randomly selected from six ears of W64A+ (top), W64A fl1-Mu1 (mto222-1) (middle), and W64A fl1-ref (bottom) and viewed on a light box.
(B) Kernel phenotypes of the wild type, fl1-Mu1 through fl1-Mu7 (1 to 7), and fl1-ref (ref). Kernel crowns were ground to reveal the thickness of the vitreous endosperm layer.
(C) Diagram of the Fl1 gene. The solid box shows the coding region, and numbered large arrowheads indicate position of Mu insertion alleles. The gray box shows the relative region coding for DUF593. The small arrowhead shows relative position of the fl1-ref point mutation (ref).
(D) FL1 amino acid sequence showing the predicted transmembrane regions (gray boxes) and the conserved DUF593 domain (underlined). The Ser-to-Gly amino acid substitution in fl1-ref is marked with an arrowhead.
To confirm that this mutation is responsible for the opaque kernel phenotype, a reverse genetics screen for additional Mu-tagged alleles was performed. This resulted in the identification of six further mutant alleles of mto222 (Mu2 through Mu7), each of which is predicted to disrupt gene function (Figure 1C). The insertion sites relative to the ATG start codon are as follows: Mu2, −194; Mu3, +7; Mu4, +190; Mu5, +5; Mu6, +849, and Mu7, +764. Although only a small quantity of seed for these mutants was available from the TUSC collection, its preliminary inspection indicated that each one segregated for an opaque kernel phenotype (data not shown). The independent mutant lines were propagated, and the resulting plants were genotyped by PCR using primers to amplify the wild-type (DO84430 and DO84431) and mutant genes (DO84430 and DO9242). Progeny plants of all six mutant lines bore ears that either segregated 3:1 vitreous to opaque, were 100% vitreous, or 100% opaque. Kernel phenotypes of the six additional Mu insertion alleles and the original Mu insertion allele are shown in Figure 1B. Mutant kernels of Mu insertion alleles 1 through 5 all displayed a similar thin layer of vitreous endosperm in comparison with the thick vitreous layer in the wild type. By contrast, mutant kernels of insertion allele 7 (within the plant-specific C-terminal conserved domain [DUF593]; see below) contained little or no vitreous endosperm. Insertion allele 6, which is downstream of the conserved domain, produced a somewhat weaker phenotype with kernels that usually had a slightly thicker layer of vitreous endosperm than alleles 1 through 5. The kernel phenotypes were entirely consistent with the parental genotypes and provided strong additional evidence that an insertional mutation in this gene causes the opaque kernel phenotype. None of the original Mu insertion alleles displayed effects on vegetative or reproductive development compared with near isogenic wild-type plants.
The mto222 and fl1 Mutations Are Allelic
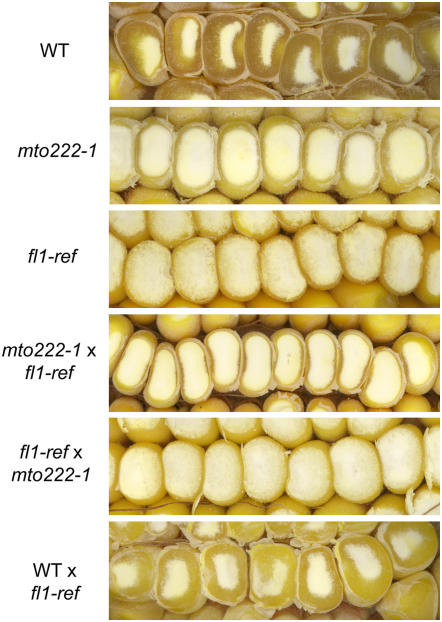
To determine the map location of the Mto222 gene, a maize BAC library array, ZMMBBb (Arizona Genomics Institute, http://www.genome.arizona.edu/orders/), was probed with a radiolabeled Mto222 DNA fragment (see Methods). Four of six strong hybridization signals identified clones b0040A17, b0116K19, b0180M24, and b0226I12, which correspond to a single BAC contig (contig 78). This contig has been mapped to BIN 2.04; thus, the Mto222 gene is also likely near this locus. It was previously reported that the o8 mutation also maps to BIN 2.04, close to marker UMC134 (Graham et al., 1993), and fl1 was shown to be allelic to o8 and o4 (Whalen, 2001). The UMC134 marker is near UMC1580, bnlg1018, bnlg1175, umc2251, and umc2249, all of which map close to the above BAC clones. Given these map locations and the fact that the mto222 and fl1 mutants display similar kernel phenotypes, it seemed possible they were allelic mutations. Fl1 mutant kernels have a similarly reduced light transmission when viewed on a light box (Figure 1A) but have an even thinner layer of vitreous endosperm than mto222 Mu insertional alleles 1 to 5 (Figure 1B). To test for allelism, crosses between homozygous mto222-Mu1 and fl1 mutants were performed. The resultant ears contained 100% opaque kernels (Figure 2), indicating that mto222 and fl1 do not complement one another and are thus allelic. Consequently, the Mto222 gene is subsequently referred to as Fl1. The original fl1 allele is referred to as fl1-ref, the original Mu insertion allele (mto222-1) is referred to as fl1-Mu1, and the additional Mu insertion alleles as fl1-Mu2 through Mu7.
Figure 2.
Kernel Phenotypes Resulting from Allelism Test Crosses between mto222-1 (Subsequently Referred to as fl1-Mu1) and fl1-ref Mutants.
The presence of starchy or vitreous endosperm was shown after removal of kernel crowns by grinding.
To identify the genetic defect in the fl1-ref allele, the gene was sequenced using genomic DNA extracted from five independent W64A fl1-ref homozygous mutant plants and five W64A wild-type plants. This analysis identified an A-to-G point mutation at position +79 relative to the ATG start codon. This changes an AGC codon to GGC, resulting in a Ser-to-Gly substitution at amino acid 27, which is the start of the first of three predicted transmembrane domains (Figure 1D).
Fl1 Encodes a Novel Membrane Protein
BLAST searches using the FL1 amino acid sequence revealed that the protein does not share significant identity with any proteins of known function. To make predictions about FL1 secondary structure and membrane topology, the FL1 amino acid sequence was subjected to a variety of analyses using publicly available bioinformatics packages (see Methods). Most programs, including DAS, Sosui, Split, Predict Protein, and TMPred, predicted the presence of three hydrophobic membrane-spanning domains in the N-terminal half of the protein. Phobius and TMHMM algorithms gave a score for the third transmembrane region that is below the cutoff threshold, and consequently they predict that only the first two transmembrane domains are real. Although there was slight variation in the predicted beginning and end of these domains, TMPred showed that they reside at amino acids 28 to 47, 86 to 105, and 124 to 142 (Figure 1D). Programs that predict overall membrane topology did not give a definitive consensus on which domains are likely to be cytoplasmic and which are noncytoplasmic. For example, TMPred derived models for both possible orientations, whereas PredictProtein suggested that the N terminus is cytoplasmic, intermembrane region one is noncytoplasmic, intermembrane region two is cytoplasmic, and the C terminus is noncytoplasmic. The latter model is supported by the fact that a removable N- terminal signal peptide was not predicted by most of these programs. According to general models for membrane topology, the absence of a signal peptide predicts that the first hydrophobic domain behaves as an internal signal peptide or start transfer sequence, the second acts as a stop transfer sequence, and the third acts as another start transfer sequence, which would direct the remainder of the C terminus out of the cytoplasm. The N terminus would thus be cytoplasmic and the C terminus would be noncytoplasmic. This hypothesis is further supported by the fact that intermembrane region two is richer in positively charged Arg residues (22%) than the region following the third putative transmembrane domain (10%). High concentrations of Arg are frequently observed in the cytosolic domains of integral membrane proteins.
