Abstract
Chromatin-based silencing provides a crucial mechanism for the regulation of gene expression. We have identified a WD40 domain cyclophilin, CYCLOPHILIN71 (CYP71), which functions in gene repression and organogenesis in Arabidopsis thaliana. Disruption of CYP71 resulted in ectopic activation of homeotic genes that regulate meristem development. The cyp71 mutant plants displayed dramatic defects, including reduced apical meristem activity, delayed and abnormal lateral organ formation, and arrested root growth. CYP71 was associated with the chromatin of target gene loci and physically interacted with histone H3. The cyp71 mutant showed reduced methylation of H3K27 at target loci, consistent with the derepression of these genes in the mutant. As CYP71 has close homologs in eukaryotes ranging from fission yeast to human, we propose that it serves as a highly conserved histone remodeling factor involved in chromatin-based gene silencing in eukaryotic organisms.
INTRODUCTION
Unlike animals that develop into adults from miniature forms (embryos), most plant organs are derived from stem cells present in the shoot apical meristem (SAM) and root apical meristem (RAM). A number of genes have been shown to regulate the activity of apical meristems, thereby determining the timing, number, size, and shape of lateral organs (Zhao et al., 2004; Cnops et al., 2006; Doerner, 2006). Some of these regulatory genes, such as WUSCHEL (WUS) and SHOOT MERISTEMLESS (STM), encode transcriptional factors that control the balance between SAM activity and lateral organ formation (Long et al., 1996; Mayer et al., 1998). Loss of function of STM, a homeodomain protein, eliminates stem cell activity in the SAM, resulting in a shoot-meristemless phenotype. In Arabidopsis thaliana, several genes, including STM, BREVIPEDICELLUS, KNAT2, and KNAT6, belong to the class I KNOTTED-like homeobox (KNOX) genes, and they are required for the initiation and maintenance of the SAM (Hake et al., 2004). A critical factor that determines the function of these regulators is their expression pattern. For example, the expression of STM in Arabidopsis is restricted to the meristem dome. Ectopic expression of these genes in other tissues has been shown to cause significant abnormalities in the organogenesis of Arabidopsis (Chuck et al., 1996; Gallois et al., 2002). Although several genes have been identified to play a role in the regulation of KNOX gene expression (Ori et al., 2000; Byrne et al., 2002; Phelps-Durr et al., 2005), the mechanism underlying the regulation is still not clear. The development of flowers, another type of lateral organ, is also regulated by the spatial and temporal expression pattern of conserved homeotic genes. For instance, the expression of the class C floral organ identity gene AGAMOUS (AG) is confined to the inner two whorls of flowers in the wild type. Many genes act to repress AG expression in the first and second whorls (Liu and Meyerowitz, 1995; Krizek et al., 2006). Ectopic expression of the AG gene in transgenic plants results in the alteration of floral organ identity (Mizukami and Ma, 1992).
Tissue-specific expression of regulatory homeotic genes is crucial for the normal pattern of organogenesis. Therefore, it is expected that the expression of KNOX, WUS, and other homeotic transcription factors must be tightly regulated. Recent studies indicate that epigenetic regulation involving chromatin remodeling may play a critical role in programming gene expression pattern in the apical meristems and during organogenesis. For instance, SNF2-class ATPase SPLAYED is specifically recruited to the promoter region of WUS and regulates WUS expression (Kwon et al., 2005). Mutations of FASCIATA (FAS) genes encoding chromatin assembly factor-1 (CAF-1) alter the expression pattern of WUS in the SAM and a regulatory gene, SCARECROW (SCR), in the RAM, leading to altered shoot and root development (Kaya et al., 2001). Another component of CAF-1, MSI1, also functions in flower homeotic gene repression (Hennig et al., 2003).
Covalent modifications of histones play a key role in regulating transcription, genome integrity, and epigenetic inheritance (Kouzarides, 2007; Li et al., 2007). A variety of modifications of histones have been characterized, of which methylation of Lys residues acts as an important component of epigenetic changes to induce transcriptional repression or activation (Richards, 2002; Kubicek and Jenuwein, 2004). The consequences of Lys methylation can be different, depending on which Lys residue is modified and the genome context (Berger, 2007). Homeotic gene silencing is associated with the methylation of histone H3 at Lys-27 (H3K27) in both animals and plants (Beuchle et al., 2001; Makarevich et al., 2006; Schonrock et al., 2006a; Schwartz and Pirrotta, 2007). It has also been shown that AG and STM loci were generally covered with the methylated H3K27, leading to repression of their expression in lateral organs (Schubert et al., 2006). Recently, the genome level distribution of trimethylation H3K27 was described in Arabidopsis (Zhang et al., 2007), and H3K27me3 appears to be associated with a large number of genes.
Chromatin consists of DNA and proteins with highly organized structure. For the assembly and maintenance of the chromatin, chaperones and protein foldases may play a critical role (Shaw, 2007). Yet, very little is understood concerning protein foldases and chaperones in gene regulation and chromatin modification. Originally discovered as a family of receptors for a group of immunosuppressants, immunophilins are protein foldases present in a broad range of organisms, from bacteria to human (Schreiber, 1991; Luan, 1998). Immunophilins consist of two subfamilies, cyclophilins (receptors for cyclosporin A) and FKBPs (for FK506 binding proteins). Immunophilins have generally retained an enzymatic activity called peptidyl prolyl isomerase (or rotamase) that catalyzes a rate-limiting step in the folding of many proteins (Shaw, 2002). In the Arabidopsis genome, at least 50 genes encode a diverse family of immunophilins. Several of these immunophilin members are known to play a role in plant developmental processes. A cyclophilin (CYP40) is required for leaf phase transition (Berardini et al., 2001). Two FKBPs (TWISTED DWARF1 and PASTICCINO1) regulate plant morphogenesis (Vittorioso et al., 1998; Geisler et al., 2003). These findings indicate that members of the immunophilin-type foldases are important for normal plant development, although the mechanism of action has not been addressed. We report here that CYP71, a highly conserved eukaryotic cyclophilin, encodes a nuclear protein and is essential for organogenesis in Arabidopsis. CYP71 functions in the maintenance of the silenced state of homeotic genes, including KNAT1 and STM, by associating with the target chromatin. Furthermore, CYP71 interacts physically with chromatin histone H3 and affects the level of histone methylation, suggesting that this cyclophilin-type foldase plays a fundamental role in the chromatin remodeling processes that influence the expression of regulatory genes.
