Abstract
In Arabidopsis thaliana and many other plant species, ovules arise from carpel tissue as new meristematic formations. Cell fate in proliferating ovule primordia is specified by particular ovule identity factors, such as the homeodomain factor BELL1 (BEL1) and MADS box family members SEEDSTICK (STK), SHATTERPROOF1 (SHP1), SHP2, and AGAMOUS. Both in the bel1 mutant and the stk shp1 shp2 triple mutant, integuments are transformed into carpelloid structures. Combining these mutants in a bel1 stk shp1 shp2 quadruple mutant, we showed that the bel1 phenotype is significantly enhanced. We also demonstrate that ovule differentiation requires the regulation of the stem cell maintenance gene WUSCHEL, repression of which is predominantly maintained by BEL1 during ovule development. Based on yeast three-hybrid assays and genetic data, we show that BEL1 interacts with the ovule identity MADS box factors when they dimerize with SEPALLATA proteins. We propose a model for ovule development that explains how the balance between carpel identity activity and ovule identity activity is established by a MADS box homeodomain protein complex.
INTRODUCTION
In Arabidopsis thaliana, cell fate in proliferating ovule primordia is specified by the ovule identity MADS box genes SEEDSTICK (STK), SHATTERPROOF1 (SHP1), and SHP2 that redundantly control ovule identity (Pinyopich et al., 2003). In the stk shp1 shp2 triple mutant, ovules are converted into leaf-like or carpel-like structures, whereas in single or double mutant combinations of these genes, ovule identity is not affected (Pinyopich et al., 2003). Furthermore, ectopic expression of these MADS box genes results in ovule formation on sepals (Favaro et al., 2003; Pinyopich et al., 2003; Battaglia et al., 2006).
Phylogenetic analysis showed that STK, SHP1, and SHP2 are all closely related to AGAMOUS (AG), which is one of the first identified MADS box factors and determines stamen and carpel identity (Bowman et al., 1989; Yanofsky et al., 1990). Evidence that AG plays a role in ovule development was obtained from investigations in which the apetala2 (ap2) single mutant was compared with the ap2 ag double mutant. In the ap2 single mutant, petals are mostly absent and sepals are converted into carpel structures on which ectopic ovules develop, some of which have been converted into carpelloid structures. Interestingly, the first-whorl organs of the ap2 ag double mutant still showed carpel identity; however, the number of ovules that lost their identity and were converted into carpel structures was significantly increased, indicating that AG activity also contributes to ovule identity (Bowman and Meyerowitz., 1991; Pinyopich et al., 2003).
Genetic and biochemical evidence has shown that MADS box transcription factors form dimers that subsequently assemble into higher-order complexes (Honma and Goto, 2001; Pelaz et al., 2001b; Immink et al., 2002; de Folter et al., 2005). Interaction studies in yeast showed that the interaction between these factors can be established by the SEPALLATA (SEP) MADS box transcription factors (Favaro et al., 2003). SEP1/2/3/4 are four closely related flower-specific proteins, and in the sep1 sep2 sep3 triple and the sep1 sep2 sep3 sep4 quadruple mutant, all floral organs are converted into sepals and leaves, respectively, showing that they are necessary for the specification of all four floral organ types (Pelaz et al., 2000; Ditta et al., 2004). Genetic evidence for the role of SEP proteins in ovule development came from the SEP1/sep1 sep2 sep3 mutant, in which SEP activity was partially reduced. This mutant showed loss in ovule identity very similar to what was observed in the stk shp1 shp2 triple mutant (Favaro et al., 2003).
Conversions of ovules into carpelloid structures have also been observed in the bell1 (bel1) mutant (Robinson-Beers et al., 1992; Modrusan et al., 1994; Reiser et al., 1995). In this mutant, the two integuments are replaced by cells that proliferate into carpelloid structures (Robinson-Beers et al., 1992). It has been proposed that in ovules, BEL1 negatively regulates AG and that therefore in the bel1 mutant, the loss of AG regulation causes the conversion of ovule integuments into carpels (Modrusan et al., 1994; Ray et al., 1994). Western and Haughn (1999) also suggested a redundant interaction between BEL1 and AG in the determination of ovule identity since they showed that loss in ovule identity was increased in the ap2 ag bel1 triple mutant relative to the ap2 ag double mutant.
Crucial factors for the control of initiation of integument development are AINTEGUMENTA (ANT), an AP2-like factor (Elliott et al., 1996; Klucher et al., 1996), and WUSCHEL (WUS) (Gross-Hardt et al., 2002; Sieber et al., 2004). WUS is a homeodomain transcription factor that plays a key role in shoot apical and floral meristem maintenance. It is expressed in the central stem cell niche and controls non-cell-autonomously the number of undifferentiated cells in the upper layers of the meristem (Laux et al., 1996; Mayer et al., 1998). Similarly, in the ovule, WUS acts non-cell-autonomously from the nucellus to induce integument growth in the underneath chalaza. Both in the ant mutant (Elliott et al., 1996; Klucher et al., 1996) and in transgenic Arabidopsis plants in which WUS was not expressed in the ovule, integuments failed to develop, showing that these factors are essential for proper ovule development (Gross-Hardt et al., 2002).
In this article, we study the relationship between the ovule identity factors BEL1, AG, STK, SHP1, and SHP2. Our data show that combining the stk shp1 shp2 triple mutant with the bel1 mutant resulted in an enhanced bel1 phenotype in which the single integument forms a green leaf-like structure. Furthermore, new ectopic ovules develop on this structure and the funiculus. Molecular analysis of the bel1 single and the bel1 stk shp1 shp2 quadruple mutant showed that WUS is ectopically expressed in the chalaza and funiculus. Furthermore, we show genetic and biochemical evidence that BEL1 interacts with the MADS box ovule identity factors to control the identity and formation of the integuments.
RESULTS
The bel1 Mutant Phenotype Is Enhanced in the bel1 stk shp1 shp2 Quadruple Mutant
Previous studies have shown that BEL1 and the ovule identity genes STK, SHP1, and SHP2 are important in determining ovule identity in Arabidopsis. Both the bel1 mutant and stk shp1 shp2 triple mutant present conversions of integuments into carpelloid structures (Robinson-Beers et al., 1992; Modrusan et al., 1994; Ray et al., 1994; Reiser et al., 1995; Favaro et al., 2003; Pinyopich et al., 2003). To understand the genetic relationship between these genes, we crossed the stk shp1 shp2 triple mutant with the bel1-1 mutant.
