Abstract
Sugars such as glucose function as signal molecules that regulate gene expression, growth, and development in plants, animals, and yeast. To understand the molecular mechanisms of sugar responses, we isolated and characterized an Arabidopsis thaliana mutant, high sugar response8 (hsr8), which enhances sugar-responsive growth and gene expression. Light-grown hsr8 plants exhibited increased starch and anthocyanin and reduced chlorophyll content in response to glucose treatment. Dark-grown hsr8 seedlings showed glucose-hypersensitive hypocotyl elongation and development. The HSR8 gene, isolated using map-based cloning, was allelic to the MURUS4 (MUR4) gene involved in arabinose synthesis. Dark-grown mur1 and mur3 seedlings also exhibited similar sugar responses to hsr8/mur4. The sugar-hypersensitive phenotypes of hsr8/mur4, mur1, and mur3 were rescued by boric acid, suggesting that alterations in the cell wall cause hypersensitive sugar-responsive phenotypes. Genetic analysis showed that sugar-hypersensitive responses in hsr8 mutants were suppressed by pleiotropic regulatory locus1 (prl1), indicating that nucleus-localized PRL1 is required for enhanced sugar responses in hsr8 mutant plants. Microarray analysis revealed that the expression of many cell wall–related and sugar-responsive genes was altered in mur4-1, and the expression of a significant proportion of these genes was restored to wild-type levels in the mur4-1 prl1 double mutant. These findings reveal a pathway that signals changes in the cell wall through PRL1 to altered gene expression and sugar-responsive metabolic, growth, and developmental changes.
INTRODUCTION
Sugars such as glucose regulate many important cellular processes and serve a fundamental role in carbon skeleton supply in animals, yeast, and plants. In plants, sugar-mediated gene expression, growth, and development are integrated with a wide range of other responses, such as light and circadian regulation, nitrogen availability, hormones, and stress (Krapp et al., 1993; Koch, 1996; Smeekens, 1998; Pego et al., 2000; Rolland et al., 2002; Gibson, 2005; Rolland and Sheen, 2005). Microarray studies also reveal widespread changes in cell function in response to carbohydrate status (Price et al., 2004; Thimm et al., 2004; Villadsen and Smith, 2004; Blasing et al., 2005; Li et al., 2006). In conditions of high carbohydrate demand, the increased expression of genes involved in photosynthesis (Koch, 1996; Smeekens, 1998; Moore et al., 2003) increases the production and mobilization of photosynthate. Conversely, when photosynthate is not immediately required, genes involved in storage and utilization (Rook et al., 2001, 2006; Baier et al., 2004) are activated to maintain a balance between photosynthate supply, demand, and storage.
Several regulatory pathways controlling sugar responses in plants have been identified by their conservation among plants, animals, and yeast (Smeekens, 1998; Rolland et al., 2002; Rolland and Sheen, 2005). Yeast hexokinase2 (Hxk2) is required for glucose-mediated catabolite repression (Entian, 1980). Human glucokinase complements glucose-signaling defects of the yeast hxk2 mutant, indicating a potential signaling function of mammalian glucokinases (Mayordomo and Sanz, 2001). In Arabidopsis thaliana, hexokinase1 (HXK1) performs dual functions as a glycolytic enzyme and a sugar response regulator (Moore et al., 2003). Members of the Arabidopsis SnRK1 family of protein kinases, which are most closely related to the yeast Sucrose-Nonfermenting1 (SNF1) protein kinase required for the derepression of glucose-repressed genes in yeast (Gancedo, 1998; Carlson, 1999), have been implicated in sugar signaling in plants (Halford and Hardie, 1998; Percell et al., 1998). The activities of two Arabidopsis SnRK1 proteins, AKIN10 and AKIN11, are regulated by PRL1 (for Pleiotropic Regulatory Locus1), an evolutionarily conserved α-importin binding nuclear WD protein (Nemeth et al., 1998; Bhalerao et al., 1999; Farras et al., 2001). Mutants in PRL1 display pleiotropic phenotypes, including transcriptional derepression of glucose-responsive genes and hypersensitivity to sugar and several hormones (Nemeth et al., 1998). G protein–coupled sugar signaling mechanisms were recently identified in Saccharomyces cerevisiae and in Arabidopsis (Lemaire et al., 2004; Huang et al., 2006). Mutant screens have also identified several other genes potentially involved in sugar signaling, such as low-β-amylase1 (Yoine et al., 2006) and impaired sucrose induction1 (Rook et al., 2001, 2006). The abscisic acid biosynthetic mutant aba2 and the abscisic acid response mutant abi4 (for ABA-insensitive4) have been consistently isolated in screens for reduced responses of seedlings to high levels of glucose or sucrose (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001; Cheng et al., 2002), demonstrating interactions between sugar-mediated and abscisic acid–mediated signaling pathways. In addition, the ethylene overproduction (eto1) and ethylene constitutive signaling (ctr1) mutants are glucose-insensitive (Zhou et al., 1998; Cheng et al., 2002). Consistent with this, several ethylene-insensitive mutants display glucose oversensitivity (Zhou et al., 1998; Leon and Sheen, 2003; Yanagisawa et al., 2003), indicating an overlap between sugar and ethylene signaling.