RESULTS
Isolation of cyp71 as a Developmental Mutant of Arabidopsis
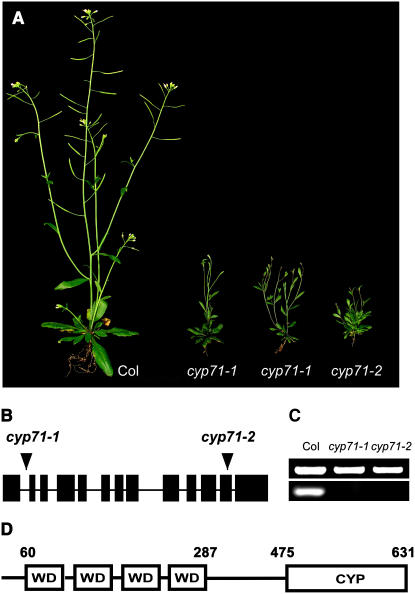
In a systematic effort to understand the function of immunophilins in the model plant Arabidopsis, we screened developmental phenotypes of T-DNA insertional mutants with disrupted immunophilin genes (Alonso et al., 2003). One of the mutants displayed bushy and stunted stature and was isolated for further analysis (Figure 1A). According to previous analysis of the immunophilin families in Arabidopsis (He et al., 2004), the corresponding gene disrupted in this developmental mutant is CYP71. Therefore, we referred to this mutant as cyp71. To confirm that the phenotypic changes were caused by the disruption of CYP71, we analyzed two independent T-DNA insertion alleles of this gene (cyp71-1 and cyp71-2) (Figure 1B). Homozygous T-DNA insertional mutant plants were isolated through PCR-based genotyping, and RT-PCR analysis indicated that both insertions eliminated the expression of CYP71 (Figure 1C). Both alleles displayed the same phenotype shown in Figure 1A, verifying the linkage between the phenotype and the disruption of CYP71. This was also confirmed by complementation using the genomic fragment containing the CYP71 gene. The complemented lines displayed a wild-type phenotype (data not shown). Because cyp71-1 and cyp71-2 displayed the same phenotype, we used cyp71-1 for further analysis, which revealed a wide range of developmental defects as described below. The CYP71 gene encodes a multidomain cyclophilin with a typical peptidyl prolyl cis-trans isomerase (PPIase) catalytic domain in the C-terminal region and four WD40 repeats in the N-terminal region (Figure 1D). The sequence comparison indicated that both the WD40 repeats and the CYP domain are highly conserved from fission yeast to human, suggesting that this protein may play a critical role in eukaryotic organisms. Yet, the function of the CYP71 homologs has not been reported in other organisms.
Figure 1.
Analysis of cyp71 Mutants and the CYP71 Protein.
(A) Wild-type and cyp71 mutant plants grown in the greenhouse for 6 weeks.
(B) Schematic presentation of T-DNA insertions in the CYP71 gene (arrowheads). Exons and untranslated regions are indicated by closed rectangles, and lines between the exons denote introns.
(C) Levels of CYP71 transcripts determined by RT-PCR. The ACTIN transcript level was taken as a control. ACTIN is shown at top and CYP71 is shown at bottom.
(D) Domain structure of the CYP71 protein with four WD40 repeats and a cyclophilin (CYP) domain. The numbers indicate amino acid positions.
CYP71 Function Is Required for the Formation and Development of Leaves and for Normal Phyllotaxy
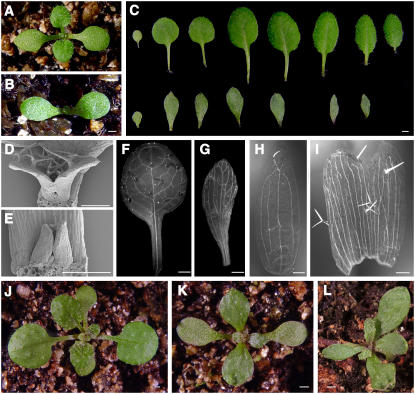
The earliest visible phenotype of the cyp71 mutant is the delayed emergence of the first pair of true leaves. A 14-d-old (14 d after germination) wild-type plant produced a pair of true leaves, whereas the mutant plant of same age did not form visible leaves yet (Figures 2A and 2B). The emergence of subsequent leaves was also delayed in the mutant. Compared with wild-type rosette leaves, mutant leaves were significantly distorted and their surface was rugged (Figure 2C). Furthermore, the mutant leaves failed to fully expand laterally and had no elongated petioles. It appears that the rosette leaves of cyp71 fail to develop normal polarities. Using scanning electron microscopy, we examined early leaf development and found that mutant leaves were defective at early developmental stages (Figures 2D and 2E).
Figure 2.
Defects of Leaf Organogenesis, Morphology, and Vein Pattern in cyp71.
(A) and (B) Fourteen-day-old seedlings of Col-0 (A) and cyp71 (B) grown in a greenhouse. The age refers to days after germination.
(C) A cotyledon and sequential rosette leaves from Col-0 (top) and cyp71 (bottom) plants.
(D) and (E) Scanning electron microscopy images of leaf primordia in Col-0 (D) and cyp71 (E) plants.
(F) and (G) Vein pattern of the first rosette leaf in the wild type (F) and the cyp71 mutant (G).
(H) and (I) Changes in the vein pattern and fusion of sepals in cyp71 (I) compared with the wild type (H).
(J) to (L) Phyllotaxy pattern of the wild type (J) and the cyp71 mutant ([K] as normal and [L] as abnormal).
Bars = 1 mm in (A) to (C), (F), (G), and (J) to (L) and 200 μm in (D), (E), (H), and (I).
Leaf deformation is also reflected by the pattern of leaf vasculature (Figures 2F and 2G). The vein pattern was altered in the mutant leaves compared with that in wild-type leaves. Secondary veins in the mutant retained a smaller angle from the main vein and tended to be parallel to the main vein. Such changes in vein patterning were also observed in other leaf-like lateral organs such as sepals (Figures 2H and 2I). One pronounced phenotype was that some adjacent sepals in the mutants were fused, reducing the count of sepals. Another significant change was in the arrangement of leaves (phyllotaxy). The normal phyllotaxy in the wild type sets the first pair of true leaves in position with an ∼90° angle from the cotyledons. The subsequent pair of true leaves would be arranged into 137° angles (Figure 2J). In the cyp71 mutant, 66% of plants showed normal phyllotaxy in the first pair of leaves, whereas 34% of plants set the first pair of true leaves with a >90° angle to the cotyledons. The data in Figure 2K show one mutant plant with normal phyllotaxy, and another mutant represents abnormal phyllotaxy (Figure 2L).
The Defective Floral Meristem in the cyp71 Mutant Reduces Flower Number and Alters Flower Morphology
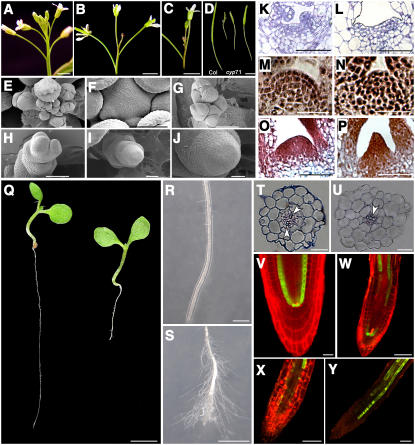
Disruption of CYP71 function also affected reproductive development. A prominent phenotypic change in the cyp71 mutant was a dramatically reduced number of flowers compared with the wild type (Figures 3A to 3C). In the cyp71 mutant, premature termination of the primary SAM caused a more bushy stature of the whole plant (Figure 1A). In some extreme cases, the primary inflorescence failed to produce any flower before terminating as a pin-like structure. We examined the young flowers and meristems in the mutants and the wild type by scanning electron microscopy. In the wild-type plants, flower buds continued to initiate at the periphery of the floral meristem (Figures 3E and 3F), whereas in cyp71, only a few flower primordia initiated around the meristem (Figures 3G and 3H). Some floral meristems did not produce any lateral organs (Figures 3I and 3J), suggesting that CYP71 is required to recruit founder cells to lateral organs. Not only was the number of flowers reduced, but floral organ numbers and morphology were also affected. Some mutant petals were elongated and deformed (see Supplemental Figure 1B online). The cyp71 mutant flower had significantly fewer sepals because of the fusion of sepals, the number of stamens was reduced, and the number of carpels was increased (see Supplemental Figure 1A online). Possibly due to these floral organ defects, the siliques in the mutant were much smaller, containing only a few viable seeds (Figure 3D).