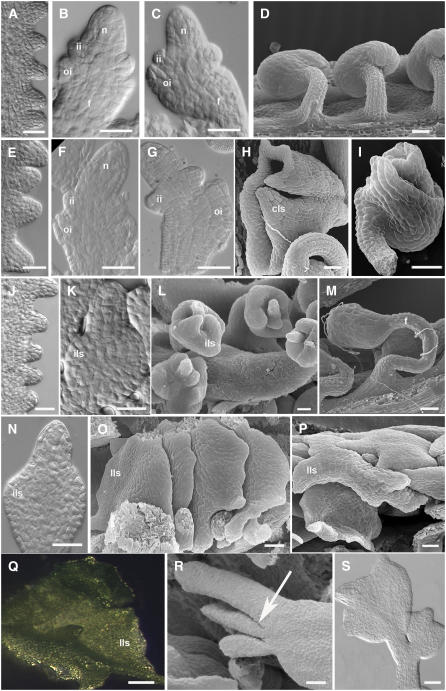
The stk shp1 shp2 triple mutant has been described previously by Pinyopich et al. (2003). To better understand the ovule defects and the developmental events in this triple mutant, we performed optical microscopy and scanning electron microscopy analyses. These analyses show that in the stk shp1 shp2 triple mutant, the number of ovule primordia that arise at stage 9 of flower development (developmental stages refer to those published in Modrusan et al., 1994) from the placental tissue is comparable to the number observed in wild-type plants (cf. Figures 1A and 1E). Subsequently, ovules developed normally until stage 11 (cf. Figures 1B and 1F), and in the nucellus, megagametogenesis proceeded through the first meiotic division as in wild-type plants. Also, in the chalazal region underneath, the two protective integuments initially grew normally. However, from stage 12, the ovule integuments began to present defects (cf. Figures 1C and 1G), and some of them clearly switched to a different developmental program and developed into wide carpel-like structures (Figure 1H). The majority of the mature ovules showed various defects in integument development (Figure 1I). In all these ovules that showed developmental defects, embryo sac maturation was arrested after megasporogenesis. Only a few ovules were able to develop normally and could be fertilized.
Figure 1.
Ovule Development in Wild-Type and Mutant Plants.
(A) to (D) Ovule development in wild-type plants.
(A) to (C) Differential interference contrast (DIC) microscopy images.
(A) Ovule primordia arise from the placenta at stage 9 of flower development.
(B) The nucellus and funiculus differentiate at stage 11. Inner and outer integument primordia are also visible.
(C) The outer integument grows asymmetrically.
(D) Megagametogenesis is initiated. Scanning electron microscopy image of wild-type mature ovules.
(E) to (I) Ovule development in the stk shp1 shp2 triple mutant.
(E) to (G) DIC microscopy images.
(E) Ovule primordia arise from the placenta at stage 9 of flower development.
(F) Inner and outer integuments develop normally until stage 11.
(G) Integument development is affected starting from stage 12.
(H) and (I) Scanning electron microscopy images of mature ovules.
(H) Ovules are converted in carpel-like structures. The funiculus is still visible, while integuments develop in a carpel-like structure.
(I) In some ovules, integuments don't show homeotic conversion; nevertheless, the growth is affected. The outer integument is only partially developed, and it doesn't overgrow the inner integument and the nucellus.
(J) to (L) Ovule development in the bel1 mutant.
(J) and (K) DIC microscopy images.
(J) Ovule primordia arising from the placenta.
(K) At stage 11, a single and thick integument-like structure differentiates in the chalaza region.
(L) Scanning electron microscopy image of bel1-1 ovules. The integument-like structures expand at the base of the nucellus.
(M) Scanning electron microscopy image of the bel1 stk mutant ovule. The ovule phenotype is additive with respect to the single mutants. A thicker and longer funiculus than the wild type is visible. Integument-like structures develop in the chalaza region.
(N) to (S) Ovule development of the bel1 stk shp1 shp2 mutant.
(N) and (S) DIC microscopy images.
(O), (P), and (R) Scanning electron microscopy images.
(N) At stage 11, integument development resembles the bel1 single mutant. A thick integument-like structure is visible.
(O) and (P) Integuments expand, forming leaf-like structures. This phenotype is fully penetrant.
(Q) Stereomicroscopy image of an ovule converted in a green leaf-like structure.
(R) The arrow indicates lateral outgrowth from the chalaza region.
(S) In some cases, secondary mutant ovule-like structures develop from the funiculus.
n, nucellus; ii, inner integument; oi, outer integument; cls, carpel-like structures; ils, integument-like structures; lls, leaf-like structures; f, funiculus. Bars = 25 μM.
Molecular and genetic analyses have demonstrated that BEL1 is required for proper morphogenesis of the ovule integuments. In the bel1 mutant, ovule development is similar to the wild type until stage 10 (cf. Figures 1A and 1J). However, at stage 11, a single integument-like structure initiates at the base of the nucellus (Figure 1K), which enlarges and converts into a carpel-like structure by late stage 12 (Figure 1L) (Modrusan et al., 1994; Ray et al., 1994). Two different bel1 alleles have been described, namely, bel1-1 and bel1-3, and in each case, the number of ovules that present thicker and larger collar tissue depends both on environmental conditions and the ecotype (Robinson-Beers et al., 1992; Modrusan et al., 1994; Western and Haughn, 1999). Furthermore, bel1 ovules do not form a mature embryo sac, since female gametophyte development is blocked after megasporogenesis (Robinson-Beers et al., 1992).
The cross between the bel1 mutant and the stk/STK shp1 shp2 triple mutant resulted in an F2 population in which all alleles segregated, which allowed us to analyze various mutant combinations. Larger numbers of specific mutant combinations were obtained by analyzing plants from F3 and F4 populations. This showed that the ovule phenotype of the bel1 stk double mutant is additive relative to the bel1 and stk single mutants, since ovules had an enlarged funiculus, as in the stk mutant, and the chalazal region developed into structures that resemble the bel1 mutant (Figure 1M). In the bel1 shp1 shp2 triple mutant, ovules developed as in the bel1 single mutant.
The formation of ovule primordia in the bel1 stk shp1 shp2 quadruple mutant was similar to the bel1 single mutant, and in the quadruple mutant, a single structure started to develop from the chalaza instead of the two integuments (cf. Figures 1K and 1N).
However, after stage 12, all the bel1 stk shp1 shp2 ovules developed differently relative to the bel1 single mutant. In bel1 ovules, the tissue that develops from the chalaza forms a collar (Figures 1L), whereas in the quadruple mutant, this structure expands into a green leaf-like structure (Figures 1O to 1Q). Furthermore, lateral outgrowing primordia were formed from the proximal region of the chalaza and from the funiculus (Figure 1R), which sometimes developed into new ovule-like structures (Figure 1S). Female gametophyte development was also arrested earlier in the quadruple mutant since tetrad formation was never observed, suggesting that development arrested before meiosis.
A similar phenotype was described previously for the ap2 bel1 mutant (e.g., in some of the ovules, the integuments were transformed into leaf-like organs) (Modrusan et al., 1994).
Molecular Analysis of Homeotic Transformations in bel1 and bel1 stk shp1 shp2 Mutant Ovules
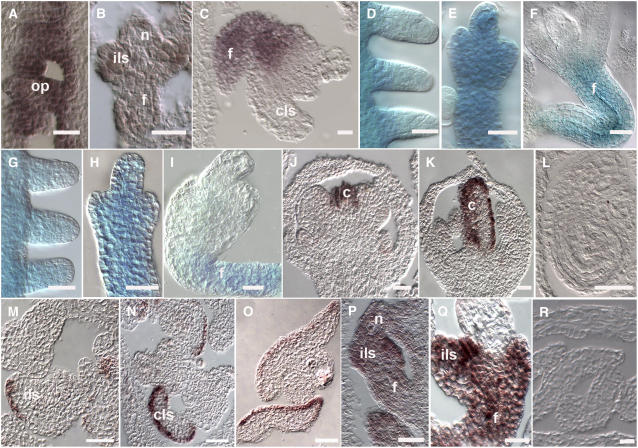
To investigate the identity of the tissue that developed from the chalaza during the different phases of ovule development in bel1 single and bel1 stk shp1 shp2 quadruple mutants, we performed both in situ hybridization and β-glucuronidase (GUS) reporter gene analyses. In situ hybridizations were used to analyze STK expression in bel1 ovules. In wild-type plants, STK starts to be expressed when ovule primordia arise from the placenta. At later stages of development, its expression is observed throughout the developing ovule and in both integuments until the ovule is mature (Rounsley et al., 1995; Kooiker et al., 2005). As shown in Figure 2A, in the bel1 mutant, STK was expressed in ovule primordia and later in the collar tissue that developed from the chalaza until stage 12 (Figure 2B). After that stage, STK transcripts were restricted to the funiculus (Figure 2C), suggesting that in the tissue that developed from the chalaza, integument identity was not maintained after stage 12.