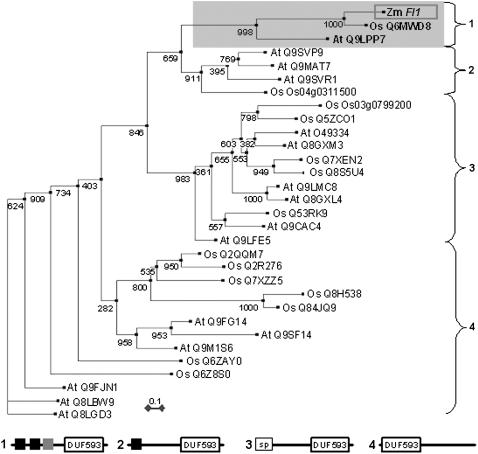
The FL1 C-terminal half contains a plant-specific domain of unknown function (DUF) 593 (pfam04576.5; Figure 1D). The Fl1 gene does not share similarity with genes outside the plant kingdom. Comparisons with other cereal species revealed proteins with a high level of overall amino acid conservation (85% in rice [Oryza sativa]), whereas in Arabidopsis thaliana, the homology is restricted to the DUF593 region (55%). We investigated the relationship between proteins containing the DUF593 domain using phylogenetic tree analysis (Figure 3). This analysis was restricted to proteins from Arabidopsis and rice since their complete genome sequence is available. The tree was constructed by comparing only the DUF593 region, since conservation outside this region was variable and often low. The complete protein sequences of the genes used in the analysis were analyzed for their general domain architecture, which is shown schematically below the tree. In general, proteins with DUF593 domains fall into two classes. The first class has DUF593 near the N terminus, and the second class has DUF593 in the C-terminal half of the polypeptide. Polypeptides of the first group (six Arabidopsis and seven rice) contain no signal peptides or transmembrane regions close to the N terminus. All sequences of the second group contain an ER signal peptide or one or more predicted transmembrane domains at or close to the N terminus (10 Arabidopsis and seven rice). Shown (boxed) at the top of the tree is a subgroup of these proteins that includes FL1, one rice protein, and one Arabidopsis protein. These three proteins contain two high-score transmembrane domains and a third, lower-score one in the N-terminal half of the polypeptide.
Figure 3.
Phylogenetic Tree of DUF593 Domain Proteins.
Maize FL1 and all currently recognized DUF593 domain proteins from Arabidopsis and rice were aligned by ClustalW; the aligned sequences were trimmed to ∼90–amino acid residues covering the DUF593 domain (see Supplemental Table 1 online). The tree was constructed using PHYLIP version 3.6 (see Methods). Distances were estimated with a neighbor-joining algorithm, and bootstrap support is indicated to the left of branches. The accession number for proteins is preceded by the species: Zm (Zea mays), At (Arabidopsis thaliana), or Os (Oryza sativa). In the case of the two rice sequences labeled with the gene name rather than accession number, protein sequences had been incorrectly predicted from the genomic sequence (public databases). In these cases, the protein sequences were curated from available maize cDNA sequences (BLAST searches against Pioneer database, and resolution was confirmed by BLAST comparisons between rice genomic sequence and maize cDNA sequence). The different general structural classes of DUF593 domain proteins are shown on the right and below the tree. FL1 belongs to a small branch with apparent single member orthologous sequences in rice and Arabidopsis (shown boxed) that contain two or three transmembrane sequences in the N-terminal half and the DUF593 domain in the C-terminal half of the protein. In the key, black and gray boxes indicate predicted highly probable and likely transmembrane sequences, respectively; boxed “sp” indicates a predicted ER signal sequence.
Mutations in Fl1 Do Not Result in Reduced Accumulation of Zeins or Constitutive UPR
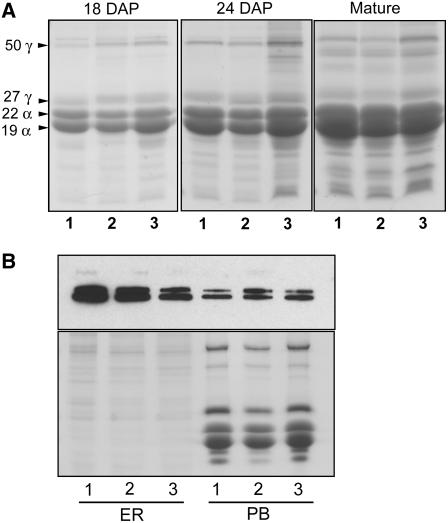
We investigated the possibility that the opaque kernel phenotype of the fl1-ref and fl1-Mu1 mutants is accompanied by differences in zein accumulation, as was observed with other opaque mutants. As shown in Figure 4A, SDS-PAGE revealed no obvious differences in the abundance of the major α- and γ-zein proteins in the wild type, fl1-Mu1, and fl1-ref during endosperm development or at maturity. Likewise, fl1-Mu2 through fl1-Mu7 also appeared to be similar to the wild type in zein accumulation (data not shown). A quantitative comparison of the accumulation of endosperm proteins is shown in Table 1, which reveals no evidence for a reduction in total zein protein in the mutants. Surprisingly, both alleles resulted in slightly elevated zein levels, while the amounts of non-zein proteins were not markedly affected (Table 1).
Figure 4.
Comparison of Zein and Bip Accumulation in fl1-Mu1 and fl1-ref Endosperm.
(A) SDS-PAGE analysis of the zein fraction from 18 and 24 DAP and mature endosperm of W64A+, W64A fl1-Mu1, and W64A fl1-ref (lanes 1 to 3, respectively). To compare protein accumulation between fresh, developing, and mature dry kernels, each lane was loaded with 1/200 of the extract from one whole endosperm.
(B) Immunoblot comparing Bip accumulation in ER and protein body fractions of W64A+, W64A fl1-Mu1, and W64A fl1-ref (lanes 1 to 3, respectively). Extract was loaded from ∼0.2 mg (fresh weight) of endosperm. Coomassie blue–stained gel compares sample loading (10-fold overloaded compared with immunoblot). ER, endoplasmic reticulum fraction; PB, protein body fraction.
Table 1.
Protein Content of Wild-Type and fl1 Mature Endosperm
| W64A Wild Type | W64A fl1-Mu1 | W64A fl1-ref | |
|---|---|---|---|
| Zeins | 60.7 ± 4.4 | 76.2 ± 10.6 | 68.3 ± 4.2 |
| Nonzeins | 19.6 ± 3.6 | 18.6 ± 2.7 | 16.7 ± 2.8 |
Values shown are milligrams of protein per gram of endosperm dry weight ± sd.
Another common feature of opaque mutants is increased expression of a variety of chaperone genes, indicative of a constitutive UPR (Hunter et al., 2002). For several of these mutants, zein defects resulting in aberrant protein body structure are believed to be the primary cause of the opaque kernel phenotype. However, the o1 mutant appears to have wild-type zeins and an elevated UPR, thus raising the possibility that the UPR itself is capable of causing an opaque kernel phenotype. To address this question for fl1 mutants, we used immunoblot analysis to compare the level of the ER lumen binding protein (Bip) in endosperm ER and protein body ER fractions of W64A+, W64A fl1-Mu1, and W64A f11-ref. Figure 4B shows that Bip levels are not markedly affected in either fl1-Mu1 or fl1-ref, which suggests that these mutations do not stimulate the UPR. Mutants such as fl2, DeB30, and Mc show elevated Bip accumulation in both ER and protein body fractions, though this is most pronounced in the protein body fraction (data not shown; Zhang and Boston, 1992).
Fl1 Transcript and Protein Accumulation in fl1-Mu1 and fl1-ref
Using RNA gel blot analysis of total RNA, a low-abundance transcript of ∼1500 bp (cDNA = 1473 bp) was detectible in wild-type endosperm but not detectible in endosperms of Mu insertion mutants (data not shown). Semiquantitative RT-PCR was used to compare Fl1 transcript levels at different stages of endosperm development relative to the RRB1 gene (Grafi et al., 1996) (Figure 5A). The Fl1 RNA increased between 10 and 24 d after pollination (DAP) in wild-type endosperm, while only a very low level of a transcript was detected in fl1-Mu1 endosperm. In fl1-ref, the transcript had only slightly lower levels than the wild type. A point mutation such as this would not be expected to result in large changes in transcript abundance.
Figure 5.
Fl1 Gene Expression and Protein Accumulation in Opaque Mutants.
(A) Semiquantitative RT-PCR analysis of Fl1 transcript in W64A+, W64A fl1-Mu1, and W64Afl1-ref endosperm. Numbers refer to DAP. Bottom panel shows expression of the RRb1 gene as a control.