Figure 3.
Analysis of the Inflorescence Apex, Siliques, SAM Structure, and Root Morphology in the Wild Type and cyp71 Mutants.
(A) A main inflorescence of a wild-type plant.
(B) Terminated main inflorescence of a cyp71 mutant plant.
(C) A main inflorescence of cyp71 with a few flowers produced before termination.
(D) Siliques from the wild type (left) and cyp71 (right).
(E) to (J) Scanning electron microscopy images of wild-type and cyp71 inflorescences.
(E) A wild-type inflorescence with a cluster of early floral buds.
(F) An enlarged view of a shoot apex with floral primordia in a wild-type plant.
(G) A cyp71 inflorescence with a few floral primordia irregularly initiated on the tip.
(H) A cyp71 shoot apex with two flower primordia.
(I) and (J) A cyp71 mutant shoot apex without floral primordium.
(K) to (P) Longitudinal SAM sections in the wild type and cyp71.
(K) and (L) SAM of a 3-d-old wild-type (K) or cyp71 (L) seedling. The age refers to days after germination.
(M) and (N) SAM of a 7-d-old wild-type (M) or cyp71 (N) seedling.
(O) and (P) SAM of a 9-d-old wild-type (O) or cyp71 (P) seedling.
(Q) to (Y) Defects in the root development of cyp71.
(Q) A typical 10-d-old wild-type (left) and cyp71 (right) seedling grown on half-strength Murashige and Skoog agar medium.
(R) and (S) Part of the primary root from a wild-type (R) or a mutant (S) seedling.
(T) and (U) Cross sections through the elongation zone of a wild-type (T) or mutant (U) root. Arrowheads point to xylem cells.
(V) to (Y) Confocal images of SCR-GFP expression pattern in the primary roots of wild-type (V) or mutant ([W] to [Y]) plants. Propidium iodide (red) was used to visualize the cell wall.
Bars = 2 mm in (A) to (D) and (Q), 100 μm in (E), (G) to (I), (R), and (S), and 50 μm in (F), (J) to (P), and (T) to (Y).
CYP71 Is Required for the Maintenance of SAM Structure
Defects of lateral organ formation in the cyp71 mutant, including delayed emergence of leaves and a reduced number of flowers, may result from altered meristem activity and structure (Laux et al., 1996; Nelissen et al., 2003). We compared the SAM structure in cyp71 and the wild type by longitudinal sectioning and microscopy and found that SAM in the mutant was broader at the early stage (3 d after germination) and that no leaf primordia were observed (Figure 3L). Cells in the SAM of cyp71 are larger compared with those in the wild type at the same stage (Figure 3K), which suggests a premature loss of meristematic activity. In a 9-d seedling, SAM structure in the mutant was much narrower than that in the wild type, and it changed into a pin-like structure, as opposed to a dome shape in the wild type (Figures 3O and 3P). In addition, the organization of the three cell layers in the SAM was also disrupted in the cyp71 mutant (Figure 3N). As the plants grew to later stages, the SAM in the mutant began to degenerate and lose its organ-forming activity, leading to a pin-like inflorescence in some extreme cases (Figure 3B). These data show that disruption of CYP71 affected both meristem cell organization and the development of organs from the meristem periphery.
To identify the earliest changes in the mutants, we compared the embryogenesis of the wild type and cyp71 by analyzing the cleared immature ovule and seeds using Normaski microscopy. There were no significant differences at most stages of embryogenesis, except that the SAM at the heart stage appeared to be broader in the mutant and the overall size of the embryo and seed of the mutant was also larger than in the wild type (data not shown).
Defects in Root Growth and Radial Patterning in the cyp71 Mutant
The developmental defects of cyp71 were not confined to the shoots but extended to the roots. The mutant seedlings germinated normally, but root growth was retarded dramatically following germination (Figure 3Q). The reduced root length of the mutant can be traced to the shortened elongation zone and a deformed root tip (Figures 3R and 3S). To further examine the defects in the roots, we obtained a cross section of the root at the elongation zone, which revealed defects in the radial patterning of the mutant. In the center, the xylem cells were arranged asymmetrically in cyp71 as opposed to a symmetrical distribution in wild-type plants. In addition, the morphology of different cell layers, especially cortex and endodermis, was not distinctive in the mutant (Figures 3T and 3U). We further investigated the RAM structure and the radial patterning of mutant roots by examining the expression pattern of the marker gene SCR. The SCR gene is expressed in the endodermis and quiescent center cells and plays a key role in the radial patterning of roots (Di Laurenzio et al., 1996). The SCR promoter–green fluorescent protein (GFP) fusion was introduced into the cyp71 mutant background by genetic crosses. The expression of SCR-GFP displayed a continuous pattern in endodermis and quiescent center cells in wild-type roots (Figure 3V), but the expression pattern was dramatically altered in the mutant and essentially lost in the RAM (Figures 3W to 3Y). In some plants, SCR-GFP was still expressed in a few cells in the RAM (Figure 3W). Because the cyp71 mutant is in the Columbia (Col-0) background and SCR-GFP plants were constructed in the Wassilewskija (Ws) ecotype, we analyzed the SCR-GFP expression pattern in 20 F2 segregates with either the cyp71 or the wild-type phenotype. The altered pattern of SCR-GFP expression was observed only in the cyp71 homozygous background, indicating that the cross of Ws and Col-0 alone did not change SCR-GFP expression (data not shown). Together, our data suggest that CYP71 function is important for the formation and activity of root meristem and radial patterning.
CYP71 Is Expressed in the Meristem and Its Protein Is Localized to the Nucleus
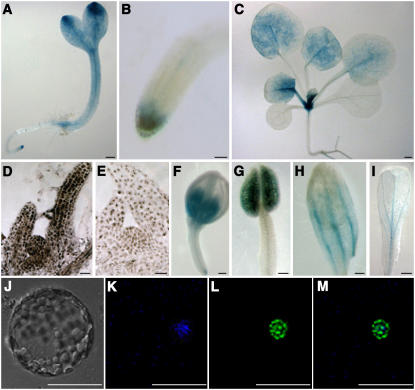
Global phenotypic changes in the cyp71 mutant suggest that CYP71 may be expressed in more than one tissue type. Our previous RT-PCR survey of CYP71 transcripts showed that this gene is expressed in roots, stems, leaves, and flowers (He et al., 2004). To examine the spatial expression pattern of CYP71 in more detail, we cloned a 4.2-kb DNA sequence upstream of the starting codon to drive GUS expression in the transgenic plants and examined GUS activity using a histochemical assay. During early seedling development, the GUS activity accumulated at high levels in the cotyledon tip and in the RAM and SAM (Figures 4A and 4B). GUS activity in the cotyledons and leaves became weak or disappeared as these tissues aged (Figure 4C). CYP71 was also expressed in the flowers (Figures 4F to 4I). In situ hybridization also showed that CYP71 was expressed in the SAM and leaf primordia (Figures 4D and 4E). The expression pattern is consistent with the function of CYP71 in influencing SAM activity, leaf morphogenesis, flower development, and root development.
Figure 4.
Expression Pattern of CYP71 and Nuclear Localization of the CYP71 Protein.
(A) GUS activity pattern in a 3-d-old seedling. The age refers to days after germination.
(B) GUS activity in the root tip.
(C) GUS activity in a 14-d-old seedling.