Figure 2.
Gene Expression in Wild-Type and in bel1, stk shp1 shp2, and bel1 stk shp1 shp2 Mutant Ovules.
(A) to (C) STK expression profile in bel1-1 mutant ovules as detected by in situ hybridization.
(A) STK mRNA is detected in the placenta and in ovule primordia.
(B) At stage 12, STK is still expressed in the funiculus and in the integument-like structure that develops from the chalaza region.
(C) STK expression is not detected in the carpel-like structures developing from the chalaza but is restricted to the funiculus.
(D) to (F) Expression of the STK:GUS reporter gene in the bel1-1 mutant.
(D) GUS activity is detected in the placenta and in ovule primordia.
(E) Until stage 12, GUS activity is detected throughout the ovule.
(F) During late stage 12, GUS activity is restricted to the funiculus.
(G) to (I) Expression of the STK:GUS reporter gene in the bel stk shp1 shp2 mutant ovules.
(G) and (H) GUS activity is detected in placenta and in developing ovules until stage 12.
(I) At later stages, GUS activity is detectable only in the funiculus.
(J) to (O) CRC expression profile as detected by in situ hybridization.
(J) to (L) Wild-type flower.
(J) At stage 7 of flower development, CRC is transcribed in developing carpels.
(K) During carpel development, CRC expression is restricted to the epidermal cell layers.
(L) CRC expression is not detected in mature ovules.
(M) bel1 ovules. CRC mRNA is detectable in the integument-like structures that develop in the chalaza region. The expression pattern resembles the situation in wild-type carpels, where only particular cell layers show the hybridization signal.
(N) stk shp1 shp2 ovules. CRC is expressed in the carpel-like structures that develop in mature mutant ovules.
(O) be1l stk shp1 shp2 ovules. CRC mRNA is detectable in the converted structure developing in the chalaza region.
(P) to (R) WUS expression profile in bel1 stk shp1 shp2 ovules as detected by in situ hybridization.
(P) Stage 11 of flower development. In the quadruple mutant, WUS mRNA is present in all the ovules.
(Q) At later stages, WUS is strongly expressed in the integument-like structure and in the funiculus.
(R) WUS mRNA is not detectable in the carpel-like structures at later stages of development.
f, funiculus; c, carpel; cls, carpel-like structure; ils, integument-like structures; op, ovule primordia; n, nucellus. Bars = 25 μM.
To study STK expression in the quadruple mutant and compare it with the expression in the bel1 mutant, we introduced a GUS reporter gene under the control of the STK promoter into the bel1 mutant and the bel1 stk shp1 shp2 quadruple mutant. This construct was previously described by Kooiker et al. (2005) and was shown to mimic the expression of the endogenous STK gene.
Analysis of GUS expression profiles showed that in the bel1 single mutant, GUS expression was detected until stage 12 in the integument structure (Figure 2E). After that, no GUS staining was observed in these structures, indicating that after this developmental stage, integument identity was not maintained (Figure 2F). These GUS expression data are confirmed by the in situ experiments using the STK probe as described above. In the bel1 stk shp1 shp2 quadruple mutant, GUS expression was detected in the structure that developed from the chalaza until stage 12 (Figures 2G and 2H). After that, GUS expression was detectable only in the funiculus, suggesting that identity was not maintained in the structures that were formed from the chalaza integument, similar to observations in the bel1 single mutant (Figure 2I).
To understand whether the integuments of these mutants are transformed into carpel-like structures at stage 12, we used CRABS CLAW (CRC) as a marker (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Lee et al., 2005). CRC is only expressed in flowers, and its expression is restricted to carpels and nectaries. CRC is expressed when the gynoecia primordium is formed (Figure 2J) and subsequently remains expressed in the epidermal cell layers of the carpel (Figure 2K) (Bowman and Smyth, 1999). CRC expression was not detected in wild-type ovules (Figure 2L). In bel1 (Figure 2M) and bel1 stk shp1 shp2 (Figure 2O) as well as in the stk shp1 shp2 (Figure 2N) mutant ovules, the CRC expression pattern resembled the profile observed in wild-type carpels: hybridization signals were clearly visible in particular cell layers of the structures that developed from the chalaza, which could correspond to the carpel epidermal cells. From these data, we concluded that in bel1 and bel1 stk shp1 shp2 ovules, the integument is transformed into a carpel-like structure after stage 12.
WUS Is Deregulated in the bel1 and bel1 stk shp1 shp2 Quadruple Mutants
In the bel1 single mutant, the structure that forms from the chalaza does not cover the nucellus as it does in the wild type. However, this structure expands more than normal integuments, forming a swollen collar-like structure. In the bel1 stk shp1 shp2 quadruple mutant, this structure expands even more drastically, forming an enormous collar structure. It has previously been shown that WUS expression in the ovule is limited to the nucellus (Gross-Hardt et al., 2002). To investigate whether the enlargement of the tissue that forms from the chalaza is due to WUS misexpression, in situ hybridization experiments were performed.
This analysis showed that in the bel1 mutant and the bel1 stk shp1 shp2 quadruple mutant, starting from stage 10, WUS transcripts were not only detected in the distal part of the ovule primordia but also in the chalaza, in structures developing from the chalaza region and in the funiculus (Figure 2P). Moreover, WUS expression was still clearly detectable in mutant ovules at late stage 12 and stage 13 (Figure 2R), while in wild-type ovules, its expression weakens from stage 11 to become undetectable at later stages (Gross-Hardt et al., 2002). These results suggest that the cell proliferations observed from the chalaza and the ectopic ovule-like structure formation from the funiculus, as observed in the quadruple mutant, might be due to a combination of ectopic WUS expression and the absence of integument identity gene activity. Interestingly, deregulation of WUS was only observed when the BEL1 gene was inactive, since in the stk shp1 shp2 triple mutant, WUS expression was not changed relative to wild-type plants.