To further understand the molecular mechanisms involved in sugar responses and carbohydrate resource allocation, we have isolated Arabidopsis high sugar response (hsr) mutants that show enhanced luciferase activity and altered sugar responses using transgenic lines containing a luciferase reporter gene driven by the highly sugar-inducible promoter of the ApL3 gene (Baier et al., 2004). Here, we describe the HSR8 gene, which encodes UDP-d-xylose-4-epimerase, which is involved in cell wall arabinose biosynthesis and is allelic to MURUS4 (MUR4) (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003). We present evidence that cell wall changes caused by mutations in HSR8/MUR4 activate a sugar-responsive pathway requiring PRL1 that mediates gene expression, cell division, and cell expansion.
RESULTS
Identification and Genetic Characterization of the hsr8 Mutant
A transgenic Arabidopsis line, A3L3 (Columbia [Col-0] background), expressing firefly luciferase cDNA under the control of the sugar-inducible ApL3 promoter (Baier et al., 2004), exhibited very low luciferase activity on sucrose- or glucose-free medium and was inducible to high levels in an almost linear response on medium containing between 1 and 3% (w/v) sucrose or glucose (Baier et al., 2004). The hsr8-1 mutant exhibiting relatively high luciferase activity was isolated from ethyl methanesulfate–mutagenized M2 A3L3 seedlings grown on medium containing 1% (w/v) glucose (Figures 1A and 1B). There was no significant difference in luciferase levels between hsr8-1 and wild type seedlings grown on glucose-free medium or medium containing 1% mannitol (data not shown), demonstrating that the hsr8-1 mutant did not have a general increase in luciferase activity and exhibited increased sugar responsiveness. The expression of both the endogenous ApL3 gene and another sugar-responsive gene encoding Arabidopsis β-amylase (Mita et al., 1995) was also increased significantly in hsr8 in response to 1% (w/v) glucose compared with the parental line (A3L3) (Figure 1C). Genetic analysis showed that all of the F1 plants from crosses between A3L3 and hsr8-1 had the wild-type phenotype and that the F2 population showed a segregation ratio of three wild type to one mutant, indicating that hsr8-1 is a single recessive mutant.
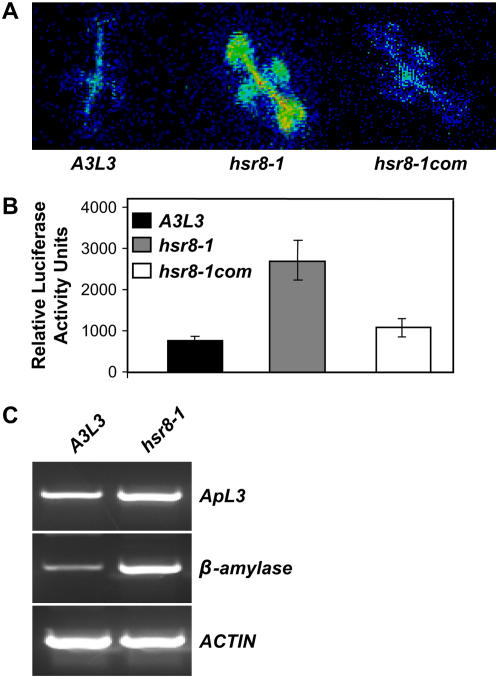
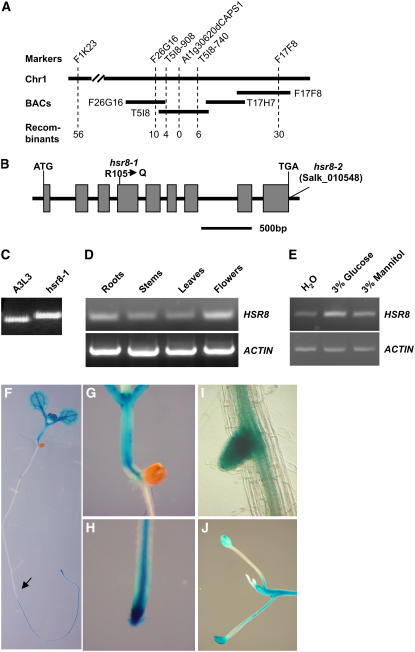
Figure 1.
Isolation of the hsr8-1 Mutant.
(A) Luciferase activity in the hsr8-1 mutant is higher than that in the wild type. A3L3 is the parental line of hsr8-1. hsr8-1com is hsr8-1 transformed with the genomic wild-type HSR8 sequence.
(B) Luciferase levels were measured in 9-d-old seedlings of the A3L3 parental line, hsr8-1, and hsr8-1com grown on Murashige and Skoog (MS) medium containing 1% glucose. Seedlings were sprayed with luciferin and analyzed luminometrically. Error bars represent se (n > 60).
(C) RT-PCR analysis of transcript levels in the hsr8-1 mutant. RT-PCR was performed on first-strand cDNA made from 9-d-old seedlings grown in constant light on medium containing 1% glucose. cDNA was standardized by reference to an actin standard.
hsr8 Plants Have Altered Starch, Chlorophyll, and Anthocyanin Levels
The ApL3 gene encodes one of four large subunits of ADPG-pyrophosphorylase (AGPase), which catalyzes the first committed step in starch synthesis. Starch levels were increased significantly in the hsr8-1 mutant compared with the parental line A3L3 (Table 1), consistent with increased expression of ApL3. Anthocyanins also accumulated to higher levels in hsr8-1 (Martin et al., 2002) compared with the parental line A3L3 (Table 1). High levels of exogenous glucose and sucrose antagonize the light-dependent induction of photogene expression (Krapp et al., 1993; Martin et al., 2002). The chlorophyll content of the hsr8-1 mutant was lower than that of the parental line A3L3 (Table 1), whereas there was no significant difference in response to 3% mannitol (data not shown), suggesting that hsr8-1 further represses chlorophyll synthesis on high-sugar medium. These results indicated that the hsr8-1 mutation enhances several glucose-mediated metabolic responses.