(B) Immunoblot comparing accumulation of FL1 protein in endosperms of various opaque mutants.
(C) Immunoblot comparing accumulation of FL1 protein in endosperms of W64A+, W64A fl1-Mu1, and W64A fl1-ref. The left panel shows a standard 2-min exposure following the chemiluminescent detection reaction, whereas the right panel shows the same blot overexposed for 30 min. In (B) and (C), each lane was loaded with non-zein protein extracts from 5 mg (fresh weight) of endosperm; arrowheads mark the predicted size of FL1 (32.6 kD).
Using affinity-purified FL1 antibody, we determined that a single protein with an expected molecular weight of 32.6 kD is present in developing wild-type endosperm, whereas this protein was not detectable in fl1-Mu1 endosperm (Figures 5B and 5C). Very low levels of the protein were detected in fl1-ref endosperm (Figure 5C). This result is consistent with the reported semidominant nature of fl1-ref (Whalen, 2001), which would be predicted to accumulate a protein that causes a dominant-negative phenotype. Immunoblot analysis revealed that the FL1 protein is present in developing endosperms of several of the previously described opaque mutants (Figure 5B). The abundance of FL1 in o1, o2, and fl2 is slightly lower than in the wild type. This may result indirectly from differences in the shape and composition of protein bodies in these mutants. Although protein bodies have not been studied at the ultrastructural level in o1, o2 possesses smaller protein bodies than the wild type, whereas fl2 has irregularly shaped protein bodies.
FL1 Localizes in Protein Body ER Membranes during Endosperm Development
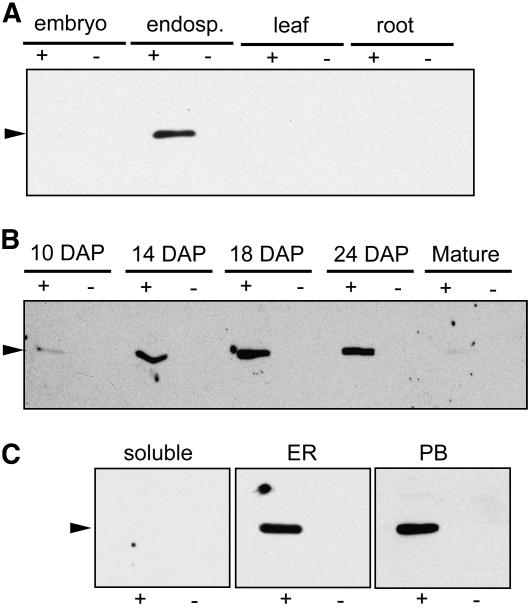
We analyzed the tissue specificity of FL1 by immunoblotting protein extracts from a variety of maize organs. In kernels, FL1 was present in the endosperm but not the embryo (Figure 6A), and FL1 could not be detected in extracts from leaf and root tissues. In endosperm, FL1 was first detectible at 10 DAP and sharply increased in abundance, peaking between 18 and 25 DAP (Figure 6B). Thus, the accumulation of FL1 largely parallels protein body formation (Lending and Larkins, 1989) and zein synthesis (Woo et al., 2001). Only a very small amount of FL1 protein was detectible in mature, dry endosperm (Figure 6B).
Figure 6.
Spatial and Temporal Expression of the FL1 Protein.
(A) Immunoblot showing the presence of FL1 in total protein extracts from various maize tissues (200 μg of protein per lane).
(B) Immunoblot showing time course of FL1 protein accumulation. To compare FL1 levels between fresh, developing, and dry, mature kernels, each lane was loaded with 1/200 of the extract from one whole endosperm.
(C) Immunoblots showing FL1 accumulation in endosperm subcellular fractions: soluble, ER membrane fraction (ER), and protein body membrane fraction (PB). Extract from 2 mg of fresh weight endosperm was loaded.
In (A) to (C), the + and – refer to W64A+ and W64A fl1-Mu1, respectively, and arrowheads represent the predicted size of FL1 (32.6 kD).
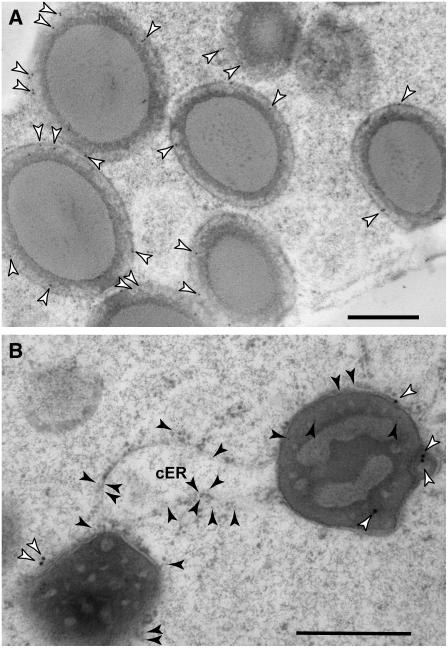
To investigate the subcellular location of the FL1 protein, endosperm cell fractions were isolated by discontinuous sucrose gradient centrifugation (Habben et al., 1993). Immunoblot analysis showed that while no FL1 protein was detected in the soluble fraction, it was present in both the ER and protein body membrane fractions (Figure 6C). Immunolocalization studies showed that FL1 is distributed in the protein body periphery, usually in close proximity to the membrane, but it was not observed in cisternal ER membranes (Figure 7). No labeling was detected in sections probed with the preimmune control (see Supplemental Figure 1 online). Double labeling with an endosperm-specific tonoplast intrinsic protein (zmTIP3-4) antibody that specifically recognizes ER and the FL1 antibody confirmed that FL1 is confined to the protein body ER (Figure 7B). In view of the developmental timescale for FL1 accumulation, this is suggestive of a function in protein body development. The presence of FL1 in the ER fraction can be explained by the fact that small, immature protein bodies are not sufficiently dense to sediment with mature protein bodies and instead partition into the ER fraction. Fragmented protein body ER membranes may also form small vesicles that partition into the ER fraction.
Figure 7.
Localization of FL1 in the Second Subaleurone Layer Endosperm Cells (22 DAP).
(A) Immunolocalization of FL1 in wild-type high-pressure frozen/freeze-substituted endosperm samples.
(B) Double labeling of FL1 (15-nm gold particles) and the ER protein zmTIP3-4 (5-nm gold particles).
The white arrowheads indicate FL1 gold labeling on the protein body ER membrane, whereas the black arrowheads point out the zmTIP3-4 labeling on both protein body ER and cisternal ER (cER). Note that no FL1 labeling was detected on cisternal ER membranes. Bars = 500 nm.
fl1 Protein Bodies Display Abnormal 22-kD α-Zein Localization
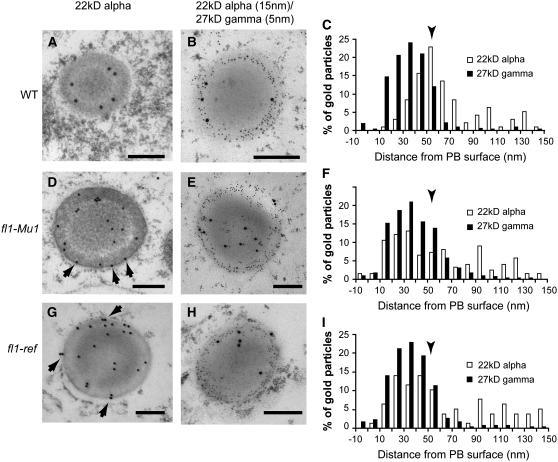
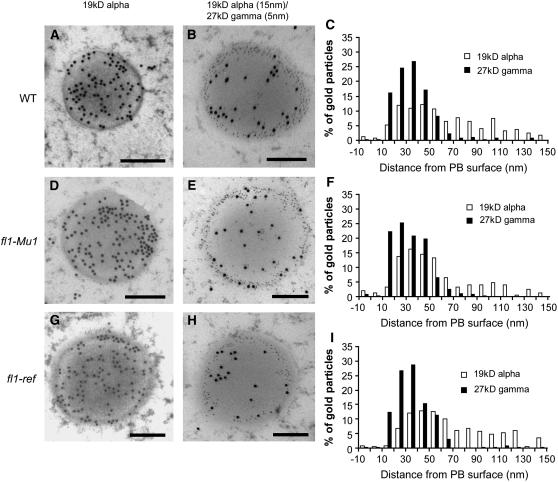
The distribution of zeins within protein bodies was investigated to determine if protein body structure is affected in fl1 mutants. Previous studies using an antiserum raised against total α-zein showed that these proteins are localized throughout the protein body core but are excluded from the peripheral, γ-zein–rich region (Lending and Larkins, 1989). In this study, the use of antibodies specifically raised against 22- and 19-kD α-zeins showed that these proteins have distinct distributions in 18 DAP, wild-type protein bodies. The 22-kD α-zein was detected in a ring at the interface between the γ-zein layer and the core and was not detected close to the ER membrane or in the center of the protein body core (Figures 8A to 8C). However, the 19-kD α-zein was dispersed more generally throughout the core and was mainly, but not entirely, excluded from the γ-zein–rich peripheral region (Figures 9A to 9C).