BEL1 Interacts with the Ovule Identity MADS Box Protein Complex
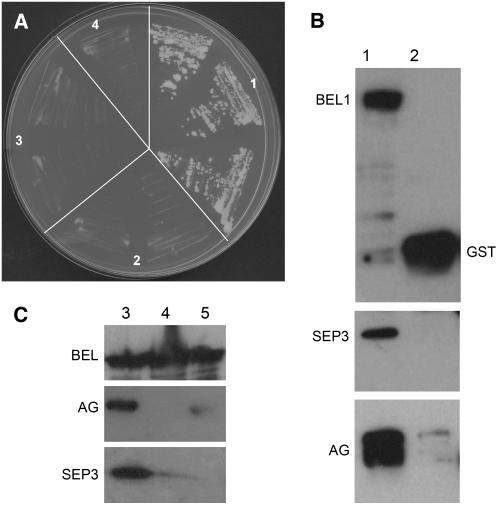
Analysis of AG expression in wild-type and bel1 ovules revealed that the function of BEL1 to regulate AG is most likely not at the level of mRNA accumulation (Reiser et al., 1995). To understand whether BEL1 might regulate AG activity at the protein level, we performed interaction assays using the GAL4-based yeast system. To test for a direct interaction between BEL1 and AG, we performed yeast two-hybrid assays. In these assays, we also tested the ovule identity MADS box factors STK, SHP1, and SHP2. The coding regions of the cDNAs corresponding to these proteins were fused to the AD and BD domains and tested for their ability to interact (see Supplemental Table 1 online). In this assay, we observed that BEL1 did not interact with any of the MADS box proteins that were tested. The MADS box proteins SEP1 and SEP3 were also tested because we have shown that SEP factors play an important role in ovule identity determination and that they are important in the establishment of MADS box complexes (Honma and Goto, 2001; Favaro et al., 2003). However, SEP proteins were unable to interact with BEL1. MADS box proteins function as dimers or higher-order complexes. STK, SHP1, SHP2, and AG do not directly interact but all form dimers with SEP1 and SEP3 (Favaro et al., 2003). To test whether BEL1 was able to interact with MADS box dimers, we performed yeast three-hybrid assays by fusing the full-length SEP1 and SEP3 proteins with the nuclear localization signal of the TFT vector (Egea-Cortines et al., 1999). Clear growth of single colonies was observed when the three proteins (BEL1 and SEP1 or SEP3 in combination with AG or STK, or SHP1 or SHP2) were expressed, whereas all controls were clearly negative (Table 1, Figure 3A; see Supplemental Table 2 online), showing that the dimer composed of SEP1 (or SEP3) and AG, SHP1, SHP2, or STK can bind BEL1. Testing these interactions on different concentrations of 3-aminotriazole showed that all of these dimers interacted strongly with BEL1, although the interaction between BEL1 and the AG-SEP1 (SEP3) dimer was significantly stronger than all other interactions (Table 1). These assays suggest that MADS box dimers establish a surface for the interaction with BEL1 and that the regulation of AG activity might be at the protein level.
Table 1.
Yeast Three-Hybrid Assays Testing BEL1 and MADS Box Protein Interactions
| −WLAH + 3AT
|
||||||||
|---|---|---|---|---|---|---|---|---|
| GAL4-AD | pTFT1 | GAL4-BD | +1 mM | +2.5 mM | +5 mM | +10 mM | +15 mM | +20 mM |
| BEL1 | SEP3 | AG | / | / | + + | + + | + + | + + |
| Empty | SEP3 | AG | / | / | + | + | + | + |
| BEL1 | empty | AG | − | / | − | − | − | − |
| BEL1 | SEP3 | empty | − | − | − | − | − | − |
| BEL1 | SEP1 | AG | + + | / | + + | + + | + + | / |
| Empty | SEP1 | AG | + + | / | + | − | − | / |
| BEL1 | SEP1 | empty | − | / | − | − | − | / |
| BEL1 | SEP3 | SHP1 | + + | + + | + + | + | / | / |
| Empty | SEP3 | SHP1 | − | − | − | − | / | / |
| BEL1 | empty | SHP1 | − | − | − | − | / | / |
| BEL1 | SEP3 | SHP2 | + + | + + | + + | + | / | / |
| Empty | SEP3 | SHP2 | + | − | − | − | / | / |
| BEL1 | empty | SHP2 | − | − | − | − | / | / |
| BEL1 | SEP3 | STK | + + | + + | + | + | / | / |
| Empty | SEP3 | STK | + + | + | − | − | / | / |
| BEL1 | empty | STK | − | − | − | − | / | / |
| BEL1 | SEP3Δ192 | AG | / | / | − | − | / | / |
| Empty | SEP3Δ192 | AG | / | / | − | − | / | / |
| BEL1 | SEP3Δ192 | empty | / | / | − | − | / | / |
| BEL1Δ396 | SEP3 | AG | / | / | + + | + + | / | + + |
| BEL1Δ396 | empty | AG | / | / | − | − | / | − |
| BEL1Δ396 | SEP3 | empty | / | / | − | − | / | − |
−, no growth; +, growth on selective media; + +, strong growth, with formation of robust yeast colonies; /, not tested. 3AT, 3-aminotriazole; W, Trp; L, Leu; H, His; A, adenine. The significant interactions in normal style controls are in bold.
Figure 3.
Interaction Assays to Test for BEL1, SEP, and AG Protein Interactions.
(A) Yeast three-hybrid assays using selective media without His. (1) Three independent yeast transformants cotransformed with pBD-AG, pTFT-SEP1, and pAD-BEL1; (2) to (4) independent yeast transformants using different empty vectors as control. (2) pAD-empty; (3) pTFT-empty; (4) pBD-empty.
(B) and (C) In vitro pull-down assays to test BEL1, SEP3, and AG protein interactions. Extracts of Escherichia coli cells expressing BEL1-GST, AG-MBP, SEP3-TRX, and only the GST tag were used to confirm the physical interactions occurring among these three transcription factors. Bound proteins were analyzed by SDS-PAGE and visualized by immunoblotting using antibodies against GST (top panels), MBP (middle panels), and TRX (bottom panels). First, we allowed the formation of the AG-MAL/SEP3-TRX heterodimer; subsequently, this complex was incubated with BEL-GST protein linked to glutathione-agarose beads (lane 1 in [B] and lanes 1 to 3 in [C]). As a negative control, the AG-MAL/SEP3-TRX heterodimer was also applied to GST bound to glutathione-agarose beads. AG and SEP3 are coimmunoprecipitated with BEL1 (lane 1 in [B] and [C]), but they do not interact with the GST tag (lane 2 in [B]). Weak interactions occur between BEL1 and SEP3 (cf. lanes 1 and 2 of [C]) or BEL1 and AG (cf. lanes 1 and 3 of [C]). Proteins are indicated close to the corresponding bands, and in each lane, equal amounts of protein were loaded.
The C-terminal part of MADS box proteins has been indicated as the domain that is important to establish higher-order MADS box protein complexes (Egea-Cortines et al., 1999; Honma and Goto, 2001; Pelaz et al., 2001a). To test whether complex formation between MADS box and homeobox domain proteins is also dependent on the C-terminal part of MADS box proteins, yeast three-hybrid assays were performed using a MADS dimer composed of full-length AG and a SEP3 protein with a C-terminal deletion (SEP3Δ192). This assay showed that no interaction could be observed, suggesting that the C-terminal domain is also important for MADS–homeodomain protein interactions (Table 1).
Bellaoui et al. (2001) identified two domains in the BEL1 protein important for protein–protein interactions. One domain is located in the first 266 N-terminal amino acids and is possibly involved in general interactions between BEL1 and KNAT proteins. The second region includes the homeodomain and is both necessary and sufficient to confer the specificity of BEL1 for interactions with some members of the KNAT family. A deleted version of the BEL1 protein named BEL1 Δ396, lacking the homeodomain at the C terminus, was still able to interact with the AG-SEP3 dimer in a three-hybrid assay, suggesting that the N-terminal region of BEL1 is sufficient to mediate the interaction with KNAT and MADS box transcription factors (Table 1).
The interactions between the MADS dimers and BEL1 were only observed when BEL1 was expressed as an AD fusion, whereas no interaction was observed when BEL1 was used as a BD fusion protein. It might well be that the BD domain influences correct folding of the BEL1 protein or causes some kind of steric hindrance.