Table 1.
Comparison of Starch, Chlorophyll, and Anthocyanin Levels between the Wild Type and hsr8-1
| Sample | A3L3 | hsr8-1 |
|---|---|---|
| Starch (mol/mg fresh weight) | 28.39 ± 5.55 | 51.11 ± 7.67 |
| Chlorophyll (mg/g fresh weight) | 0.75 ± 0.01 | 0.53 ± 0.05 |
| Anthocyanin (E530 nm/g fresh weight) | 12.38 ± 0.07 | 16.38 ± 0.24 |
hsr8 Seedlings Exhibit Glucose-Hypersensitive Dark Development and Hypocotyl Elongation
Dark-grown Arabidopsis seedlings develop leaf- and flower-like organs when exogenous sugars are supplied directly to the developing shoot meristem (Roldan et al., 1999). Seedlings of the A3L3 parental line grown on medium containing 0.05% (w/v) glucose for 3 weeks did not develop beyond a slight opening of the cotyledonary petioles and expansion of the cotyledon (Figure 2A). At 0.5% (w/v) glucose, the cotyledonary petioles were fully expanded and true leaves had just started to develop (Figure 2B), and at 1.0% (w/v) glucose, the first pair of true leaves had developed and a clear internode was apparent (Figure 2C). Propidium iodide staining revealed an increase in leaf primordia size and cell number in 8-d dark-grown A3L3 seedlings in response to increasing concentrations of glucose (Figures 2G to 2I). These results show that dark development is a progressive response to low levels of exogenous glucose. Dark development was then compared in 14-d-old dark-grown A3L3 and hsr8-1 seedlings grown on medium containing 1% glucose. A3L3 seedlings had just started to develop the true leaves in these conditions (Figures 2D and 5A), while at this stage most hsr8-1 seedlings had fully expanded cotyledonary petioles, the first true leaves had formed, and an internode was visible (Figures 2D and 5A). This enhanced dark development in hsr8-1 was also observed when seedlings were grown on medium containing increasing concentrations of glucose for 8 d (Figures 2J to 2L). The increased dark development in the hsr8-1 mutant was not a result of earlier germination (data not shown) or an osmotic effect, because seedlings never developed beyond a slight opening of the cotyledonary petioles and expansion of the cotyledon on medium without sugar (Figure 2E) or containing 1% mannitol (Figure 2F).
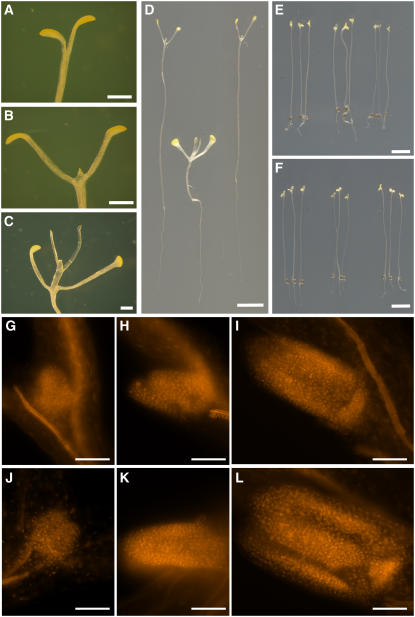
Figure 2.
Glucose-Hypersensitive Dark Development in the hsr8-1 Mutant.
(A) to (C) Dark development of Col-0 seedlings grown on medium containing 0.05% glucose (A), 0.5% glucose (B), and 1% glucose (C) for 21 d.
(D) to (F) Fourteen-day-old dark-grown seedlings of the wild type (right), hsr8-1 (middle), and hsr8-1com (left) grown on MS medium (E) and MS medium containing 1% glucose (D) and 1% mannitol (F).
(G) to (I) The leaf primordium of 8-d-old dark-grown seedlings of A3L3 grown on MS medium (G) and MS medium containing 0.5% glucose (H) and 1% glucose (I).
(J) to (L) The leaf primordium of 8-d-old dark-grown hsr8-1 seedlings on MS medium with no glucose (J), 0.5% glucose (K), and 1% glucose (L).
Bars = 1 mm in (A) to (C), 0.5 cm in (D), 0.2 cm in (E) and (F), and 0.1 μm in (G) to (L).
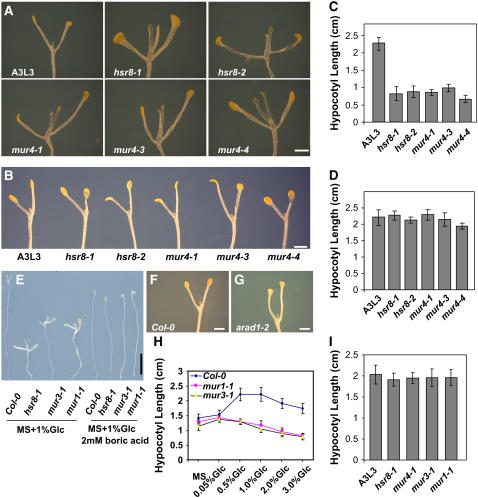
Figure 5.
Dark Development Phenotypes of Cell Wall Biosynthetic Mutants.
(A) Dark development phenotypes of 14-d-old wild-type Col-0, mur4, and hsr8 alleles grown on MS medium supplemented with 1% glucose. All hsr8 alleles exhibit increased dark development compared with the wild type.