Figure 8.
Immunolocalization of 22-kD α-Zein and 27-kD γ-Zein in Wild-Type, fl1-Mu1, and fl1-ref Mutant Endosperm Cells (22 DAP).
The double labeling of the 22-kD α- and 27-kD γ-zeins was performed using anti-22-kD α-zein polyclonal antibodies raised in rabbit and anti-27-kD γ-zein polyclonal antibodies raised in chicken. For gold particle quantification, the distance from the protein body surface to the center of the gold particle was measured using ImageJ. Arrows indicate labeling on or very close to the protein body membrane. Arrowheads in (C), (F), and (I) indicate the area of maximal 22-kD α-zein signal in wild-type protein bodies. 103 gold particles in 41 protein bodies were counted in the labeled wild-type samples, 104 gold particles in 30 protein bodies in the fl1-Mu1 samples, and 78 gold particles in 28 protein bodies in the fl1-ref samples. Bars = 200 nm.
Figure 9.
Immunolocalization of 19-kD α-Zein and 27-kD γ-Zein in Wild-Type, fl1-Mu1, and fl1-ref Mutant Endosperm Cells (18 DAP).
The double labeling of the 19-kD α- and 27-kD γ-zeins was performed using anti-19-kD α-zein polyclonal antibodies raised in rabbit and anti-27-kD γ-zein polyclonal antibodies raised in chicken. For gold particle quantification, the distance from the protein body surface to the center of the gold particle was measured using ImageJ. 273 gold particles in 12 protein bodies were counted in the labeled wild-type samples, 263 gold particles in 24 protein bodies were counted in the fl1-Mu1 samples, and 260 gold particles in 20 protein bodies were counted in the fl1-ref samples. Bars = 200 nm.
In fl1-Mu1 and fl1-ref protein bodies, 22-kD α-zein showed an altered distribution. Whereas 27-kD γ-zein localization in the outer shell was not affected, 22-kD α-zein was uniformly distributed throughout the protein body, occurring in the central core (Figures 8D to 8I). In addition, fl1 mutant protein bodies accumulated 22-kD α-zein in close proximity to the ER membrane in the region occupied by γ-zein (Figures 8D to 8I). Quantitation of gold particle distribution showed that in wild-type protein bodies, there is a peak of 22-kD α-zein abundance ∼50 nm from the membrane (arrowhead in Figure 8C). However, in fl1 mutants, this peak was not observed (arrowheads in Figure 8F and 8I), and the majority of gold particles were distributed between 10 and 50 nm from the membrane. We did not observe differences in the distribution of 19-kD α-zein between the wild type and the fl1mutants (Figures 9D to 9I).
The C-Terminal Conserved Domain of FL1 Interacts with α-Zeins in a Yeast Two-Hybrid Assay
The yeast two-hybrid assay was used to search for putative FL1-interacting proteins. Four different regions of the FL1 ORF were used to create in-frame translational fusions with the GAL4 DNA binding domain in pGBKT7 (BD Biosciences). These were as follows: (1) the full-length ORF; (2) the region encoding the N-terminal half of the protein, including the hydrophobic membrane-spanning regions; (3) the region encoding the C-terminal half of the protein, including DUF593; and (4) a short fragment encoding just the conserved domain (DUF593). These were used in mating experiments with a 10, 14, 18, and 24 DAP mixed stage, endosperm yeast-two hybrid library. Baits 1, 2, and 3 exhibited slight cytotoxicity in yeast strain Y187, resulting in inadequate mating efficiencies. However, experiments with bait 4 (the conserved domain) produced optimal mating efficiencies and resulted in the identification of a variety of interacting proteins, including α-zeins.
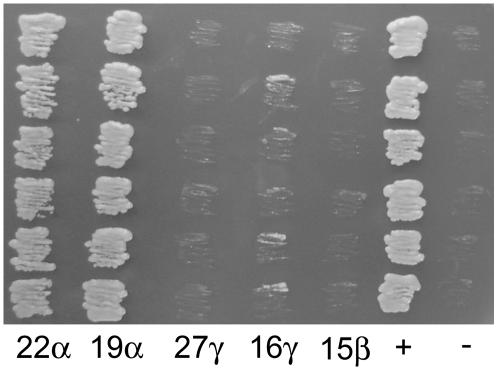
Since FL1 is associated with the surface of zein protein bodies, we examined FL1–zein interactions in more detail. Pairwise mating experiments between all four FL1 baits in pGBKT7 and zein preys in pGADT7 were conducted. In these experiments, diploid colonies were first selected on -Leu/-Trp dropout media and then screened for interaction on quadruple dropout media. This media selects for only those diploid clones in which reporter genes for the synthesis of adenine and His are activated by a functional GAL4 transcription factor. Again, baits 1 to 3 exhibited poor mating efficiencies indicated by low numbers of diploid clones capable of growth on -Leu/-Trp dropout media, and bait 4 produced optimal mating efficiencies with all the zein preys tested. In pairwise mating, 100% of diploid colonies should grow on stringent quadruple dropout media if the bait and prey proteins produce a functional interaction. This was true for both a 22-kD α-zein and a 19-kD α-zein, both of which exhibited similar numbers of vigorously growing colonies on -Leu/-Trp media and quadruple dropout media. One hundred percent of diploid colonies exhibited vigorous growth when restreaked from -Leu/-Trp media onto quadruple dropout media, indicating a functional interaction (Figure 10). By contrast, while matings between bait 4 and the 27-kD γ-zein, 16-kD γ-zein, and 15-kD β-zein prey constructs resulted in vigorous growth of diploid colonies on -Leu/-Trp media, little or no growth was observed on quadruple dropout medium, suggesting no significant functional interactions (Figure 10).
Figure 10.
Yeast Two-Hybrid Interactions between FL1 and Different Types of Zeins.
Six representative colonies of pGADT7-zein × pGBKT7-DUF593 diploid strains were streaked on -Leu/-Trp/-Ade/-His dropout media. Control two-hybrid matings were performed as suggested in the BD Matchmaker library construction and screening manual. Diploid colonies from the positive interaction control mating (+) (pGADT7-RecT × pGBKT7-53) and from the noninteraction control mating (-) (pGADT7-RecT × pGBKT7-Lam) were also streaked.
DISCUSSION
Using a series of Mu insertion mutants, we identified and characterized the gene responsible for the starchy endosperm phenotype of fl1, the earliest described maize opaque mutant. FL1 protein is not detectible in the fl1-Mu1 mutant endosperm, and this mutation results in the formation of only a thin layer of vitreous endosperm. Other Mu insertion alleles in this region of the Fl1 gene result in kernels with similar phenotypes. This suggests that vitreous endosperm formation is partially dependent on FL1 function.
The fl1-ref mutation is semidominant. The likely cause of this is the accumulation of a small amount of a full-length protein that bears a single amino acid substitution in close proximity to the first transmembrane domain. There is no absolute consensus between the various prediction programs for the first amino acid of this domain. As a result, the amino acid substitution could be either immediately adjacent to or within the transmembrane domain. In either case, an alteration in secondary structure could disrupt FL1 membrane localization. A failure of FL1 to anchor to the ER membrane would be expected to result in its entrance into the ER lumen and its eventual degradation. However, the transient presence of FL1 protein, perhaps in an inappropriate ER luminal location, could result in a dominant-negative effect that increases the apparent severity of the phenotype. The fl1-Mu7 allele also has a more severe phenotype than the other Mu insertion alleles. This could be explained by the fact that the Mu element is inserted into DUF593, and our data suggest that this region specifically interacts with 22-kD α-zein. It is possible that the fl1-Mu7 insertion allele also has the potential to produce a protein with a dominant-negative function.