To confirm the observed interactions in yeast, we performed glutathione S-transferase (GST) pull-down assays. The BEL1 protein was tagged with GST and served as bait by binding it to an immobilized glutathione support. SEP3 was tagged with thioredoxin (TRX) and AG with the maltose binding protein (MBP). These assays showed that after washing, the SEP3 and AG proteins (when applying both proteins) were retained by the BEL1-GST resin (Figure 3B). When adding only AG or SEP3 to the BEL1-GST resin, a weak interaction was observed since significantly less protein was retained on the BEL1-GST column (Figure 3C).
Reducing AG Activity Enhances the stk shp1 shp2 Triple Mutant Phenotype
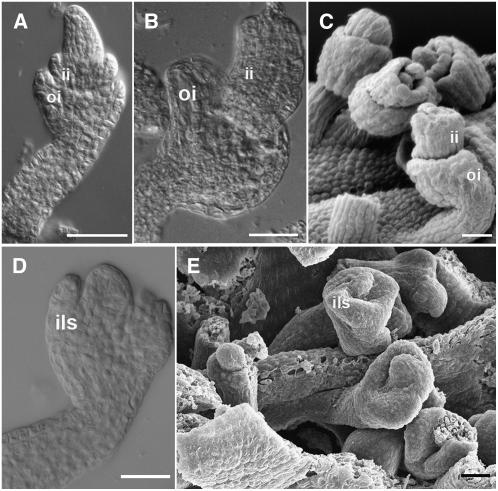
The yeast three-hybrid analysis showed that BEL1 is not only interacting with the ovule identity factors STK, SHP1, and SHP2 but also with AG. To study the effect on the complex when the quantity of AG protein is reduced, we combined the triple mutant stk shp1 shp2 with the ag-3 allele. Of course, in the homozygous quadruple mutant, the effects on ovule development cannot be studied since AG is essential to specify stamen and carpel identity; complete inhibition of its function results in flowers that are only composed of sepals and petals. However, in plants that were homozygous for stk shp1 and shp2 and heterozygous for the ag-3 allele, we observed that the reduction in AG activity has a clear affect on integument development. The most striking observation was that the development of the outer integument was precociously arrested and no longer developed into an extended carpel-like structure in the ovules of this quadruple mutant (Figures 4A to 4C). Interestingly, when these quadruple mutant plants were grown at 30°C, the phenotype of the ovules clearly resembled bel1 mutant ovules, with only one integument developing from the chalaza (Figures 4D and 4E).
Figure 4.
Ovule Morphology in the ag/AG stk shp1 shp2 Mutant.
(A) to (C) ag/AG stk shp1 shp2 mutant grown at 21°C.
(A) and (B) DIC microscopy images.
(A) Integument development proceeds like in the wild-type plant until stage 10. Inner and outer integument primordia emerge from the chalaza.
(B) At later stages, integument development is arrested in all the ovules.
(C) Scanning electron microscopy image. Outer and inner integuments do not overgrow the nucellus.
(D) and (E) ag/AG stk shp1 shp2 mutant grown at 30°C.
(D) DIC microscopy image. An integument-like structure develops at the base of the nucellus resembling bel1 ovules.
(E) Scanning electron microscopy image of ag/AG stk shp1 shp1 ovules that resemble bel1 ovules.
ii, inner integument; oi, outer integument; ils, integument-like structure. Bars = 25 μM.
DISCUSSION
The AG-SEP3 Dimer Interacts with the Homeodomain Protein BEL1 to Control Integument Development
Various experiments performed in the last decade using different plant species suggest that correct ovule development is dependent on a proper balance between the activity of proteins controlling carpel identity and proteins controlling ovule identity. An example is the Arabidopsis stk shp1 shp2 triple mutant, in which the integuments are transformed into carpel-like structures. The same phenotype was obtained by increasing carpel identity activity relative to ovule identity activity. An example is the overexpression of the AG homolog of Brassica napus in tobacco (Nicotiana tabacum; Mandel et al., 1992) or the ectopic expression of AG in Arabidopsis (Ray et al., 1994). In both cases, ovule integuments were transformed into carpelloid structures.
Previous studies have suggested that BEL1 may act as a negative regulator of AG, since in the bel1 mutant there is an excess in AG activity, which causes the homeotic transformation of integuments into carpelloid structures (Ray et al., 1994; Western and Haughn, 1999). However, expression analysis of BEL1 and AG indicated that the regulation of AG by BEL1 is probably not at the transcription level (Reiser et al., 1995). To understand whether balancing AG activity by BEL1 occurs at the protein level, we performed interaction studies in yeast and pull-down experiments. These analyses showed that BEL1 does not directly interact with AG. However, since MADS box proteins do not seem to function as monomers but as dimers (de Folter et al., 2005), we tested if the AG-SEP3 (or AG-SEP1) dimer could interact with BEL1. SEP proteins were chosen since it has previously been shown that these proteins interact with AG and are important components for higher-order MADS box complex formation; furthermore, they play a critical role in ovule development (Honma and Goto, 2001; Favaro et al., 2003). The yeast three-hybrid interaction and in vitro pull-down studies showed that the AG-SEP1 and AG-SEP3 dimers strongly interact with BEL1. The observed interaction together with the information from previous studies suggest that AG may lose its carpel identity activity when the AG-SEP dimer is bound to BEL1.
However, this MADS box homeodomain protein complex might not only be formed to regulate AG activity. Since integuments do not develop correctly in bel1 mutant ovules even in the absence of AG (Western and Haughn, 1999), it is very likely that this complex is also needed to control more downstream genes for normal integument development. A candidate gene might be INO, whose activity is dependent on BEL1 expression (Balasubramanian and Schneitz, 2002).
Our experiments suggest that the C terminus of the MADS box protein is the interaction domain that establishes the interaction with the homeodomain protein. This part of the protein also establishes the interaction between other MADS box proteins to form higher-order MADS box complexes (Egea-Cortines et al., 1999). These higher-order complexes have been shown to be important for MADS box function in floral organ identity determination (Honma and Goto, 2001; Pelaz et al., 2001b). It will be interesting to investigate whether the interaction between the AG-SEP dimer and BEL1 inhibits the formation of higher-order MADS box protein complexes and if this results in the loss of AG function in carpel identitiy determination. Of course it might also be that BEL1 simply attracts the AG-SEP dimer to the ovule identity complex.
AP2 is another repressor of AG in developing flowers. Ovules of plants carrying the ap2-6 allele have carpelloid features, indicating that AP2 also represses AG in the ovule. Combining the ap2 and the bel1 mutant resulted in a severe phenotype with ovules transformed in leaf-shaped structures with cells that resembled carpel cells (Modrusan et al., 1994). We observed a similar phenotype in the bel1 stk shp1 shp2 quadruple mutant, which suggests that when the imbalance between ovule identity and carpel identity activity is increased, a more complete transformation of ovules into carpels is obtained.
BEL1 Controls WUS Expression during Ovule Development
Recessive mutations in bel1 affect integument development. At early stages of bel1 ovule development from the chalaza, a single outgrowth of cells develops that enlarges and generates an irregular collar-like tissue at the base of the nucellus (Modrusan et al., 1994).
In wild-type ovules, WUS expression is restricted to the nucellus. From here, the WUS-induced signaling pathway activates integument formation in the underlying chalaza (Gross-Hardt et al., 2002). Loss- and gain-of-function analysis showed that WUS is not only necessary but also sufficient for integument formation from the chalaza, since ectopic WUS expression in the chalaza resulted in the formation of multiple outer integument primordia (Gross-Hardt et al., 2002). Our in situ analysis of bel1 ovules showed that in addition to the nucellus, WUS was also expressed in the chalaza and in the funiculus, which might cause the enlargement and irregular shape of the integument structure.