(B) Dark development phenotypes of 14-d-old wild-type Col-0 and hsr8 alleles grown on MS medium supplemented with 1% glucose and 30 mM l-arabinose. The increased dark development phenotypes of hsr8 mutants in response to glucose were restored to wild-type levels by exogenous l-arabinose.
(C) Hypocotyl lengths of 14-d-old dark-grown seedlings of the wild type and hsr8 mutants grown on MS medium supplemented with 1% glucose.
(D) Hypocotyl lengths of 14-d-old dark-grown seedlings of the wild type and hsr8 mutants grown on MS medium supplemented with 1% glucose and 30 mM l-arabinose. The reduced hypocotyl elongation of hsr8 mutants was rescued by l-arabinose.
(E) Dark development phenotypes of 14-d-old wild type Col-0, hsr8-1, mur3-1, and mur1-1 alleles grown on MS medium supplemented with 1% glucose (left) or with 1% glucose + 2 mM boric acid (right). Dark development and hypocotyl length of hsr8-1, mur4-1, mur3-1, and mur1-1 were restored to wild-type levels (see [I]).
(F) and (G) Dark development phenotypes of 14-d-old wild type Col-0 (F) and arad1-2 (G) alleles grown on MS medium supplemented with 1% glucose.
(H) Hypocotyl lengths of 14-d-old dark-grown seedlings of Col-0, mur1-1, and mur3-1 grown on MS medium supplemented with different glucose concentrations.
(I) Hypocotyl lengths of 14-d-old dark-grown seedlings of Col-0, mur1-1, and mur3-1 grown on MS medium supplemented with 1% glucose + 2 mM boric acid.
Error bars represent sd (n > 30). Bars = 0.5 mm in (E) and 1 mm in (A), (B), (F), and (G).
In dark-grown A3L3 seedlings, hypocotyl length increased in response to 0.05 and 0.5% glucose and elongation was progressively inhibited at glucose concentrations between 1 and 3% (Figure 3A). Hypocotyl elongation in hsr8-1 seedlings was similar to that of wild-type seedlings at no or low glucose levels (Figure 3A) but became progressively shorter and thicker than in A3L3 (Figures 2D and 3A to 3D) at higher glucose levels. Thus, hsr8-1 seedlings displayed glucose-hypersensitive inhibition of hypocotyl elongation. hsr8-1 hypocotyl epidermal cells were significantly shorter and fatter than wild-type cells (Figures 3B and 3C). There were no significant differences in the number of epidermal cells of hypocotyls between wild-type and hsr8-1 seedlings (data not shown), showing that the inhibition of hypocotyl elongation by glucose in hsr8-1 was largely attributable to reduced hypocotyl cell elongation rather than reduced cell division.
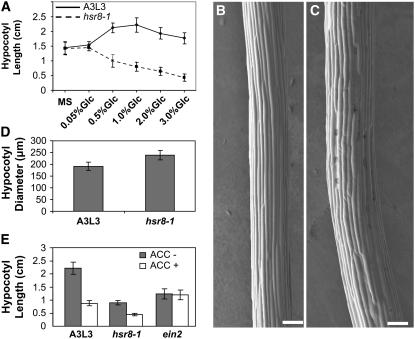
Figure 3.
Glucose-Hypersensitive Cell Elongation in the hsr8-1 Mutant.
(A) Hypocotyl lengths of 14-d-old dark-grown seedlings of A3L3 and hsr8-1 grown on MS medium supplemented with increasing glucose concentrations. Error bars represent sd (n > 30).
(B) and (C) Scanning electron microscope images of 4-d-old dark-grown hypocotyls of the parental line A3L3 (B) and hsr8-1 (C) grown on MS medium supplemented with 1% (w/v) glucose. Bars = 100 μm.
(D) Hypocotyl diameters of 4-d-old dark-grown seedlings of A3L3 and hsr8-1 grown on MS medium supplemented with 1% (w/v) glucose. Error bars represent sd (n > 10).
(E) Hypocotyl elongation in response to ACC treatment. Hypocotyl lengths of 14-d-old dark-grown seedlings of A3L3, hsr8-1, and ein2 mutants grown on MS medium supplemented with 1% glucose in the presence and absence of 10 μM ACC. Error bars represent sd (n > 30).
Ethylene inhibits the elongation of dark-grown hypocotyls. The reduction in length of dark-grown hypocotyls in response to 1-aminocyclopropane-carboxylic acid (ACC; an ethylene precursor) was not significantly different between A3L3 and the hsr8-1 mutant (Figure 3E), indicating normal ethylene response in the hsr8-1 mutant. The ethylene-resistant mutant ein2 was not responsive to ACC (Alonso et al., 1999), as expected. Therefore, hsr8-1 does not display altered ethylene responses previously associated with sugar response mutants (Yanagisawa et al., 2003).
hsr8 Has Normal Sugar Levels and Uptake
To rule out the possibility that increased cellular levels of sugars contribute to enhanced responses of hsr8-1 plants to exogenous glucose, glucose, fructose, sucrose, and total sugar levels were measured in 7-d-old seedlings of the hsr8-1 mutant and the parental line A3L3. No significant differences were seen (see Supplemental Figure 1A online). We tested whether altered glucose uptake contributes to the enhanced glucose responses observed in the hsr8-1 mutant. The uptake of [14C]glucose into 7-d-old seedlings of the hsr8-1 mutant, its parental line A3L3, a transgenic line expressing the glucose transporter STP1 under the control of the 35S promoter (Baier et al., 2004), and its parental Wassilewskija line was measured. There were no detectable differences in 14C uptake between the A3L3 parental line and the hsr8-1 mutant (see Supplemental Figure 1B online). The line expressing 35S:STP1 showed greatly elevated levels of [14C]glucose uptake, as expected. We concluded that neither altered intracellular sugar levels nor altered glucose uptake accounts for the enhanced glucose responses seen in the hsr8-1 mutant.