The specific location of FL1 in the protein body membrane is suggestive of a function at some stage during the formation of zein protein bodies. However, our data do not support a role in protein body initiation. First, FL1 only begins to accumulate at 10 DAP, by which time the 27-kD γ-zein is abundant (Woo et al., 2001). Second, protein body number and distribution appear to be unaffected in fl1-Mu1 and fl1-ref. Third, our data indicate that FL1 is not an abundant protein, as might be expected if it were a structural protein involved in protein body nucleation.
Could FL1 play a role in recruiting zein polyribosomes, aiding in their translation or the association of the signal peptide with a signal recognition particle? Our data do not support this hypothesis either, because the levels of zein proteins in fl1 are not reduced and the protein body size and shape are normal. Also, several factors suggest that FL1 functions on the luminal side of the membrane, although this is not proven. A removable N-terminal signal peptide was not predicted in FL1 by most of the available topology programs. According to general models for membrane protein structure, this implies that the C-terminal domain is luminal. This hypothesis is also supported by the fact that intermembrane region two is rich in positively charged Arg residues, which is characteristic of cytosolic domains.
Results from yeast two-hybrid assays involving FL1 and zein proteins also support a luminal location for the C-terminal domain of FL1. Although FL1 bait constructs coding for the entire C-terminal half or the full-length protein, including the transmembrane domains, did not result in adequate mating efficiencies and so did not allow us to detect protein interactions, a strong interaction was demonstrated between the FL1 C-terminal conserved domain and α-zeins. For such interactions to occur, the FL1 C terminus would have to be in the lumen. Furthermore, such interactions would be predicted to be transient, since FL1 is in the protein body membrane and α-zeins are transported through the γ-zein layer and away from the membrane. Also, FL1 has a low abundance compared with α-zeins. Together, these factors imply that other, unidentified factors are likely involved in α-zein targeting. Perhaps FL1 promotes the formation of a complex with chaperone-type molecules, the identity of which could be revealed by the characterization of additional opaque mutants, work that is underway in our lab.
Due to the inherent insolubility of both FL1 and α-zein proteins, it was not possible to obtain suitable endosperm extracts to confirm their interaction using techniques such as coimmunoprecipitation and glutathione S-transferase (GST) pull-down analysis. However, the validity of the interaction between FL1 and 22-kD α-zeins is supported by the results of immunolocalization studies. We found that 22-kD α-zein is distributed throughout the protein body in fl1 mutants, including the γ-zein–rich periphery and ER membrane region and the center of the core. In wild-type protein bodies, the 22-kD α-zein is restricted to a ring at the interface between the γ-zein layer and the core (Figure 8). These results demonstrate that in wild-type protein bodies, the 22-kD α-zeins have a more discrete location than the 19-kD α-zeins, which are found throughout the core and encroach into the γ-zein layer. The FL1 protein may participate in the localization of 22-kD α-zein, such that in its absence, 22-kD α-zein deposition is more random. However, since the distribution of 19-kD α-zeins in f11 is similar to the wild type, FL1 appears not to be necessary for their correct localization. In light of the distinct patterns of 22- and 19-kD α-zein deposition, one could predict differences in their targeting mechanisms. If FL1 is not required for 19-kD α-zein location, this makes their yeast two-hybrid interaction somewhat puzzling. Perhaps their interaction is physiologically relevant but other factors can compensate for the absence of FL1 activity in the mutants.
Our results indicate that DUF593 is central to the function of FL1. Using phylogenetic analysis, we investigated the relationship between FL1 and other proteins that contain DUF593. This domain was found in several plant species, but we used only proteins from rice and Arabidopsis in the analysis due to the completeness of their genomic information. There are at least 16 unique proteins containing this domain in Arabidopsis and at least 14 in rice. The major branches on the tree correspond to different classes of proteins based on predicted structural characteristics, such as the presence or absence of transmembrane domains or signal peptide and the position of DUF593. Each major branch of the tree has representatives from both Arabidopsis and rice, suggesting that these proteins were present in a common ancestor and that members in each branch are probably involved in indispensable functions in the plant cell. FL1 belongs to a small group with a single member each from Arabidopsis and rice that have three transmembrane domains in the N-terminal half and DUF593 in the C-terminal half of the protein. It is possible that the proteins in this group all have a function in ER-derived protein body formation. Experiments that involved the transgenic expression of zeins in tobacco (Nicotiana tabacum) showed that ER-localized protein bodies can form, at least temporarily, in dicotyledons (Bagga et al., 1995, 1997; Coleman et al., 1996, 2004), and it is tempting to speculate that this is also possible in Arabidopsis. The group of proteins that are characterized by the presence of a signal peptide could function in protein scaffolding processes in other parts of the secretory system.
We observed a slight reduction in the abundance of FL1 protein in the endosperms of both the o2 and fl2 mutants (Figure 5B). This could result indirectly from differences in the shape, composition, and structure of protein bodies in these mutants. o2 possesses smaller protein bodies than the wild type, while fl2 has irregularly shaped protein bodies; the extent to which the total protein body surface area is altered is unknown. Any such change might be expected to affect the level of FL1. Alternatively, the lower level of FL1 could reflect the reduction in α-zein accumulation in o2 and fl2 endosperm. If FL1 is required to ensure the correct localization of 22-kD α-zein, its expression could be downregulated, perhaps by a feedback mechanism linked with 22-kD α-zein expression.
Despite progress in understanding the mechanism of zein protein body formation, the nature of the interactions between protein bodies and other cellular components responsible for a vitreous endosperm are unknown. The structure of the vitreous endosperm was compared with a box of white marbles (starch grains) densely interspersed with buckshot (protein bodies) and coalesced by a transparent glue (clear viscous cytoplasm), forming a rigid, glassy conglomerate when dry (Duvick, 1961). Duvick also proposed that in the central, starchy endosperm, smaller protein bodies, larger starch grains, and a reduced proportion of cytoplasm results in a matrix that shatters upon desiccation. In scanning electron micrographs, protein bodies appear to make contact with starch grains (Gibbon et al., 2003), and although the nature of these interactions is unknown, they appear to influence endosperm texture. It is possible that the hydrophilic γ-zeins at the periphery of the protein body are important for generating surface characteristics that will ultimately result in an interaction between protein bodies and starch granules. In the maturing kernel, endosperm cells die as a result of programmed cell death and begin to dehydrate. During this process, ER membranes break down and this would leave γ-zeins exposed at the protein body surface where they could make physical contact with starch granules. Such contact could be central to the formation of vitreous endosperm. In fl1, the ectopic deposition of 22-kD α-zeins at the protein body surface could alter its hydrophobicity/hydrophilicity characteristics, and this could disrupt the interactions with starch grains and affect the formation of vitreous endosperm. Indeed, this could explain the phenotype of other endosperm mutants. For example, in fl2, the signal peptide defect results in accumulation of the preprotein at the surface of the protein body (Gillikin et al., 1997). As a consequence, zein distribution in fl2 protein bodies is altered such that γ-zeins are found in small locules instead of at the periphery, and α-zeins occur at the surface. The De-B30 mutation results in failure to cleave the signal peptide in a 19-kD α-zein, with similar results (Kim et al., 2004). Like fl2, De-B30 shows a constitutive UPR (Kim et al., 2004). The starchy endosperm texture of fl2 and De-B30 could result from altered zein distribution, altered protein body shape, elevated UPR, or a combination of these factors. The opaque kernel phenotype of fl1, with its normal protein body size and shape and wild-type UPR, implies that the organization of 22-kD α-zeins and their possible influence on protein body surface characteristics could be an important factor influencing the formation of vitreous endosperm.