In the bel1 stk shp1 shp2 quadruple mutant, the phenotype was even more drastic. The single integument showed a more drastic enlargement compared with the bel1 single mutant, and occasionally ectopic lateral ovule-like structures developed from this structure and from the funiculus. This could be due to the absence of integument identity gene expression that might contribute to the maintenance of the cells in a more indeterminate developmental state.
Is the Single bel1 Integument an Outer Integument?
Detailed morphological analysis suggests that in the bel1 mutant, the development of the inner integument is blocked and only one enlarged outer integument develops (Robinson-Beers et al., 1992; Modrusan et al., 1994; Ray et al., 1994). A well-known marker for the outer integument is INO, a gene that encodes a transcription factor belonging to the YABBY family. INO expression has been detected in cells that give rise to the outer integument before its initiation (Villanueva et al., 1999). INO is necessary for outer integument development since only the inner integument develops in the loss-of-function mutant (Baker et al., 1997; Schneitz et al., 1997). BEL1 activity is needed to activate and maintain INO expression. Balasubramanian and Schneitz (2002) were unable to detect INO expression in the bel1 mutant using in situ hybridization experiments, whereas the more sensitive reporter gene assay using an INO:GUS transgene showed that INO appears to be transiently active in bel1 mutants (Meister et al., 2004). Furthermore, this reporter gene study showed that most of the aberrant growth in the bel1 mutant derives from cells distal to the INO expression and the INO expressing cells do not seem to participate in the formation of the outgrowth.
Using STK as a marker, we showed that until stage 12 the single structure that develops from the chalaza has integument identity. Our analysis also showed that in the bel1 and the bel1 stk shp1 shp2 mutants, WUS expression is highly upregulated in the chalaza. Previously, it has been shown that this ectopic WUS expression causes the formation of multiple outer integuments in wild-type plants. This phenotype was not observed in the bel1 and the bel1 stk shp1 shp2 quadruple mutants.
Based on these data, we concluded that in the bel1 mutant, the structure that develops from the chalaza has integument identity until stage 12 of flower development. However, due to the observation that INO is not expressed in this region and that there is no multiple integument formation due to WUS expression in the chalaza, this structure is most likely the inner integument and not the outer as was suggested previously.
BEL1, STK, SHP1, and SHP2 Are Needed for Megasporogenesis
In the bel1 mutant, embryo sac development is arrested at a late stage of megagametogenesis. The linear tetrad of megaspores is formed, but shortly after, embryo sac development is arrested, and a reduced number of postmitotic cells and remnants of degenerated megaspores are observed (Modrusan et al., 1994). In the stk shp1 shp2 triple mutant, the same situation was observed: meiosis takes place and tetrads are formed, but after meiosis, megagametogenesis is blocked. Interestingly, in the bel1 stk shp1 shp2 quadruple mutant, megasporogenesis is arrested prior to meiosis. This could point to a redundant function of the MADS box and homeodomain factors in megasporogenesis or it might be a pleiotropic effect due to the more severe defects in ovule development in this quadruple mutant.
The ag/AG stk shp1 shp2 Quadruple Mutant Phenocopies the bel1 Mutant at 30°C
Ovule development cannot be studied in the ag mutant since in this mutant, the fourth floral whorl that normally forms the pistil is transformed into a new ag flower composed of sepals and petals. Therefore, the role of AG in ovule development was until now studied in the ap2 mutant, which forms ectopic ovules on carpelloid sepals in whorl 1.
We investigated the role of AG in ovule development by reducing AG activity in the stk shp1 shp2 mutant. This allowed us to study the role of AG in ovules that developed in their normal environment, the carpel. The reduction of AG activity in the triple mutant resulted in outer integument defects. Interestingly, when we incubated the ag/AG stk shp1 shp2 mutant at 30°C, the phenotype resembled the bel1 mutant. These genetic data confirmed our yeast three-hybrid and in vitro pull-down results and suggest that AG, STK, SHP1, and SHP2 all form a complex with BEL1 to control integument identity and development.
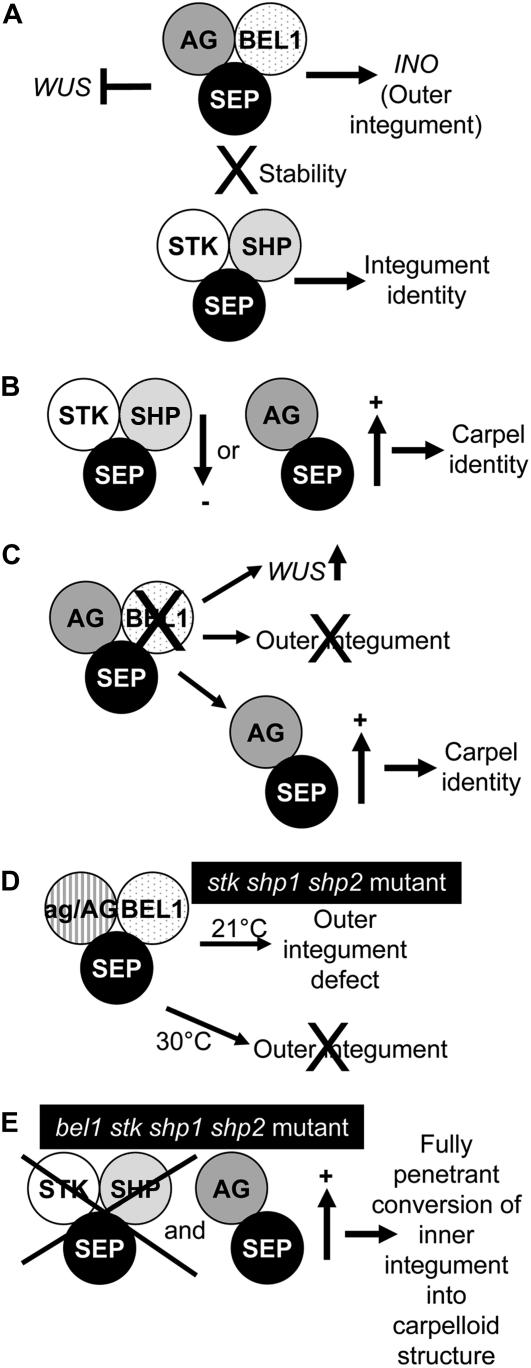
A Model for Integument Identity Determination in Arabidopsis
The data obtained from our experiments show that the ovule identity MADS box factors are all involved in the determination of integument identity. However, previously published data (from our group and others) also show that when the balance between carpel identity activity (mainly established by AG) and ovule integument activity (mainly supplied by STK, SHP1, and SHP2) is disturbed by increasing carpel activity or by reducing integument identity activity, integuments develop into carpelloid structures. Furthermore, we discovered that the integument identity factor BEL1 interacts with AG, SHP1, SHP2, and STK when they are present as dimers with SEP proteins.