Positional Cloning and Expression Patterns of HSR8
The HSR8 gene was identified by map-based cloning in an F2 population of a cross between hsr8-1 and Landsberg erecta. The HSR8 gene was mapped into a 48-kb interval between markers T5I8-908 and T5I8-740 (Figure 4A) on chromosome 1. DNA sequencing revealed that hsr8 has a G-to-A transition in gene At1g30620, leading to a change from Arg-105 (CGG) to Gln-105 (CAG) (Figure 4B). At1g30620 is MUR4, encoding UDP-d-xylose-4-epimerase (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003). The derived cleaved-amplified polymorphic sequence (dCAPS) marker At1g30620dCAP1 was developed based on this mutation in hsr8-1, and it cosegregated with the hsr8-1 phenotypes (Figures 4A and 4C). A plasmid containing the entire At1g30620 open reading frame, the 2.6-kb promoter sequence, and the 1.5-kb downstream sequence was introduced into the hsr8, mur4-1, and mur4-3 mutants. Nearly all transgenic lines exhibited complementation of hsr8 phenotypes (Figures 1A, 1B, and 2D). Therefore, At1g30620 is the HSR8 gene. A T-DNA insertion into the 3′ untranslated region of At1g30620 (Salk_010548) was called hsr8-2 (Figure 4B), and the original ethyl methanesulfate–induced allele is henceforth called hsr8-1. All mutants (hsr8-2, mur4-1, mur4-3, and mur4-4) in the At1g30620 gene show the hsr8-1 dark development phenotypes (Figure 5A). F1 progeny of crosses of the five lines (hsr8-1, hsr8-2, mur4-1, mur4-3, and mur4-4) all exhibited a similar dark development phenotype, showing that these lines were allelic (data not shown).
Figure 4.
Map-Based Cloning and Expression Patterns of the HSR8 Gene.
(A) Fine-mapping of the HSR8 locus. The HSR8 locus was mapped to chromosome 1 (Chr1) between markers F1K23 and F17F8. The HSR8 locus was further refined to a 48-kb genomic DNA region between CAPS markers T5I8-908 and T5I8-740 and cosegregated with dCAPS marker At1g30620dCAPS1. The numerals at bottom indicate the number of recombinants identified from F2 plants.
(B) HSR8 gene structure, showing the mutated sites of the two hsr8 alleles. The start codon (ATG) and the stop codon (TGA) are indicated. Closed boxes indicate the coding sequences, and lines between boxes indicate introns. The mutation site in hsr8-1 and the T-DNA insertion site in hsr8-2 also are shown.
(C) The mutation in hsr8-1 was measured with the dCAPS marker At1g30620dCAPS1.
(D) RT-PCR analysis of HSR8/MUR4 gene expression. Total RNA was isolated from roots, stems, leaves, and flowers.
(E) Expression of HSR8 in response to glucose after 6 h of treatment.
(F) to (J) Histochemical analysis of GUS activity in a HSR8pro:GUS seedling (F), hypocotyl and shoot apices (G), a primary root (H), a lateral root (I), and a dark-grown seedling (J).
HSR8 transcripts were detected in various tissues by RT-PCR analysis, including roots, stems, leaves, and flowers (Figure 4D), consistent with previous RNA gel blot analysis (Burget et al., 2003). Figure 4E shows that HSR8 expression in light-grown seedlings is elevated in response to glucose, consistent with previous microarray data (Li et al., 2006). The tissue-specific expression patterns of HSR8 were assessed using histochemical assay of β-glucuronidase (GUS) activity of transgenic plants containing a HSR8 promoter:GUS fusion (HSR8pro:GUS). High levels of GUS activity were detected in the shoot and root apices and hypocotyls of both light- and dark-grown seedlings (Figures 4F to 4J). These expression patterns and induction by glucose are consistent with the dependence of glucose-responsive hypocotyl elongation and shoot apical development on HSR8 function in many actively growing tissues (Figure 2).
Altered Sugar Responses in hsr8 Are Rescued by Arabinose
Mutations in MUR4 lead to a reduction in l-arabinose levels in most organs and affect arabinose-containing pectic cell wall polysaccharides and arabinogalactan proteins (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003). Cell wall monosaccharide composition in the parental line A3L3, hsr8-1, and mur4-1 was determined by gas chromatography of alditol acetate derivatives. Table 2 shows that the amounts of arabinose in hsr8-1 and mur4-1 were reduced to 54.9 and 50.2% of A3L3 levels, respectively, consistent with previously reported results (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003). Feeding l-arabinose to mur4 plants restored cell wall monosaccharide composition to wild-type levels (Burget and Reiter, 1999), showing that l-arabinose can be used directly by plants through a salvage pathway as a source of arabinosyl units for polymer synthesis. Treatment with 1% glucose and 30 mM arabinose fully restored the enhanced dark development and glucose-hypersensitive hypocotyl elongation phenotypes of hsr8-1 and mur4 seedlings to that of wild-type seedlings (Figures 5B to 5D).
Table 2.