(D) and (E) Expression of CYP71 mRNA in the meristem and leaf primordia as indicated by in situ hybridization using an antisense probe. Color development in the cyp71 mutant (E) served as a background control.
(F) to (I) GUS activity was detected in a young flower bud (F), a stamen (G), and the vascular tissues of sepal (H) and petal (I).
(J) to (M) Nuclear localization of the CYP71-GFP fusion protein transiently expressed in Agrobacterium-infiltrated tobacco leaf protoplast.
(J) Bright-field image of a protoplast.
(K) 4′,6-Diamidino-2-phenylindole fluorescence.
(L) GFP fluorescence.
(M) Merged image of (K) and (L).
Bars = 0.2 mm in (A) to (C) and (F) to (I) and 10 μm in (D) to (E) and (J) to (M).
Disruption of CYP71 appears to affect a number of plant developmental processes. To investigate further regarding its mode of action, we examined the subcellular localization of the CYP71 protein. We constructed a plasmid with GFP fused to CYP71 and transferred it into tobacco (Nicotiana benthamiana) leaf protoplast. The CYP71-GFP fusion protein was specifically located in the nucleus, and the green fluorescence essentially overlapped with 4′,6-diamidino-2-phenylindole staining (Figures 4J to 4M). We also produced stable transgenic plants expressing the CYP71-GFP fusion protein driven by the 35S promoter. In these plants, we found that CYP71-GFP was also localized in the nucleus (data not shown).
Disruption of CYP71 Causes Ectopic Expression of Meristem and Flower Homeotic Genes in Leaves
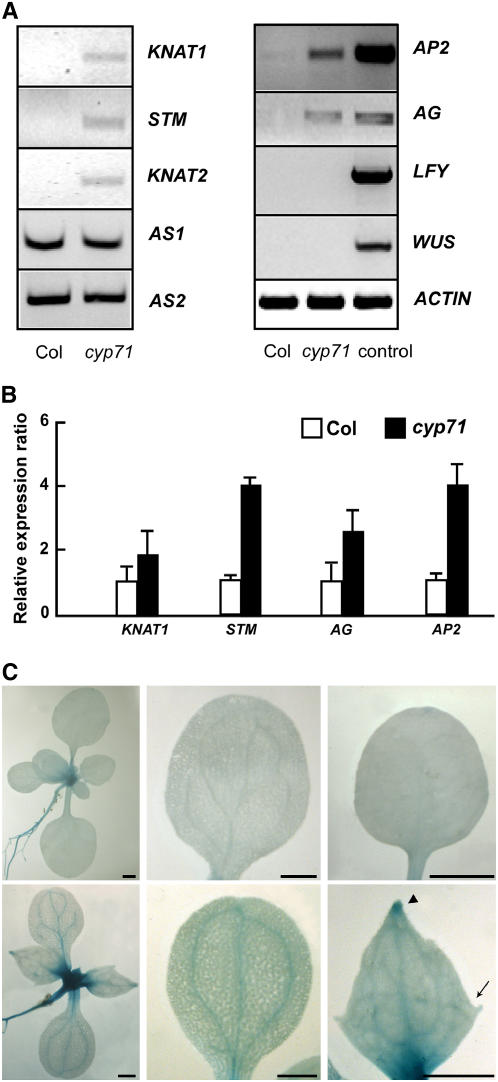
Nuclear localization of the CYP71 protein and pleiotropic changes in developmental programs in cyp71 plants indicate that CYP71 may function as a global regulator of gene expression in the nucleus. In particular, the regulatory genes for SAM activity, leaf development, and floral development could be altered in this mutant. Previous work showed that class I KNOX genes are expressed specifically in the confined area of SAM and repressed in lateral organs. It has been described that the ectopic expression of class I KNOX genes such as KNAT1 and KNAT2 causes defects in leaf morphology (Lincoln et al., 1994; Chuck et al., 1996). Disruption of CYP71 led to lobed and asymmetric leaves with short petioles, mimicking some of the phenotypes caused by the ectopic expression of class I KNOX genes (arrow in Figure 5C). To investigate whether the expression of class I KNOX genes was affected by CYP71 mutation, we performed both semiquantitative and real-time RT-PCR analyses of several class I KNOX genes in true leaves of wild-type and cyp71 mutant plants. As described previously, transcripts of class I KNOX genes were not detected in leaves of wild-type plants. However, these genes, including KNAT1, KNAT2, and STM, were ectopically expressed in the leaves of cyp71 plants (Figures 5A and 5B).
Figure 5.
Class I KNOX Genes and Floral Homeotic Genes Are Ectopically Expressed in cyp71 Mutant Leaves.
(A) Left, RT-PCR analysis of KNAT1, KNAT2, STM, AS1, AS2, and ACTIN in rosette leaves of the wild type and cyp71. Right, RT-PCR analysis of mRNA levels of AG, AP2, LFY, WUS, and ACTIN in rosette leaves of Col-0 and cyp71. The Col-0 seedling was used as a control.
(B) Quantitative PCR analysis of KNAT1, STM, AG, and AP2 expression in leaves of the wild type and cyp71. The results are shown as means ± se from three experiments.
(C) KNAT1-GUS expression in the wild type (top) and cyp71 (bottom). From left to right are seedlings, cotyledons, and the first pair of true leaves. Note that the leaf of cyp71 is lobed (arrow) and that GUS activity typically accumulated at the tip of cyp71 leaves (arrowhead). Bars = 1 mm.
To further analyze the expression pattern of class I KNOX genes in the mutant, we introduced KNAT1-GUS (the KNAT1 promoter fused to the GUS reporter) into the cyp71 mutant by genetic crosses. While GUS activity was not detected in the wild-type cotyledons and leaves, it was expressed at a significant level in the mutant cotyledons and leaves. GUS activity was particularly high in the vascular tissues of both cotyledons and true leaves and in the tip region of the true leaves (Figure 5C). In addition, GUS activity appeared to be higher also in the shoot meristem region of the mutant plants, indicating that a generally higher level of KNAT1 expression may occur in the mutant plants (Figure 5C). Because class I KNOX genes are negatively regulated by ASYMMETRIC LEAVES1/2 (AS1/2) genes that are involved in leaf morphogenesis (Byrne et al., 2000; Semiarti et al., 2001), we examined the expression of these two genes in the cyp71 mutant and found no difference from the wild type (Figure 5A). We also analyzed the expression of several class II KNOX genes and found no difference between wild-type and mutant plants (data not shown).
The defects in flower development in the mutant plants prompted the analysis of several floral regulatory genes, such as AG, AP2, and LFY. These genes determine floral identity and are expressed mainly in the floral apex in wild-type plants. We examined the transcript levels of AG, AP2, LFY, and WUS mRNAs in the leaves of wild-type and cyp71 plants by RT-PCR and found that AG and AP2 mRNAs accumulated to significantly higher levels in cyp71 leaves compared with wild-type leaves, but no change was detected for LFY and WUS (Figures 5A and 5B).