From the obtained data, we propose the following model (Figure 5). BEL1 interacts with the AG-SEP3 dimer to control outer integument growth probably by activating INO (Figure 5A). When BEL1 is missing, this complex is not formed and a single integument develops that after stage 12 is transformed into carpelloid tissue. This is due to the fact that the AG-SEP dimer is not linked to the BEL1 complex and therefore has carpel identity activity (Figures 5B and 5C). The ovule identity factors STK, SHP1, and SHP2 stabilize the AG-SEP-BEL1 complex. When STK, SHP1, and SHP2 are not present, the AG-SEP dimer is also less tightly bound to BEL1 and the carpel pathway is activated, although the phenotype is not completely penetrant. When AG activity is further reduced in this triple mutant, there is still enough AG-SEP-BEL complex available to establish inner and outer integument development. However, when this mutant is grown at a higher temperature, the integument identity complex becomes less stable due to the absence of STK, SHP1, and SHP2; the complex AG-SEP-BEL is not made and only one integument develops as in the bel1 single mutant (Figure 5D).
Figure 5.
A Model for Integument Identity Determination and Development.
(A) In wild-type ovules, the BEL1 protein interacts with the AG-SEP dimer to repress WUS in the chalaza and to regulate outer integument development. This complex is stabilized by the ovule identity complex.
(B) When the ovule identity complex is reduced or the AG-SEP dimer is increased, integuments acquire carpel identity.
(C) In the bel1 mutant, the amount of AG-SEP dimer is increased. In this case, WUS is ectopically expressed in the chalaza and the outer integument does not develop. Moreover, the inner integument develops like a carpel structure.
(D) In the stk shp1 shp2 ag/AG mutant, there is still enough AG-BEL-SEP complex to regulate inner and outer integument development. However, at higher temperatures, this complex is very unstable due to the absence of the ovule identity complex, and the outer integument does not develop.
(E) In the stk shp1 shp2 bel quadruple mutant, only the SEP-AG dimer is available, resulting in the conversion of the inner integument into a carpelloid structure.
METHODS
Plant Material
The plants were grown at 21 or 30°C under short-day (8 h light/16 h dark) or long-day (16 h light/ 8 h dark) conditions. The ag-3 mutant ecotype Landsberg was kindly provided by R. Sablowsky, and the Arabidopsis thaliana stk shp1 shp2 mutant was kindly provided by M. Yanofsky. Seeds from the bel1-1 (ecotype Landsberg) Arabidopsis mutant were obtained from the Nottingham Arabidopsis Stock Centre. The bel1 stk shp1 shp2 quadruple mutant was obtained by pollinating the stk/STK shp1 shp2 triple mutant with bel1-1 homozygous pollen. The quadruple mutant was maintained by selfing the stk/STK,bel1/BEL1 shp1shp2 plants of the F2 segregating population. The ag-3/AG stk shp1 shp2 mutant was obtained by pollinating ag3/AG mutant plants with stk shp1 shp2 triple mutant pollen. In F2, F3, and F4 populations, 80 stk shp1 shp2 plants (five carpels per plant) were analyzed, and for the bel1 shp1 shp2 stk quadruple mutant, 30 plants were analyzed (five carpels per plant).
Analysis of STK Expression in the bel1 stk shp1 shp2 Quadruple Mutant
The STK:GUS reporter construct is a translation fusion of 2.9 kb upstream of the STK start codon with the GUS (uidA) gene cloned into pCAMBIA 1300-H (Kooiker et al., 2005). Pollen from bel1 stk shp1 shp2 homozygous mutants was used to pollinate wild-type Columbia plants carrying the STK:GUS reporter construct. GUS assays were performed overnight as described previously (Liljegren et al., 2000). Samples were incubated in clearing solution, dissected, and observed.
Mutant Genotyping Using PCR
Genotyping of the ag-3 mutation was described previously (Favaro et al., 2003).
Genotyping the stk shp1 shp2 mutant was done by PCR. The stk-2 allele contains a 74-bp insertion near the splice site of the third intron and was genotyped with the primers Atp 560 (5′-CACTGTCCAAGAAATCAATGCCGC-3′) and Atp 561 (5′-GGAACTCAAAGAGTCTCCCATCAG-3′). The shp1-1 and shp2-1 alleles were identified as described previously (Liljegren et al., 2000). The bel1-1 allele contains a C-to-T transition at nucleotide 497, which introduces a BsaAI restriction site. Bel1-1 alleles were identified by BsaAI digestion of PCR products amplified with the primers Atp 407 (5′-GAGAGACATGGCAAGAGATCAG-3′) and Atp 408 (5′-GAGCATGGAGAGCAACTTGG-3′).
Morphological Analysis by DIC Microscopy
To analyze ovule development in mutant and transgenic plants, flowers at different developmental stages were cleared from overnight to a week using a solution composed of 160 g of chloral hydrate (C-8383; Sigma-Aldrich), 100 mL of water, and 50 mL of glycerol. After clearing, ovules were dissected from premature pistils on a slide under a stereomicroscope, mounted with a cover slip, and subsequently observed using a Zeiss Axiophot D1 microscope equipped with DIC optics. Images were captured on an Axiocam MRc5 camera (Zeiss) using the Axiovision program (version 4.1).
The observed ovule phenotypes were consistent in the F2, F3, and F4 segregating populations and the backcross population that was made to introduce a reporter gene construct, which indicates that the observed phenotypic effects are not due to differences in the ecotype background of our mutants.
Morphological Analysis by Scanning Electron Microscopy
Plant material was fixed at 4°C overnight in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde in 0.025 M phosphate buffer, pH 7.0. Samples were subsequently washed two times (20 minutes) in 70% ethanol in 0.025 M phosphate buffer, pH 7.0. Ovules where extracted from pistils immersed in the same solution under a stereomicroscope. After dissection, isolated ovules were rehydrated through an ethanol series and then postfixated for 4 h in 1% osmic acid in 0.05 M phosphate buffer, pH 7.0, at 4°C. Samples were washed in 0.05 M phosphate buffer, pH 7, dehydrated gradually in ethanol series to 100% ethanol, and dried in liquid carbon dioxide at critical point. Samples were subsequently covered with gold using a sputter coater (SEMPREP2; Nanotech) and observed with a LEO 1430 scanning electron microscope (LEO Electron Microscopy).
In Situ Hybridization Analysis
Arabidopsis flowers were fixed and embedded in paraffin as described previously (Huijser et al., 1992). Sections of plant tissue were probed with digoxigenin-labeled STK, CRC, and WUS antisense RNA. The antisense STK probe corresponds to nucleotides 455 to 818, the CRC probe from nucleotides 444 to 878 and for the WUS probe, the cDNA sequence without the homeobox domain was used. The probe was subsequently hydrolyzed in carbonate buffer at pH 10.2 for 75 min. Hybridization and immunological detection were performed as described by Coen et al. (1990).
Yeast Two- and Three-Hybrid Assays
The two- and three-hybrid assays were performed in the yeast strain AH109 (Clontech). pBD, pAD, and pTFT1 (Egea-Cortines et al., 1999) vector constructs were selected on yeast synthetic dropout (YSD) medium lacking Leu, Trp, and adenine, respectively. Three-hybrid interactions were assayed on selective YSD medium lacking Leu, Trp, and adenine and His supplemented with different concentrations of 3-aminotriazole (1, 2.5, 5, 10, 15, and 20 mM). Selections were performed at 28°C. Genes used for the yeast two- and three-hybrid assays were cloned in the Gateway vector GAL4 system (pDEST32 for binding domain fusions and pDEST22 for activation domain fusions) or pTFT1 Gateway (kindly provided by G. Angenent) passing through pDONOR201 (Life Technologies). Cloning of AG, SHP1, SHP2, and STK proteins in AD or BD vectors and SEP1 and 3 in pTFT were reported by Favaro et al. (2003). The coding sequences of SEP3Δ192, BEL1, and BEL1Δ396 were amplified using primers AtP1234 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGGAAGAGGGAGAGTAGAAT-3′) and AtP1236 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTAAACCTCTTCTTGGTTAGG-3′) for SEP3Δ192, primers AtP1347 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGCAAGAGATCAGTTCTATG-3′) and AtP1081 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCAAACAATATCATGAAGTAATTG-3′) for BEL1, and AtP1347 and AtP1348 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTCAGCGTTGTGGACGCCAAGG-3′) for BEL1Δ396.