METHODS
Fl1 Cloning
The selected amplification of insertion flanking fragments method was used to clone Fl1. Genomic DNA (∼0.5 μg) from 10 true breeding fl1 Mu-1 and 10 wild-type plants was digested with BfaI in 1× RL buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgOAc, 50 mM KOAc, and 5 mM DTT) at 37°C for 6 h in a final volume of 25 μL. Following denaturation at 80°C for 20 min, 5 μL of ligation mix containing 0.3 μL of 100 mM rATP, 0.5 μL of 10× RL buffer, 1 μL of 40 μM adaptor, 1 μL of Promega T4 ligase (3 units/μL), and 2.2 μL of water was added to each digestion reaction. The BfaI adaptor is a mixture of 5′-TACTCAGGACTCATCGACCGT-3′ and 5′-GTGAACGGTCGATGAGTCCTGAG-3′. After overnight incubation at 4°C, the ligation reaction products were isolated with a Qiagen PCR purification kit to remove excess adaptor.
The Mu-flanking fragments were amplified with the Mu-TIR primer, MuExt22D (5′-CCAACGCCAWSGCCTCYATTTC-3′), and the BfaI adaptor primer, BfaExt18 (5′-GTGAACGGTCGATGAGTC-3′), with HotStartTaq DNA polymerase (Qiagen). A 2-μL aliquot of the purified ligation reaction was used in a 10-μL PCR reaction containing 5% DMSO. The cycling conditions were as follows: 95°C for 15 min (94°C for 30 s, 55°C for 30 s, and 72°C for 2 min and 30 s) for 25 cycles and 72°C for 7 min. The PCR reaction was diluted 1:10 by adding 90 μL of water. Equal volumes from each reaction were bulked to make the wild-type and mto222 pools (10 plants/pool).
Mu-flanking fragments from the wild type and mutant pools were amplified with nested Mu-TIR primer, MuInt19 (5′-GCCTCYATTTCGTCGAATC-3′), and +2 selective adaptor primers with Ex Taq DNA polymerase (Takara). A total of 16 +2 selective primers was used (see Supplemental Table 2 online). A 1-μL aliquot of the pooled PCR reaction was used in a 10-μL PCR reaction containing 5% DMSO. The touchdown cycling conditions were as follows: 95°C for 2 min (94°C for 30 s, 65 to 0.8°C/cycle for 30 s, and 72°C for 2 min and 30 s) for 11 cycles, 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min and 30 s for 24 cycles, and 72°C for 7 min.
The PCR products of MuInt19 paired with 16 +2 selective adaptor primers were separated on 1.5% agarose gels. A fragment segregating with the mutant phenotype was identified with the +2 primer BfaIntGCT. The nested PCR was repeated with BfaIntGC for each individual plant, and perfect cosegregation was observed for a fragment of 945 bp. The cosegregating fragment was sequenced, and a fragment-specific primer, Tuschg11bfa1 hR (5′-CTCCAGCATTGTCCTCTTCC-3′), was designed to pair with the Mu-TIR primers. A Mu insertion in the candidate gene was confirmed in 22 W64Afl1Mu-1 kernels and 16 B73fl1-Mu1 kernels.
Identification of Additional fl1-Mu Alleles
Using the Mu-flanking sequence and a 5.2-kb GSS contig, two primers, DO84430 (5′-AACCCTACGCTACGGCAAGCTATTCCA-3′) and DO84431 (5′-AACACGGTATGACGGATATCAGACACCC-3′), were designed that spanned the entire coding sequence. These primers were each tested in conjunction with the Mu-TIR primer, DO9242 (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′), for mto222-1 and gave the expected size fragments. To identify additional fl1-Mu alleles, the TUSC population was screened with DO84430 or DO84431 in conjunction with the DO9242 primer. Six additional Mu insertion alleles that were predicted to alter gene function were selected for detailed analysis. The Mu-flanking regions were cloned into pCR2-1 TOPO (Invitrogen) and sequenced at the University of Arizona Sequencing Facility to determine the insertion site. These sites, relative to the ATG initiation codon, are as follows: fl1-Mu2, -194; fl1-Mu3, +7; fl1-Mu4, +190; fl1-Mu5, +5; fl1-Mu6, +849; and fl1-Mu7, +764.
For mapping the Mto222 gene (Fl1), a maize (Zea mays) genomic DNA BAC library array, ZMMBBb (Arizona Genomics Institute, http://www.genome.arizona.edu/orders/), was probed with a 32P dCTP-labeled (Perkin-Elmer) Fl1 DNA fragment made with the Invitrogen random prime labeling kit and the hybridization conditions recommended on the Arizona Genomics Institute website. The template for the labeling reaction was the full-length Fl1 coding sequence, which was amplified from genomic DNA using primers 5′-AAAAGGATCCATGGGCGGCAATAATGACGGCGTTTCC-3′ and 5′-AAAAGAATTCCTACTTCTTATCTCTGTCATCCTCGCTTG-3′ using GC2 Advantage polymerase (BD Biosciences Clontech).
For sequencing of the W64Afl1-ref mutant allele, the full-length coding sequence was PCR amplified from genomic DNA of five independent W64A+ and W64Afl1-ref plants using Phusion polymerase in conjunction with the supplied GC buffer and DMSO (Finnzymes). The primers used were 5′-ATGGGCGGCAATAATGACGGCGTTTCC-3′ and 5′-CTACTTCTTATCTCTGTCATCCTCGCTTG-3′. PCR fragments were gel purified and cloned into pCR2-1 TOPO (Invitrogen) before being sequenced at the University of Arizona Sequencing Facility from both ends using M13 forward and reverse primers. W64A+ and W64Afl1-ref sequences were aligned, along with the B73 wild-type sequence using Sequencher.
Bioinformatics
Phylogenetic analysis was performed on the Fl1 gene and all nonredundant DUF593 domain proteins publicly available. Sequences were obtained from the Pfam database (June 7, 2007; http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF04576). The tree was constructed from only Arabidopsis thaliana (16) and rice (Oryza sativa) (14) sequences because of the complete genome sequence availability in these species. For this reason, the three DUF593 proteins identified in other species were not used. Since overall protein similarity outside the DUF593 domain was low, protein sequences were trimmed to the conserved region around the DUF593 domain (∼90 amino acid residues). Supplemental Table 1 online presents information that collates gene accession number, species, predicted protein length, position, and sequence of the DUF593 domain for all the proteins used to construct the tree. Sequences were aligned and a bootstrap tree was built by the neighbor-joining procedure and bootstrapping (1000 times) using PHYLIP version 3.6. (http://evolution.genetics.washington.edu/phylip.html).
The following websites were used for analysis of FL1 topology: DAS transmembrane prediction server (http://www.sbc.su.se/∼miklos/DAS/tmdas.cgi), Sosui Classification and Secondary Structure Prediction of Membrane Proteins (http://bp.nuap.nagoya-u.ac.jp/sosui/), Split Membrane Protein Secondary Structure Prediction Server (http://split.pmfst.hr/split/), TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), Phobius combined transmembrane topology and signal peptide predictor (http://phobius.cgb.ki.se/), and Predict Protein (http://www.predictprotein.org).