CYP71 Is Associated with the Target Gene Chromatin
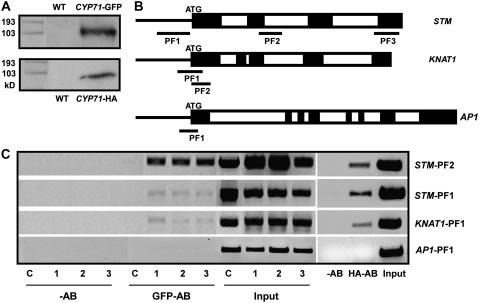
The nuclear localization of CYP71 and the ectopic expression of some meristem-specific genes in the leaves of cyp71 suggest that the CYP71 protein may be required for the silencing of these genes in the leaves. To investigate whether CYP71 represses the expression of target genes through direct interaction with the chromatin, we performed chromatin immunoprecipitation (ChIP) using transgenic plants expressing tagged CYP71 driven by the 35S promoter. An obvious concern is that the 35S promoter may induce high levels of CYP71 protein and that the association of CYP71 tag with chromatin may not reflect the function of endogenous CYP71 protein. To minimize this possibility, we used two types of transgenic plants, expressing CYP71-GFP and hemagglutinin (HA)-tagged CYP71, respectively. The fusion constructs were introduced into the cyp71 mutant background to confirm that the fusion protein functionally replaced the wild-type protein in complementing the mutant. The expression of CYP71-GFP and CYP71-HA in the plants was confirmed by protein gel blot analysis (Figure 6A), and both types of transgenic plants showed the wild-type phenotype, indicating that fusing GFP or HA to the C terminus of CYP71 did not affect the function of the protein (see Supplemental Figure 2 online; data not shown). The ChIP procedure was performed using antibodies to GFP and HA to examine the possible association of CYP71 with the chromatin of various genes, including KNAT1, STM, and AP1. Coprecipitated DNA was analyzed by PCR using primer pairs that amplify regions of the target genes (Figure 6B). The GFP antibody coprecipitated the promoter and coding regions of KNAT1 and STM genes in three independently transformed lines, and the same results were confirmed using a 35S-CYP71-HA transgenic line. On the contrary, the AP1 expression level was not changed in the mutant and could not be coprecipitated in these lines (Figure 6C). Furthermore, we used wild-type plants and transgenic plants expressing GFP-tagged FKBP53, another immunophilin localized in the nucleus (H. Li and S. Luan, unpublished data), in the ChIP experiment as controls and found that the same GFP antibody did not precipitate DNA at the KNAT1 and STM loci in these plants (Figure 6C; data not shown). These results indicate that CYP71 was associated with the chromatin at the STM and KNAT1 loci.
Figure 6.
Association of CYP71 with the STM and KNAT1 Chromatin Loci.
(A) Protein gel blot analysis of total protein in wild-type and transgenic plants using anti-GFP or anti-HA serum.
(B) Schematic presentation of the KNAT1, STM, and AP1 genes, indicating the positions of the PCR fragments amplified from the ChIP products. PF1 to PF3, PCR fragments 1 to 3. Exons and introns are indicated by closed and open rectangles, respectively.
(C) ChIP assay revealed the association of CYP71 with the KNAT1 and STM loci, but not with AP1. Results from three lines of 35S-CYP71-GFP and one line of 35S-CYP71-HA transgenic plants are presented. ChIP assay did not detect an association of FKBP53 with KNAT1 and STM loci in the 35S-FKBP53-GFP plants. Samples were from leaves of transgenic plants for 28 d. The input is chromatin before immunoprecipitation. C, 35S-FKBP53-GFP transgenic plants; 1 to 3, three lines of 35S-CYP71-GFP plants; −AB, ChIP product with no antibody; GFP-AB, ChIP product with antibody to GFP; HA-AB, ChIP product with antibody to HA. The experiments were repeated three times using independent materials, and results from one experiment are shown.
CYP71 Interacts with Histone H3 and Affects the Level of H3K27 Methylation at the Target Loci
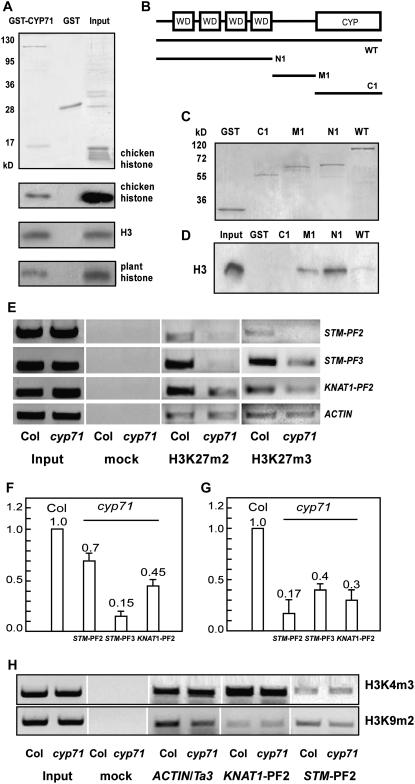
The ectopic expression of homeotic genes in the cyp71 mutants and the chromatin association of CYP71 suggest that CYP71 may regulate gene silencing by interacting with chromatin structure. Recent studies have shown that some WD40 domains can create the surface for binding histones (Schuetz et al., 2006), supporting the possibility of CYP71 interacting physically with the core histones through its WD40 domain. We tested this idea by a pull-down assay using a recombinant glutathione S-transferase (GST)–CYP71 fusion protein expressed in Escherichia coli and commercially available core histones. A histone component corresponding to H3 was copurified with GST-CYP71 but not with GST. Protein gel blot analysis using H3 antibody identified the CYP71-associated histone as histone H3 (Figure 7A). Furthermore, GST-CYP71 interacted with the recombinant H3 protein in the absence of other components in the core histones, indicating that H3 is sufficient for the interaction with CYP71 (Figure 7A). We also performed further experiments using histone-enriched nuclear extract from Arabidopsis plants. Again, H3 was copurified with GST-CYP71 immobilized to the glutathione–Sepharose beads (Figure 7A). In the experiments to identify the domain in CYP71 responsible for H3 interaction, we found that the WD40 repeat domain was sufficient for interacting with H3 (Figures 7B to 7D). In addition, the domain between the WD40 domain and the PPIase also showed interaction with H3. The PPIase domain alone did not interact with H3 under the same conditions (Figure 7D). We found that the interaction of H3 with the WD40 domain alone was stronger than that of the full-length CYP71 protein. It is possible that the WD40 domain was partially masked by other modules in the full-length protein, reducing the affinity of interaction with H3. It is interesting to find that the domain between WD40 repeats and the PPIase region also interacted with H3. This domain does not appear to have any recognizable motif with known function and may represent a new histone binding motif.
Figure 7.
CYP71 Interacts with H3 and Is Required for the Methylation of H3K27.
(A) Protein pull-down assays were performed using recombinant GST or GST-CYP71 with different inputs (chicken histones, recombinant H3, or histone-enriched nuclear extract from Arabidopsis). Polypeptides were resolved by SDS-PAGE and visualized by Coomassie blue staining (for core histones in the top panel) or protein gel blot with anti-H3 antibody (for the bottom three panels with chicken histones, H3, or histone-enriched nuclear extract).
(B) Schematic representation of full-length and truncated forms of CYP71.
(C) Coomassie blue staining of GST-CYP71 and GST-truncated forms (N1, M1, and C1) that were expressed and purified from E. coli and used to examine the interaction with the H3 peptide in (D).
(D) The H3 peptide pulled down by full-length or truncated forms (N1, M1, and C1) of CYP71 was analyzed by protein gel blot with anti-H3 antibody.
(E) ChIP analysis using antibodies against H3K27 (me2 and me3). Samples were from leaves of 28-d-old wild-type and cyp71 mutant plants. Representative images from three independent experiments are shown. The positions of amplified PCR fragments are shown in Figure 6. Input is described in the Figure 6C legend, and mock indicates the control sample without antibody.