Pull-Down Assays
The BEL1 open reading frame was amplified with primers AtP1347 and AtP1081 and cloned by Gateway recombination in pGEX-2T (Amersham Biosciences). The AG coding sequences was amplified with primer 5′-GGAATTCACGGCGTACCAATCGGAGCTAGGAGG-3′ and with primer 5′-GCGGTCGACTTACACTAACTGGAGAGCGG-3′. The PCR product was digested with EcoRI and SalI and ligated into the pMALC2 vector (New England Biolabs). The SEP3 coding sequence was amplified with primer 5′-GGAATTCG ATGGGAAGAGGGAGAGTAG-3′ and primer 5′-GCGGTCGACTCAAATAGAGTTGGTGTCATAAGG-3′. The PCR product was digested with EcoRI and SalI and ligated in pet32b (Novagen). All the heterologous proteins were induced in the BL21-Gold strain (Stratagene), BEL1-GST and SEP3-TRX were partially soluble at 37°C, and AG-MBP was partially soluble at 16°C. For the binding experiments, Escherichia coli lysis was obtained in 140 mM NaCl, 2.7 KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.5% Triton X-100, and 5 mM DTT, with 1 mM PMSF and protease inhibitors. First, we allowed the formation of the SEP3-TRX/AG-MBP heterodimer; subsequently, this dimer was left to interact with a mix of BEL-GST bound to the immobilized glutathione support (GSH-Sepharose) or with GST bound to the glutathione support as control and incubated overnight at 4°C. The beads were precipitated by microcentrifugation, washed three times with lysis buffer, and resuspended in 5× SDS-PAGE sample buffer. Proteins were eluted by boiling for 5 min and separated on a 12% SDS-polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and probed with anti-MBP antibodies (New England Bioloabs), anti-GST antibodies (Amersham), and anti-TRX antibodies (Sigma-Aldrich). Primary antibodies were detected with horseradish peroxidase–coupled anti-rabbit or anti-goat antibodies using the ECL detection system (Amersham Biosciences).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Two-Hybrid Interactions among BEL1 and MADS Box Proteins in the Yeast GAL4 System.
Supplemental Table 2. Three-Hybrid Interactions of BEL1 and MADS Box Proteins in the Yeast GAL4 System.
Supplementary Material
Acknowledgments
We thank Robert Sablowski for providing the ag mutant and Martin Yanofsky for providing the stk shp1 shp2 triple mutant. Scanning electron microscopy analysis was performed with the help of Giulio Melone at the Centro Interdipartimentale Microscopia Avanzata, an advanced microscopy laboratory established by the University of Milan. This research was supported by the European Union Marie Curie Research and Training Network Project Transistor and by the Ministero dell'Istruzione, dell'Università e della Ricerca Cofinanziamento 2005 Protocollo 2005054223_002.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lucia Colombo (lucia.colombo@unimi.it).
Online version contains Web-only data.
References
- Alvarez, J., and Smyth, D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126 2377–2386. [DOI] [PubMed] [Google Scholar]
- Baker, S.C., Robinson-Beers, K., Villanueva, J.M., Gaiser, J.C., and Gasser, C.S. (1997). Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, S., and Schneitz, K. (2002). NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana. Development 129 4291–4300. [DOI] [PubMed] [Google Scholar]
- Battaglia, R., Brambilla, V., Colombo, L., Stuitje, A.R., and Kater, M.M. (2006). Functional analysis of MADS-box genes controlling ovule development in Arabidopsis using the ethanol-inducible alc gene-expression system. Mech. Dev. 123 267–276. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., and Meyerowitz, E.M. (1991). Genetic control of pattern formation during flower development in Arabidopsis. Symp. Soc. Exp. Biol. 45 89–115. [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen, E.S., Romero, J.M., Doyle, S., Elliot, R., Murphy, G., and Carpenter, R. (1990). Floricaula a homeotic gene required for flower development in Antirrhinum majus. Cell 63 1311–1322. [DOI] [PubMed] [Google Scholar]
- de Folter, S., Immink, R.G., Kieffer, M., Parenicova, L., Henz, S.R., Weigel, D., Busscher, M., Kooiker, M., Colombo, L., Kater, M.M., Davies, B., and Angenent, G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17 1424–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M.F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 1935–1940. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., Yanofsky, M.F., Kater, M.M., and Colombo, L. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser, P., Klien, J., Lonnig, W.E., Meijer, H., Saedler, H., and Sommer, H. (1992). Bracteomania, an inflorescence anomaly is caused by the loss of function of the MADS-box gene SQUAMOSA in Antirrhinum majus. EMBO J. 11 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. [DOI] [PubMed] [Google Scholar]
- Immink, R.G., Gadella, T.W., Jr., Ferrario, S., Busscher, M., and Angenent, G.C. (2002). Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99 2416–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiker, M., Airoldi, C.A., Losa, A., Manzotti, P.S., Finzi, L., Kater, M.M., and Colombo, L. (2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Baum, S.F., Alvarez, J., Patel, A., Chitwood, D.H., and Bowman, J.L. (2005). Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Bowman, J.L., Kempin, S.A., Ma, H., Meyerowitz, E.M., and Yanofsky, M.F. (1992). Manipulation of flower structure in transgenic tobacco. Cell 71 133–143. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Meister, R.J., Williams, L.A., Monfared, M.M., Gallagher, T.L., Kraft, E.A., Nelson, C.G., and Gasser, C.S. (2004). Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37 426–438. [DOI] [PubMed] [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohalmi, S.E., Crosby, W.L., and Yanofsky, M.F. (2001. a). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26 385–394. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Tapia-Lopez, R., Alvarez-Buylla, E.R., and Yanofsky, M.F. (2001. b). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11 182–184. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. [DOI] [PubMed] [Google Scholar]
- Ray, A., Robinson-Beers, K., Ray, S., Baker, S.C., Lang, J.D., Preuss, D., Milligan, S.B., and Gasser, C.S. (1994). Arabidopsis floral homeotic gene BELL1 (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl. Acad. Sci. USA 91 5761–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83 735–742. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruitt, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley, S.D., Ditta, G.S., and Yanofsky, M.F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., Hulskamp, M., Kopczak, S.D., and Pruitt, R.E. (1997). Dissection of sexual organ ontogenesis: a genetic analysis of ovule development in Arabidopsis thaliana. Development 124 1367–1376. [DOI] [PubMed] [Google Scholar]
- Sieber, P., Gheyselinck, J., Gross-Hardt, R., Laux, T., Grossniklaus, U., and Schneitz, K. (2004). Pattern formation during early ovule development in Arabidopsis thaliana. Dev. Biol. 273 321–334. [DOI] [PubMed] [Google Scholar]
- Villanueva, J.M., Broadhvest, J., Hauser, B.A., Meister, R.J., Schneitz, K., and Gasser, C.S. (1999). INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription. Nature 346 35–39. [DOI] [PubMed] [Google Scholar]
- Western, T.L., and Haughn, G.W. (1999). BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J. 18 329–336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.