Comparison of Cell Wall Monosaccharide Composition in A3L3 and the hsr8-1, mur4-1, prl1-3, and mur4-1 prl1-3 Mutants
| Genotype | Arabinose | Rhamnose | Fucose | Xylose | Mannose | Galactose | Glucose |
|---|---|---|---|---|---|---|---|
| A3L3 | 17.28 ± 0.31 | 12.33 ± 0.10 | 3.51 ± 0.07 | 39.38 ± 0.12 | 17.35 ± 0.08 | 27.47 ± 0.43 | 173.68 ± 5.11 |
| hsr8-1 | 9.49 ± 0.36 | 12.58 ± 0.14 | 3.27 ± 0.03 | 39.27 ± 0.45 | 17.76 ± 0.27 | 30.11 ± 1.01 | 194.21 ± 3.28 |
| mur4-1 | 8.68 ± 0.33 | 12.57 ± 0.36 | 3.52 ± 0.00 | 37.47 ± 0.33 | 19.21 ± 0.08 | 27.62 ± 0.16 | 195.56 ± 2.50 |
| prl1 | 14.81 ± 0.03 | 12.49 ± 0.41 | 3.84 ± 0.14 | 36.34 ± 0.83 | 17.51 ± 0.38 | 25.35 ± 0.11 | 174.63 ± 1.82 |
| mur4-1 prl1 | 8.53 ± 0.03 | 12.90 ± 0.06 | 3.16 ± 0.01 | 37.58 ± 0.88 | 18.62 ± 0.42 | 29.27 ± 0.15 | 188.66 ± 0.83 |
The sugar content of cell walls are means ± se of three independent assays. Each wall component was calculated as milligrams per gram of total cell wall residues.
l-Arabinose is found mainly in the arabinogalactan side chains of pectins, glucuronoarabinoxylans, Hyp-rich glycoproteins, and cell wall arabinogalactan proteins (AGPs) (Carpita and Gibeaut, 1993). Mutations in the ARAD1 gene (At2g35100) cause a specific decrease in arabinan side chains of rhamnogalacturonan I (RG-I), a major component of pectins (Harholt et al., 2006). To test whether specific changes in RG-I cause sugar responses in hsr8/mur4, we tested the dark development responses of the arad1-2 mutant. Figures 5F and 5G show no significant differences in dark development phenotype between the wild type and arad1-2, suggesting that alterations in arabinan side chains of RG-I did not significantly affect the increased dark development phenotype in the hsr8/mur4 mutant. mur4 mutations reduce the arabinose content of both cell wall polysaccharides and AGPs (Burget and Reiter, 1999). AGPs have been implicated in many aspects of cell development, including cell proliferation, cell expansion, organ extension, apoptosis, germination, and somatic embryogenesis (Willats and Knox, 1996; Majewska-Sawka and Nothnagel, 2000; Gaspar et al., 2001; Lee et al., 2005). We tested whether altered AGP function could account for altered sugar responses in hsr8-1 using β-d-glucosyl Yariv (βGlcY) (Yariv et al., 1962, 1967) to disrupt AGP function (Willats and Knox, 1996). Roots of wild-type seedlings grown for 14 d in 50 μM βGlcY were significantly shorter than those of untreated seedlings, while the growth of roots of seedlings grown in the same concentration of β-d-mannosyl Yariv (βManY), which does not bind to AGPs, showed no significant difference compared with that of untreated seedlings (see Supplemental Figures 2E and 2G online). In addition, reduction in root growth was proportional to concentrations of βGlcY supplied in the medium (see Supplemental Figure 2G online). This indicated that AGP function was altered by βGlcY. However, sugar-responsive phenotypes such as dark development and hypocotyl elongation were not affected significantly by βGlcY (see Supplemental Figures 2A to 2D and 2F online), suggesting that altered AGP function did not significantly affect the enhanced sugar responses in hsr8-1.
Dark-Grown mur1 and mur3 Seedlings Exhibit Similar Sugar Responses to hsr8/mur4
We investigated whether other cell wall biosynthetic mutants also affect dark development. mur1-1 and mur2-1 have decreased cell wall fucose (Bonin et al., 1997; Vanzin et al., 2002), mur3-1 has altered xyloglucan structure (Madson et al., 2003), mur5-1, mur6-1, and mur7-1 have decreased cell wall arabinose (Reiter et al., 1997), mur8-1 has reduced cell wall rhamnose (Reiter et al., 1997), mur9-1 has reduced cell wall xylose and fucose (Reiter et al., 1997), radial swelling phenotype1 (rsw1) and isoxaben resistance2 (ixr2) have reduced cellulose synthesis (Arioli et al., 1998; Fagard et al., 2000; Desprez et al., 2002), and irx4 has 50% less lignin than wild-type plants (Jones et al., 2001). mur2-1, mur5-1, mur6-1, mur7-1, mur8-1, mur9-1, rsw1, ixr2, and irx4 seedlings showed similar glucose-responsive dark development to the wild type. By contrast, mur1-1 (in At3g51160) and mur3-1 (in At2g03220) seedlings had similar levels of enhanced dark development in response to glucose as the hsr8-1 mutant (Figure 5E) and similar glucose-hypersensitive reduction in hypocotyl elongation (Figures 5E and 5H). These data showed that several different cell wall changes, including reduced arabinose and fucose content and altered xyloglucan structure, can lead to altered glucose-responsive growth and development. Furthermore, the strong alleles mur4-1, mur4-3, and mur4-4, which have less cell wall arabinose than the weaker alleles mur4-2, mur5-1, mur6-1, and mur7-1 (Reiter et al., 1997), had a greater extent of glucose-responsive dark development and hypocotyl elongation than the weaker allele (Figures 5A and 5C; see Supplemental Figure 3 online), suggesting that only major changes in the cell wall may lead to sugar-hypersensitive development and hypocotyl elongation.