(F) and (G) Relative levels of H3K27me2 (F) or H3K27me3 (G) in the cyp71 mutant versus the wild type, normalized by the level of ACTIN2. The se of three experiments is indicated.
(H) ChIP analysis using antibodies against H3K4me3 and H3K9me2. ACTIN served as an inner control for H3K4me3; Ta3 served as an inner control for H3K9me2.
The interaction of CYP71 with histone H3 provides a mechanism for chromatin association that in turn may lead to the modification of chromatin structure. As mentioned above, chromatin remodeling and histone modification are critical mechanisms for gene regulation in both plants and animals. Studies show that the methylation of histone H3 (H3K27) in the STM coding region is essential for the repression of this gene in lateral organs (Schubert et al., 2006). Our study here showed that class I KNOX genes, including STM and KNAT1, were derepressed in cyp71 leaves. Together with the finding that CYP71 interacts with histone H3, these results suggest that histone modification of these target loci may have changed in the mutant. We determined the levels of both H3K27me2 (dimethylation) and H3K27me3 (trimethylation) at STM and KNAT1 loci in wild-type and mutant plant leaves by ChIP experiments. We found that H3K27me2 and H3K27me3 levels were significantly reduced in the mutant background compared with the wild type (Figures 7E to 7G). This result suggests that CYP71 reinforces H3K27 methylation, thereby maintaining the silenced state of regulatory genes, including STM and KNAT1, that play a large role in controlling organogenesis in Arabidopsis. We also examined the methylation of histone H3 at Lys-4 (H3K4) and Lys-9 (H3K9) through ChIP and found no significant changes at STM and KNAT1 loci in the mutant (Figure 7H).
DISCUSSION
Cyclophilins are ubiquitous proteins of the immunophilin superfamily with proposed functions in facilitating protein folding and trafficking (Bose et al., 1996). Some immunophilins may serve as scaffolding proteins for the organization of supermolecular structures (Buchner et al., 1998; Goel et al., 2001). Despite extensive research on their biochemical properties, little is known of the physiological functions of immunophilins. cyp71 was among the most drastically altered mutants of the immunophilin genes we examined, and its analysis provides new insights into the mechanism of epigenetic gene silencing and organogenesis in plants. In this study, we showed that CYP71 directly associated with the chromatin histone H3 and mediated the methylation of H3K27 at some regulatory genes, including KNAT1 and STM, that regulate meristem activity and organogenesis in Arabidopsis. Because CYP71 is highly conserved among eukaryotes ranging from fission yeast to mammals, we expect that CYP71 homologs may function as universal chromatin remodeling factors required for epigenetic gene silencing.
In both animals and plants, many genes involved in developmental regulation are expressed in specific cell types and remain silenced when they are not required in other cells. Previous studies have revealed a critical role for regulation at the level of chromatin structure, namely chromatin-based gene activation or silencing (Wolffe and Guschin, 2000; Kadam and Emerson, 2002). In particular, chromatin-based gene silencing has been shown to be a universal mechanism for gene repression (Berger and Gaudin, 2003; Rando and Paulsson, 2006). The class I KNOX genes in Arabidopsis are specifically expressed in SAM and silenced in differentiated tissues. The transcriptional repression of class I KNOX genes is crucial to plant development, as ectopic expression of these genes leads to severe abnormalities. In several developmental mutants, KNOX genes were ectopically expressed in leaves, because the mutations may have disrupted the regulatory circuit for KNOX gene expression. For example, mutations of AS1 and AS2, genes that encode transcriptional factors, resulted in asymmetric and lobed leaves that ectopically express KNOX genes (Byrne et al., 2000; Iwakawa et al., 2002). Disruption of SERRATE or PICKLE, which encode C2H2 zinc finger and CHD3 proteins, respectively, enhanced the ectopic expression of KNOX genes in lateral organs of as1 and as2 mutants (Ogas et al., 1999; Ori et al., 2000; Prigge and Wagner, 2001). A recent study showed that AS1 can interact physically with HIRA, a candidate histone chaperone with a WD40 repeat domain involved in chromatin remodeling (Phelps-Durr et al., 2005). When HIRA was suppressed in transgenic Arabidopsis plants, KNOX genes were derepressed in the leaves, leading to phenotypic changes similar to those in as1 and as2 mutants. These results suggest that KNOX gene silencing in the leaves is partially maintained by AS1 or HIRA by an epigenetic mechanism.
Deformed rosette leaves with short petioles and parallel veins in cyp71 plants indicated that CYP71 functions in leaf differentiation and leaf shape establishment. The finding of ectopic expression of KNOX genes in cyp71 mutant leaves suggested that the mutant phenotype may have been caused, in part, by disruption of the repressed state of KNOX genes in the leaves. Compared with the mutants mentioned above, cyp71 displayed much broader defects in SAM structure, flowers, and roots, consistent with the altered expression of a wider range of regulatory genes. Studies indicate that the SET domain proteins CLF and SWN also redundantly modulate H3K27me3 level and repress the activity of the STM gene. However, these factors often have a broad range of target genes and could differ significantly in other target genes in addition to the STM gene. Therefore, it is not surprising to find that cyp71 displayed a different phenotype from the clf swn mutant. The identification of CYP71 as a factor for the repression of target genes such as KNAT1 and STM has provided evidence for a previously unidentified mechanism for the regulation of homeobox genes through chromatin-based gene silencing. As CYP71 is directly associated with histone H3 and is involved in histone methylation of target chromatin, it may function as a component of a large molecular complex involved in the assembly and/or maintenance of chromatin structure that is required for gene activation or repression. This hypothesis will need to be tested by further biochemical analysis.
The SAM and the RAM are responsible for the development of the aerial organs and roots, respectively. Phenotypic analysis of SAM and RAM tissue in cyp71 showed that CYP71 was essential for maintaining the meristem function both in the shoot and in the root. Several genes have been identified that regulate both SAM and RAM. For example, FAS1 and FAS2 encode subunits of CAF-1, and mutations in these genes caused fasciated SAM and abnormal RAM, leading to short roots and enlarged inflorescences (Kaya et al., 2000, 2001). A recent study showed that CAF-1 regulated a large number of genes involved in developmental regulation in Arabidopsis (Schonrock et al., 2006b), further revealing the importance of the chromatin assembly process in regulating gene expression. It is worth noting that the maize homolog of MSI1, another subunit in the CAF-1 complex, could bind to histone deacetylase and help to anchor the complex to histones (Rossi et al., 2003; Hennig et al., 2005). Interestingly, two subunits of CAF-1, FAS2 and MSI1, each contain WD40 repeats similar to those found in CYP71. Although the function of WD40 repeats in the CAF-1 subunit remains unknown and it is not clear how CAF1 regulates gene expression, our study of CYP71 provides evidence that these WD40 repeat proteins may be functionally related in the chromatin remodeling processes by interacting with histones and modifying histone codes, such as the methylation status of some Lys residues. However, it is not known whether CYP71 directly or indirectly modulates the histone modification process.
In addition to the WD40 repeats, CYP71 contains a typical cyclophilin catalytic domain that functions as a PPIase, a protein-folding catalyst. A study of an FBKP-type PPIase in yeast indicates that PPIase can regulate the level of H3K36 methylation by adjusting the conformation of Pro residues near Lys-36 (Nelson et al., 2006), implicating the potential of prolyl isomerization as a new mechanism for chromatin modification. Although it remains unknown whether H3 methylation is also regulated by PPIases in higher eukaryotes, including plants and animals, which display a more complex pattern of histone methylation than yeast, the functional analysis of CYP71 in this report reveals a new link between a cyclophilin-type PPIase and histone modification and epigenetic gene silencing in plants. We propose that CYP71 targets histone H3 through the WD40 repeat domain and may modulate the structure of the histones by the protein foldase domain. Further structural analyses will elucidate the mechanism of CYP71 action.