RNA Extraction and RT-PCR
For endosperm RNA extraction, 10 kernels were defrosted sufficiently to remove the pericarp and embryo and subsequently refrozen in liquid nitrogen. Endosperms were ground for 30 s in 3 mL of NTES (20 mM Tris-HCl, pH 8, 100 mM NaCl, 10 mM EDTA, and 1% SDS), which does not solubilize starch. Three milliliters of TRIS-buffered phenol/chloroform (1:1), pH 8, was added, and grinding continued for 1 to 2 min until homogenization was complete. The aqueous phase was separated by centrifugation at 3500 rpm in a Sorval SH-3000 rotor at 4°C for 15 min and then reextracted with phenol/chloroform. Nucleic acids were precipitated using 2.5 volumes of ethanol and 1/10 volume of 3 M sodium acetate, pH 5.3, and resuspended in 1 mL of RNase-free deionized water. RNA was precipitated using 3 volumes of 4 M LiCl at −20°C for 1 h as above. Following resuspension in 500 μL of RNase-free water, RNA was further purified using Trizol (Invitrogen) according to the manufacturer's instructions. In the RRB1 control gene amplification, the primers spanned an intron (Grafi et al., 1996) and no genomic amplification products were detected. RNA isolated in this manner did not give rise to genomic amplification products when testing the expression of other genes (data not shown). For first-strand reactions, 2 μg of RNA samples were primed using oligo(dT) (Invitrogen), and cDNA was synthesized using Superscript 2 (Invitrogen) according to the manufacturer's instructions. The primers for Fl1 allowed the amplification of the 3′ half of the ORF, downstream of the Mu insertion site in fl1-Mu1. Two-microliter samples of the first-strand reactions were added to 50 μL of PCR reactions that were subsequently divided and PCR amplified under identical conditions for 15 cycles and 30 cycles using GC2 Advantage polymerase (BD Biosciences Clontech). Products of the 30-cycle, nonquantitative reactions were separated by electrophoresis in 1% agarose gels and visualized with ethidium bromide to ensure that the reactions had worked. Bands were then excised and purified for use as a labeling template. Products of the 15-cycle quantitative PCR reactions, which were not visible with ethidium bromide staining, were blotted from 1% agarose gels onto nylon membranes and hybridized with the 32P dCTP-labeled (Perkin Elmer) probe fragment, according to standard protocols. For radioactive labeling, a random prime kit (Invitrogen) was used. Bands were visualized using autoradiography for 2 to 4 h. The primers used for amplifying the 747-bp fragment corresponding to the 3′ end of the maize RRB1 gene (Grafi et al., 1996) were RRB1-6 (5′-CTAAAGATTCATTAGCACTCTACACAGC-3′) and Zm RBF1 (5′-CGAACCGTAATGGGGTATTAGTATCG-3′) (Sabelli et al., 2005).
Zein Protein Extraction and Quantification
Developing endosperms were dissected as described above, whereas for mature endosperms, the germ was drilled out and the remainder of the pericarp left in place. Zein and non-zein proteins were extracted from endosperm flour according to the method of Wallace et al. (1990) and were stored at −20°C. For quantitative measurements of zein and non-zein proteins, six independent ears of each genotype, W64A+, W64Afl1-Mu1, and fl1-ref, were sampled. Five kernels from each ear were randomly selected and their endosperm flours pooled. Proteins were extracted from 50 mg of each pooled sample. A BCA protein assay kit (Pierce) was used for measurement of total zein and non-zein proteins, according to the manufacturer's instructions.
FL1 Antibody Preparation
The full-length FL1 ORF was amplified from genomic DNA using primers 5′-AAAAGGATCCATGGGCGGCAATAATGACGGCGTTTCC-3′ and 5′-AAAAGAATTCCTACTTCTTATCTCTGTCATCCTCGCTTG-3′ with GC2 Advantage polymerase (BD Biosciences Clontech). The fragment was restricted with BamHI and EcoRI and ligated into pGEX-4T-3 (Pharmacia) at these sites to create an in-frame fusion with GST. After transformation into BL21 (DE3) Codonplus Escherichia coli, small-scale induction experiments were conducted on several colonies to identify clones that gave the best yield of soluble, full-length fusion protein that was cleavable with thrombin. The selected clones were sequenced at the University of Arizona Sequencing Facility to exclude the possibility of nonsilent PCR-induced errors, and a single clone was subsequently propagated and used for large-scale induction of the fusion protein. This was typically performed by inducing 3 liters of mid-log phase cells in Luria-Bertani broth with 0.1 mM isopropylthio-β-galactoside for 2 h at 30°C. Fusion protein was purified using a standard technique (details of which can be found at http://www.ag.arizona.edu/research/larkinslab/protocols/GST%20Purification%20Protocol.pdf). The FL1 moiety was cleaved from the column-bound GST moiety using thrombin, and the eluate was concentrated with Amicon Ultra-15 centrifugal filter devices (10,000 nominal molecular weight limit) (Millipore) before being further purified from SDS-PAGE gels. Antisera were raised in rabbits by Strategic Biosolutions. Crude antisera were immunoaffinity purified as follows. Sera were first subjected to ammonium sulfate precipitation, and the resulting crude antibody fraction was affinity purified according to standard protocols (Harlow and Lane, 1988). The column was prepared by binding concentrated FL1 protein, prepared as described above, to Reactigel 6X (Pierce) using the protocol found at http://www.ag.arizona.edu/research/larkinslab/protocols/Coupling%20of%20proteins%20to%20Pierce%20Reactigel.pdf.
Cell Fractionation and Immunoblot Analysis
Endosperm cell fractions were prepared according to the method of Habben et al. (1993) and were stored at −80°C. SDS gels (12.5% acrylamide) were prepared and run using the Mini-protean system (Bio-Rad) and blotted onto nitrocellulose using the Mini-transblot system (Bio-Rad) according to standard protocols. Blots were blocked in TBST (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) with 3% nonfat dried milk for 1 h at root temperature. Purified FL1 antibody was used at 1/200 dilution in TBST with 3% milk for 2 h at room temperature or overnight at 4°C. After extensive washes in TBST, membranes were briefly reblocked, and secondary antibody (goat anti-rabbit IgG conjugated to horseradish peroxidase [Pierce]) was applied at 1/20,000 dilution in TBST with 3% milk for 2 h at room temperature or overnight at 4°C. After extensive washes in TBST, secondary antibody was detected with the Super Signal West Pico chemiluminescent substrate kit (Pierce) according to the manufacturer's instructions.
A Bip antiserum (supplied by Rebecca Boston, North Carolina State University) was used at 1/10,000 dilution. This antibody typically recognizes a doublet at 75 kD, which may correspond to BiP and the cytosolic hsc70 proteins (J. Gillikin, personal communication). Both bands in this doublet are equally elevated in fl2, DeB30, and Mc mutant endosperms (data not shown).
Structural Analysis and Immunolabeling
Thin slices of endosperm tissue from developing kernels of wild-type, fl1-ref, and fl1-Mu1 plants were loaded into sample holders that were filled with 0.1 M sucrose, frozen in a Baltec HPM 010 high-pressure freezer (Technotrade), and transferred to liquid nitrogen for storage. Substitution was performed in 2% OsO4 in anhydrous acetone at −80°C for 72 h and followed by slow warming to room temperature over a period of 2 d. After several acetone rinses, samples were teased from the holders and infiltrated in Epon resin (Ted Pella) according to the following schedule: 5% resin in acetone (4 h), 10% resin (12 h), 25% resin (12 h), and 50, 75, and 100% (24 h each concentration). Polymerization was performed at 60°C. Sections were stained with 2% uranyl actetate in 70% methanol for 10 min followed by Reynold's lead citrate (2.6% lead nitrate and 3.5% sodium citrate, pH 12) and observed in a Zeiss EM 109. All images shown depict 2nd or 3rd subaleurone layer endosperm cells.
For immunolabeling, high-pressure frozen samples were substituted in 0.2% uranyl acetate (Electron Microscopy Sciences) plus 0.2% glutaraldehyde (Electron Microscopy Sciences) in acetone at −80°C for 72 h and warmed to −50°C for 24 h. After several acetone rinses, these samples were infiltrated with Lowicryl HM20 (Electron Microscopy Sciences) for 72 h and polymerized at −50°C under UV light for 48 h. Sections were mounted on formvar-coated nickel grids and blocked for 20 min with a 5% (w/v) solution of nonfat milk in PBS containing 0.1% Tween 20. The sections were incubated with the primary antibodies (1:10 in PBS-Tween 20) for 1 h, rinsed in PBS containing 0.5% Tween 20, and then transferred to the secondary antibody (anti-rabbit IgG 1:10) conjugated to 15-nm gold particles for 1 h. Controls omitted either the primary antibodies or used the preimmune serum.
For double-labeling experiments of α- and γ-zeins, plastic sections were first blocked with 5% (w/v) milk, incubated with the first rabbit primary antibody specific for either 22- or 19 kD α-zeins, followed by incubation in the goat anti-rabbit IgG conjugated to 15-nm gold particles. The second labeling step was performed in the same way but using chicken anti-27 kD γ-zein antibodies followed by incubation in rabbit anti-chicken IgG conjugated to 5-nm gold particles. For the double labeling of FL1 and zmTIP3-4, the first labeling step with the anti-FL1 antibodies was done as described. Following a fixation step (5% glutaraldehyde, 30 min) and a second blocking step with 5% milk, the grids were incubated with either no antiserum or the rabbit anti-zmTIP3-4 primary antibody for 1 h, followed by 1 h of incubation with secondary antibody linked directly to 5-nm colloidal gold particles.