Altered Sugar Responses in hsr8/mur4, mur1, and mur3 Are Rescued by Boric Acid
Boron, an essential element for higher plants, maintains the integrity of cell walls (Takano et al., 2002) by cross-linking hydroxyl groups of cell wall carbohydrates such as pectins (Hu and Brown, 1994) and RG-II (O'Neill et al., 2001) with borate diester bonds. As mur1-1 has altered borate (O'Neill et al., 2001) and sugar responses, and treatment of mur1-1 plants with low concentrations of borate rescued their growth defects, the effect of borate treatment on sugar-dependent dark development and hypocotyl elongation was tested. In medium containing 1% glucose and 2 mM boric acid, the glucose-hypersensitive dark development and hypocotyl elongation phenotypes of hsr8-1, mur4-1, mur3-1, and mur1-1 seedlings were restored to those of wild-type seedlings (Figures 5E and 5I). This suggested that changes in the cell wall, such as those mediated by borate cross-linking of several cell wall components such as RG-II, xyloglucan, and arabinan-containing polymers, may lead to the sugar-dependent dark development phenotypes of hsr8-1, mur4-1, mur3-1, and mur1-1 seedlings.
Glucose-Responsive Growth and Development in hsr8 Plants Is Suppressed by prl1
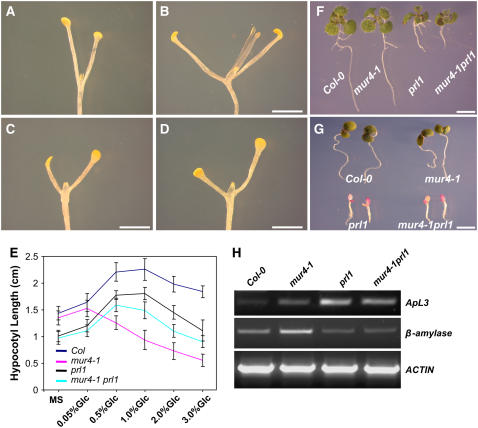
Mutations in PRL1 (At4g15900) lead to transcriptional derepression of glucose-responsive genes and increase the sensitivity of plants to growth hormones, including cytokinin, ethylene, abscisic acid, and auxin (Nemeth et al., 1998). Since the Arabidopsis prl1 mutant shows hypersensitivity to sugars (Nemeth et al., 1998), we tested the dark development responses of prl1 seedlings in response to glucose. Dark development (Figures 6A and 6C) and the pattern of hypocotyl elongation responses (Figure 6E) were essentially similar in prl1 and wild-type Col-0 seedlings, although hypocotyl length was slightly and equally reduced in prl1 compared with Col-0. This suggested that the sugar-responsive pathways controlling dark development and hypocotyl elongation in wild-type seedlings have little or no dependence on PRL1. To investigate whether PRL1 is involved in hsr8/mur4-mediated sugar responses, dark development and hypocotyl elongation phenotypes were assessed in a mur4-1 prl1 double mutant and compared with those of its parents. The mur4-1 prl1 double mutant exhibited reduced dark development (Figure 6D) compared with mur4-1 (Figure 6B) and similar to that seen in prl1 (Figure 6C). The double mutant also displayed similar glucose-responsive hypocotyl growth to prl1 (Figure 6E), indicating that prl1 suppressed the hsr8/mur4 glucose-hypersensitive dark development and hypocotyl elongation phenotypes. Therefore, the enhanced sugar responses seen in hsr8/mur4 seedlings require PRL1 function. Furthermore, light-grown mur4-1 prl1 double mutant seedlings had similar phenotypes to prl1 (Figures 6F and 6G), and the expression patterns of the ApL3 and At β-amylase genes in prl1 (Figure 6H) suggested that prl1 is also epistatic to hsr8/mur4 in the light. Analysis of cell wall monosaccharide composition revealed that the mur4-1 prl1 double mutant had similar reduced cell wall arabinose levels as mur4-1 (Table 2), indicating that the suppression of hsr8/mur4 growth and developmental phenotypes by prl1 is not due to an alteration of cell wall arabinose content seen in the mur4-1 mutant. Analysis of mur4-1 gin2-1 and hsr8-1 abi4-1 double mutants indicated that GIN2 and ABI4, which have well-established roles in sugar signaling (Rolland et al., 2006), were not required for the dark development phenotype in hsr8/mur4 seedlings (data not shown).
Figure 6.
The Glucose-Responsive Phenotypes of Dark-Grown mur4-1 Seedlings Are Suppressed by prl1.
(A) to (D) Fourteen-day-old dark-grown seedlings of the wild type (A), mur4-1 (B), prl1 (C), and mur4-1 prl1 (D) grown on MS medium supplemented with 1% glucose.
(E) Hypocotyl lengths of 14-d-old dark-grown seedlings of the wild type, prl1, mur4-1, and mur4-1 prl1 grown on MS medium supplemented with different glucose concentrations. Error bars represent sd (n > 30).
(F) and (G) Constant light–grown seedlings of the wild type, mur4-1, prl1, and mur4-1 prl1 grown on MS medium supplemented with 1% glucose (F) and 3% glucose (G).