METHODS
Plant Materials and Growth Conditions
The seeds of cyp71-2 (accession number SALK_050092; background, Col), SCR-GFP (accession number CS3999; background, Ws), KNAT1-GUS (accession number CS6141; background, Col) Arabidopsis thaliana plants were provided by the ABRC. The seeds of cyp71-1 (accession number JP69.6C06; background, Col) were provided by the Salk Institute. The SCR-GFP and KNAT1-GUS transgenic lines were crossed to cyp71, and homozygous lines were identified from the F2 populations. Plants were grown in a greenhouse at 22°C with a photon flux density of 180 μmol·m−2·s−1 under a 16-h-light/8-h-dark cycle.
Plasmid Construction and Transgenic Plant Analysis
For complementation by genomic DNA, an 8.2-kb genomic DNA fragment containing 3.9 kb upstream of ATG and 470 bp downstream of the stop codon of the gene was amplified by PCR from BAC clone F14L2 and subcloned into pCAMBIA1300 vector. To express the CYP71-GFP fusion protein in transgenic plants, the coding region of the gene without the terminator codon was amplified and cloned into pCAMBIA1302 with GFP fused in-frame at the C terminus of the CYP71 protein under the control of the cauliflower mosaic virus 35S promoter. The HA tag was added to the C terminus of the CYP71 protein by PCR and constructed into pCAMBIA1390 containing a 35S promoter. For the CYP71 promoter–GUS reporter analysis, a 4.5-kb genomic DNA fragment upstream of the CYP71 start codon was amplified by PCR and cloned into the pBI121 binary vector. The constructs were transformed into Arabidopsis plants using the Agrobacterium tumefaciens–mediated floral dip procedure. For transient protein localization analysis, the coding region of CYP71 was cloned into the pMDC83 vector (Sokolov et al., 2006), resulting in C-terminal fusion with GFP. For CYP71 protein expression, full-length CYP71 and truncated forms of CYP71 were amplified and cloned into pGEX-4T-3 vector.
Photography, Microscopy, GUS Detection, Histological Sectioning, and in Situ Hybridization
Fresh materials from the wild-type and mutant plants were photographed using a Canon Power Shot Pro1 digital camera, and images were processed with Adobe Photoshop software. For scanning electron microscopy, wild-type and mutant plant materials were processed and examined with a Phillips XL30 field emission gun scanning electron microscope as described previously (Chen et al., 2000; Sokolov et al., 2006). The expression pattern of SCR-GFP in the root was analyzed by a procedure described previously (Gallagher et al., 2004). Confocal images were obtained on a Zeiss LSM-510 laser-scanning confocal microscope.
GUS activity measurement followed a previously described procedure (Ori et al., 2000; Li et al., 2005). Tissue sections were examined as described by Ruzin (1999). In situ hybridization was performed as described previously (Coen et al., 1990; Jackson, 1991) using RNA probes generated from a CYP71-specific PCR fragment.
RNA and Protein Expression Analysis
Total RNA was extracted by Trizol from plant materials, and cDNA was synthesized by SuperScript II RNA polymerase. RT-PCR was conducted using Taq polymerase to examine the expression levels of various marker genes. Real-time PCR was performed according to previous methods (Li et al., 2005).
Protein was extracted from 14-d-old seedlings in extraction buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 0.1% Tween, and 1× protease inhibitor cocktail) and precipitated by acetone. Total protein (30 μg) was loaded for SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Bio-Rad). The blot was probed with anti-GFP (Santa Cruz Biotechnology) or anti-HA (Sigma-Aldrich) antibody and was visualized using the ECL system (Amersham Biosciences).
ChIP
ChIP experiments were performed as described previously (Gendrel et al., 2002; Chua et al., 2005) with some modifications. Approximately 1.5 g of rosette leaves from 28-d-old plants grown in soil was fixed with 1% formaldehyde (EMD Biosciences). The chromatin extract was pretreated with protein A agarose beads/salmon sperm DNA slurry (16-157; Upstate) to reduce nonspecific binding before adding specific antibodies for immunoprecipitation. For detection of the CYP71 association with chromatin, 8 μL of anti-GFP (Santa Cruz Biotechnology) or 10 μL of anti-HA (Sigma-Aldrich) antibody was added to the chromatin mixture. For the measurement of histone methylation level, chromatin from wild-type or cyp71 mutant leaves was immunoprecipitated with 10 μL of antibodies against dimethylated H3K27 (07-322; Upstate), trimethylated H3K27 (07-449; Upstate), trimethylated H3K4(ab8580; Abcam), or dimethylated H3K9 (ab1220; Abcam). The precipitated DNA was dissolved in 20 μL of TE buffer (10 mM Tris and 1mM EDTA, pH 8.0), and 1 μL was used for PCR analyses using specific primer pairs at 32 to 38 cycles with Ex-Tag polymerase (Takara).
Protein–Protein Interaction Assay
CYP71 and deletion forms of the construct were transformed into Escherichia coli Rosetta cells (Novagen) for protein expression in E. coli. Protein expression was induced with 100 M isopropylthio-β-galactoside at 25°C for 3 h. The culture was centrifuged at 6500 rpm for 15 min at 4°C, and the pellet was resuspended in 20 mL of 4°C PBS. Cells were sonicated for 2 min on ice (output power, 4; duty cycle, 50%; Branson Sonifier 250). The lysate was centrifuged at 10,000 rpm for 15 min at 4°C, and the supernatant was collected. The truncated and full-length fusion proteins were purified following the manufacturer's protocol through glutathione–Sepharose beads (Amersham Pharmacia Biotech). Protein concentration was determined by the Bradford method using the protein assay kit (Bio-Rad Laboratories), and proteins were stored at −20°C until use. Pull-down assays were performed as described previously with modifications (Shibahara et al., 2000). In the assay, 1.5 μg of purified recombinant GST-CYP71 or GST was immobilized to glutathione–Sepharose beads that were incubated with 15 μg of core histones (13-107; Upstate) or 0.5 μg of recombinant histone H3 peptide (M2503S; New England Biolabs) for 6 or 1 h at 4°C in 400 μL of buffer A300 or A500. Beads were washed six times with 1000 μL of buffer A300 or A500. The amount of histone H3 bound to the beads was determined by SDS-PAGE and Coomassie Brilliant Blue staining or immunoblotting. Histone-enriched nuclear extract was prepared using Arabidopsis plants as described earlier (Yu et al., 2004), and the pull-down assay was performed in A300 buffer as described above. Buffer A300 or A500 contained 25 mM Tris-HCl, pH 8.0, 10% glycerol, 0.01% Nonidet P-40, 1 mM EDTA, and either 300 or 500 mM NaCl. The histone H3 antibody (ab1791; Abcam) was used at a dilution of 1:5000 for protein gel blot analysis.
Accession Numbers
Sequence data for CYP71, KNAT1, and STM can be found in GenBank under the nucleotide accession numbers AT3G44600, AT4G08150 and AT1G62360, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Disruption of CYP71 Alters the Number and Morphology of Flower Organs.
Supplemental Figure 2. Complementation of the cyp71 Mutant by 35S-CYP71-GFP.