Yeast Two-Hybrid Assay
A BD Matchmaker library construction and screening kit (BD Biosciences Clontech) was used for yeast two-hybrid assays. All protocols were performed according to the user manual. FL1 bait constructs were made in pGBKT7. For the FL1 DUF593 bait, a fragment that extended 30 bp either side of this conserved region was amplified from genomic DNA using GC2 Advantage polymerase (BD Biosciences Clonetech) and ligated into pGBKT7 at the EcoRI and BamHI sites. Primers were 5′-AATTGAATTCACGAGGCCATGTCCAAGATTGCGTGCC-3′ and 5′-TATAGGATCCGCTCCCAGCCTTGCATTCCAGACTTCATG-3′. Clones were sequenced to ensure that an in-frame fusion with the GAL4 DNA binding domain had been created and that there were no mutations that would cause amino acid substitutions.
For pairwise mating between pGBKT7-FL1 DUF593 and pGADT7-zein constructs, zein coding regions were amplified using Phusion polymerase (Finnzymes) either from genomic DNA (zein genes do not have introns) or from cDNA constructs available in our lab. Primers were designed to exclude the 5′ region encoding the signal peptide and incorporate an EcoRI site at the 5′ end and a BamHI at the 3′ end for cloning into pGADT7, with the exception that the 3′ primer for the 16-kD γ-zein was designed with an XhoI site, since BamHI cuts within this gene. The primers used for zein amplification are listed in Supplemental Table 3 online.
All pGADT7-zein clones were sequenced at the University of Arizona Sequencing Facility to ensure that in-frame fusions with the GAL4 DNA AD domain had been created and that there were no mutations that would cause amino acid substitutions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunogold Labeling of Wild-Type High-Pressure Frozen/Freeze-Substituted Endosperm Cells (22 DAP) Using Preimmune Serum as a Negative Control for the Immunolocalization of FL1.
Supplemental Table 1. DUF593-Containing Proteins Used for Construction of the Phylogenetic Tree.
Supplemental Table 2. The +2 Selective Adaptor Primers for SAIFF Cloning of Fl1.
Supplemental Table 3. Primers for Zein/pGADT7 Constructs.
Supplementary Material
Acknowledgments
We thank Craig Coleman and Dwight Bostwick for their participation in the screen that identified fl1-Mu1. This research was supported by grants to B.A.L. from the Department of Energy (DE-96ER20242), the National Science Foundation (DBI-0077676), and Pioneer Hi-Bred International.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in Instructions for Authors (www.plantcell.org) is: Brian A. Larkins (larkins@ag.arizona.edu).
Online version contains Web-only data.
References
- Bagga, S., Adams, H., Kemp, J.D., and Senguptagopalan, C. (1995). Accumulation of 15-kilodalton zein in novel protein bodies in transgenic tobacco. Plant Physiol. 107 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga, S., Adams, H.P., Rodriguez, F.D., Kemp, J.D., and Sengupta-Gopalan, C. (1997). Coexpression of the maize delta-zein and beta-zein genes results in stable accumulation of delta-zein in endoplasmic reticulum-derived protein bodies formed by beta-zein. Plant Cell 9 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen, R.J., Johal, G.S., Crane, V.C., Tossberg, J.T., Schnable, P.S., Meeley, R.B., and Briggs, S.P. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., Clore, A.M., Ranch, J.P., Higgins, R., Lopes, M.A., and Larkins, B.A. (1997). Expression of a mutant alpha-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94 7094–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., Herman, E.M., Takasaki, K., and Larkins, B.A. (1996). The maize gamma-zein sequesters alpha-zein and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. Plant Cell 8 2335–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., Yoho, P.R., Escobar, S., and Ogawa, M. (2004). The accumulation of alpha-zein in transgenic tobacco endosperm is stabilized by co-expression of beta-zein. Plant Cell Physiol. 45 864–871. [DOI] [PubMed] [Google Scholar]
- Dombrink-Kurtzman, M.A., and Bietz, J.A. (1993). Zein composition in hard and soft endosperm of maize. Cereal Chem. 70 105–108. [Google Scholar]
- Duvick, D.N. (1961). Protein granules of maize endosperm cells. Cereal Chem. 38 374–385. [Google Scholar]
- Gibbon, B.C., Wang, X.L., and Larkins, B.A. (2003). Altered starch structure is associated with endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. USA 100 15329–15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillikin, J.W., Zhang, F., Coleman, C.E., Bass, H.W., Larkins, B.A., and Boston, R.S. (1997). A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol. 114 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi, G., Burnett, R.J., Helentjaris, T., Larkins, B.A., DeCaprio, J.A., Sellers, W.R., and Kaelin, W.G. (1996). A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 93 8962–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, G.I., Suresh, J., and Phillips, R.L. (1993). Preliminary mapping and biochemical analysis of opaque8. Maize Newsl. 67 102. [Google Scholar]
- Habben, J.E., Kirleis, A.W., and Larkins, B.A. (1993). The origin of lysine-containing proteins in opaque-2 maize endosperm. Plant Mol. Biol. 23 825–838. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hayes, H.K., and East, E.M. (1915). Further experiments on inheritance in maize. Conn. Agric. Exp. Stn. Bull. 188 1–31. [Google Scholar]
- Hunter, B.G., Beatty, M.K., Singletary, G.W., Hamaker, B.R., Dilkes, B.P., Larkins, B.A., and Jung, R. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14 2591–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.S., Gibbon, B.C., Gillikin, J.W., Larkins, B.A., Boston, R.S., and Jung, R. (2006). The maize Mucronate mutation is a deletion in the 16-kDa gamma-zein gene that induces the unfolded protein response. Plant J. 48 440–451. [DOI] [PubMed] [Google Scholar]
- Kim, C.S., Hunter, B.G., Kraft, J., Boston, R.S., Yans, S., Jung, R., and Larkins, B.A. (2004). A defective signal peptide in a 19-kD alpha-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiol. 134 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending, C.R., and Larkins, B.A. (1989). Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending, C.R., and Larkins, B.A. (1992). Effect of the floury-2 locus on protein body formation during maize endosperm development. Protoplasma 171 123–133. [Google Scholar]
- Mertz, E.T., Nelson, O.E., and Bates, L.S. (1964). Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145 279–280. [DOI] [PubMed] [Google Scholar]
- Muszynski, M.G., Dam, T., Li, B., Shirbroun, D.M., Hou, Z., Bruggemann, E., Archibald, R., Ananiev, E.V., and Danilevskaya, O.N. (2006). Delayed Flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, O.E., Mertz, E.T., and Bates, L.S. (1965). Second mutant gene affecting the amino acid pattern of maize endosperm proteins. Science 150 1469–1470. [DOI] [PubMed] [Google Scholar]
- Sabelli, P.A., Dante, R.A., Leiva-Neto, J.T., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (2005). RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc. Natl. Acad. Sci. USA 102 13005–13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R.J., Burr, F.A., Aukerman, M.J., and Burr, B. (1990). Maize regulatory gene Opaque-2 encodes a protein with a leucine-zipper motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.Y., Huber, D.M., and Warren, H.L. (1978). Relationship of kernel sink for N to maize productivity. Crop Sci. 18 399–404. [Google Scholar]
- Wallace, J.C., Lopes, M.A., Paiva, E., and Larkins, B.A. (1990). New methods for extraction and quantitation of zeins reveal a high content of gamma-zein in modified ppaque-2 maize. Plant Physiol. 92 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, R. (2001). Allelism of chromosome 2 endosperm mutants. Maize Newsl. 75 6. [Google Scholar]
- Woo, Y.M., Hu, D.W.N., Larkins, B.A., and Jung, R. (2001). Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13 2297–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., and Boston, R.S. (1992). Increases in binding-protein (BiP) accompany changes in protein body morphology in three high-lysine mutants of maize. Protoplasma 171 142–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.