Supplementary Material
Acknowledgments
We thank Steven E. Ruzin for advice and assistance with the histological section procedure. This work was supported by grants from the U.S. Department of Energy and the National Science Foundation (to S.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sheng Luan (sluan@nature.berkeley.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Berardini, T.Z., Bollman, K., Sun, H., and Poethig, R.S. (2001). Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291 2405–2407. [DOI] [PubMed] [Google Scholar]
- Berger, F., and Gaudin, V. (2003). Chromatin dynamics and Arabidopsis development. Chromosome Res. 11 277–304. [DOI] [PubMed] [Google Scholar]
- Berger, S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412. [DOI] [PubMed] [Google Scholar]
- Beuchle, D., Struhl, G., and Muller, J. (2001). Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128 993–1004. [DOI] [PubMed] [Google Scholar]
- Bose, S., Weikl, T., Bugl, H., and Buchner, J. (1996). Chaperone function of Hsp90-associated proteins. Science 274 1715–1717. [DOI] [PubMed] [Google Scholar]
- Buchner, J., Weikl, T., Bugl, H., Pirkl, F., and Bose, S. (1998). Purification of Hsp90 partner proteins Hop/p60, p23, and FKBP52. Methods Enzymol. 290 418–429. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chen, C., Wang, S., and Huang, H. (2000). LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26 42–54. [DOI] [PubMed] [Google Scholar]
- Chua, Y.L., Channeliere, S., Mott, E., and Gray, J.C. (2005). The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev. 19 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops, G., et al. (2006). The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell 18 852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen, E.S., Romero, J.M., Doyle, S., Elliott, R., Murphy, G., and Carpenter, R. (1990). Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63 1311–1322. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433. [DOI] [PubMed] [Google Scholar]
- Doerner, P. (2006). Plant meristems: What you see is what you get? Curr. Biol. 16 R56–R58. [DOI] [PubMed] [Google Scholar]
- Gallagher, K.L., Paquette, A.J., Nakajima, K., and Benfey, P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14 1847–1851. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L., Woodward, C., Reddy, G.V., and Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129 3207–3217. [DOI] [PubMed] [Google Scholar]
- Geisler, M., et al. (2003). TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14 4238–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Goel, M., Garcia, R., Estacion, M., and Schilling, W.P. (2001). Regulation of Drosophila TRPL channels by immunophilin FKBP59. J. Biol. Chem. 276 38762–38773. [DOI] [PubMed] [Google Scholar]
- Hake, S., Smith, H.M., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20 125–151. [DOI] [PubMed] [Google Scholar]
- He, Z., Li, L., and Luan, S. (2004). Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 134 1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, L., Bouveret, R., and Gruissem, W. (2005). MSI1-like proteins: An escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15 295–302. [DOI] [PubMed] [Google Scholar]
- Hennig, L., Taranto, P., Walser, M., Schonrock, N., and Gruissem, W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130 2555–2565. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Jackson, D.P. (1991). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach. (Oxford, UK: Oxford University Press), pp. 157–181.
- Kadam, S., and Emerson, B.M. (2002). Mechanisms of chromatin assembly and transcription. Curr. Opin. Cell Biol. 14 262–268. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Sato, S., Tabata, S., Kobayashi, Y., Iwabuchi, M., and Araki, T. (2000). Hosoba toge toge, a syndrome caused by a large chromosomal deletion associated with a T-DNA insertion in Arabidopsis. Plant Cell Physiol. 41 1055–1066. [DOI] [PubMed] [Google Scholar]
- Kaya, H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104 131–142. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128 693–705. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A., Lewis, M.W., and Fletcher, J.C. (2006). RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J. 45 369–383. [DOI] [PubMed] [Google Scholar]
- Kubicek, S., and Jenuwein, T. (2004). A crack in histone lysine methylation. Cell 119 903–906. [DOI] [PubMed] [Google Scholar]
- Kwon, C.S., Chen, C., and Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Li, B., Carey, M., and Workman, J.L. (2007). The role of chromatin during transcription. Cell 128 707–719. [DOI] [PubMed] [Google Scholar]
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., Zhang, D., Xu, Y., and Huang, H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., and Meyerowitz, E.M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121 975–991. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Luan, S. (1998). Immunophilins in animals and higher plants. Bot. Bull. Acad. Sin. 39 217–223. [Google Scholar]
- Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., Grossniklaus, U., and Kohler, C. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71 119–131. [DOI] [PubMed] [Google Scholar]
- Nelissen, H., Clarke, J.H., De Block, M., De Block, S., Vanderhaeghen, R., Zielinski, R.E., Dyer, T., Lust, S., Inze, D., and Van Lijsebettens, M. (2003). DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, C.J., Santos-Rosa, H., and Kouzarides, T. (2006). Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126 905–916. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Phelps-Durr, T.L., Thomas, J., Vahab, P., and Timmermans, M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, O.J., and Paulsson, J. (2006). Noisy silencing of chromatin. Dev. Cell 11 134–136. [DOI] [PubMed] [Google Scholar]
- Richards, E.J. (2002). Chromatin methylation: Who's on first? Curr. Biol. 12 R694–R695. [DOI] [PubMed] [Google Scholar]
- Rossi, V., Locatelli, S., Lanzanova, C., Boniotti, M.B., Varotto, S., Pipal, A., Goralik-Schramel, M., Lusser, A., Gatz, C., Gutierrez, C., and Motto, M. (2003). A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51 401–413. [DOI] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microcopy. (Oxford, UK: Oxford University Press).
- Schonrock, N., Bouveret, R., Leroy, O., Borghi, L., Kohler, C., Gruissem, W., and Hennig, L. (2006. a). Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock, N., Exner, V., Probst, A., Gruissem, W., and Hennig, L. (2006. b). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281 9560–9568. [DOI] [PubMed] [Google Scholar]
- Schreiber, S.L. (1991). Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251 283–287. [DOI] [PubMed] [Google Scholar]
- Schubert, D., Primavesi, L., Bishopp, A., Roberts, G., Doonan, J., Jenuwein, T., and Goodrich, J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25 4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz, A., Allali-Hassani, A., Martin, F., Loppnau, P., Vedadi, M., Bochkarev, A., Plotnikov, A.N., Arrowsmith, C.H., and Min, J. (2006). Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 25 4245–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, Y.B., and Pirrotta, V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8 9–22. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shaw, P.E. (2002). Peptidyl-prolyl isomerases: A new twist to transcription. EMBO Rep. 3 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P.E. (2007). Peptidyl-prolyl cis/trans isomerases and transcription: Is there a twist in the tail? EMBO Rep. 8 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara, K., Verreault, A., and Stillman, B. (2000). The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1-mediated nucleosome assembly onto replicated DNA in vitro. Proc. Natl. Acad. Sci. USA 97 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov, L.N., Dominguez-Solis, J.R., Allary, A.L., Buchanan, B.B., and Luan, S. (2006). A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. USA 103 9732–9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorioso, P., Cowling, R., Faure, J.D., Caboche, M., and Bellini, C. (1998). Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol. Cell. Biol. 18 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe, A.P., and Guschin, D. (2000). Review. Chromatin structural features and targets that regulate transcription. J. Struct. Biol. 129 102–122. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Dong, A., and Shen, W.H. (2004). Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 40 699–711. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y.V., Pellegrini, M., Goodrich, J., and Jacobsen, S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5 e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Medrano, L., Ohashi, K., Fletcher, J.C., Yu, H., Sakai, H., and Meyerowitz, E.M. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.