(H) RT-PCR analysis of transcript levels in Col-0, mur4-1, prl1, and mur4-1 prl1. RT-PCR was performed on first-strand cDNA made from 9-d-old seedlings grown in constant light on medium containing 1% glucose. cDNA was standardized by reference to an actin standard.
Bars = 2 mm in (A) to (D) and 1 cm in (F) and (G).
Gene Expression Profiles in mur4-1, prl1, and mur4-1 prl1
Genome-wide gene expression analysis using ATH1 Arabidopsis whole genome microarray chips was conducted on RNA isolated from seedlings of Col-0, mur4-1, prl1, and mur4-1 prl1 6-d-old dark-grown seedlings. At this early stage of dark development, all seedlings showed a similar degree of growth and development. Compared with wild-type Col-0, expression levels of 92 genes were increased and those of 97 genes were reduced in mur4-1. Approximately one-third (30) of the genes with increased expression in mur4-1 encoded enzymes involved in cell wall metabolism, such as xyloglucan endotransglycosylase, pectin acetylesterase, and glycosyl hydrolase, proteins associated with structural constituents of the cell wall (e.g., Pro-rich extensin proteins), enzymes involved in primary metabolism, transcription factors, and regulatory proteins (see Supplemental Table 1 online). Consistent with the sugar-hypersensitive phenotypes of mur4-1, the expression levels of several classes of sugar-regulated genes (Li et al., 2006) were also increased in mur4-1, such as lipid transfer and seed storage proteins, enzymes of carbohydrate metabolism, transcription factors, and regulatory proteins. The main class of genes with reduced expression in mur4-1 (10%) were also those involved in cell wall–related functions, such as AGP14 and AGP22. A significant proportion were also involved in disease and stress responses (e.g., RD22), transcription, and regulatory processes (see Supplemental Table 1 online).
Compared with wild-type Col-0, expression levels of 74 genes were increased and those of 112 genes were repressed in prl1 (see Supplemental Table 2 online). Consistent with PRL1 function in derepressing sugar-repressed genes (Nemeth et al., 1998), the expression of a set of light-activated and sugar-repressed genes, such as genes encoding photosystem subunits (e.g., psbA photosystem II protein and photosystem II G protein), and a/b binding proteins, such as LHCB3 and LHCB6, was increased in prl1. The expression of genes involved in sugar metabolism and transport was also upregulated in prl1, including genes encoding a glucose-6-phosphate/phosphate translocator, glycerol-3-phosphate transporter, pyruvate kinase, and 2-isopropylmalate synthase2, consistent with the accumulation of free sugars in prl1 (Nemeth et al., 1998). Among genes with reduced expression in prl1 were those involved in stress responses, such as RD21A, RD29B, RAB18, low-temperature-induced LTI30, and drought-induced Di21, consistent with altered abscisic acid responses reported in prl1 (Nemeth et al., 1998). Other major classes of genes differentially regulated in prl1 were those encoding proteins and enzymes involved in cell wall modification, transcription factors, and protein kinases.
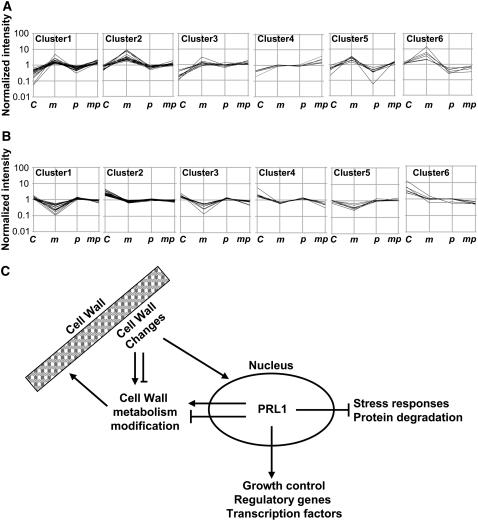
To gain insight into the mechanisms by which prl1 may suppress the sugar-responsive phenotypes of mur4-1, we classified the genes with increased and reduced expression in mur4-1 into different clusters using the quality threshold method (Li et al., 2006) according to their expression profiles in Col-0, mur4-1, prl1, and the mur4-1 prl1 double mutant (Figures 7A and 7B). These expression profiles were confirmed by quantitative RT-PCR analysis of eight selected genes (see Supplemental Figure 4 online). In the 92 genes upregulated in mur4-1, the expression of 31 genes in cluster 2 and cluster 6 was suppressed to levels seen in the wild type in mur4-1 prl1 (see Supplemental Table 3 online). A significant proportion of these genes (nine) encoded proteins associated with cell wall cell metabolism and modification, including xyloglucan endotransglycosylase, putative pectinesterases, and Pro-rich extensin proteins. Other classes of genes encoding lipid transfer proteins, storage proteins, and regulatory proteins, such as nonsymbiotic hemoglobin1 (GLB1) and PSK5, were also suppressed to levels seen in Col-0. Plants overexpressing GLB1 have considerably faster growth of both roots and shoots compared with control plants (Hunt et al., 2002), suggesting that GLB1 may be required for enhanced dark development in mur4-1. PSK-α, a peptide growth factor, has several biological activities, including promoting plant cell proliferation (Yang et al., 2001). In the set of 97 genes downregulated in mur4-1, the expression of 41 genes in cluster 1, cluster 3, and cluster 5 was restored to wild-type levels in mur4-1 prl1 (see Supplemental Table 3 online). Genes encoding stress-responsive proteins (RD22) and disease-responsive proteins, cell wall–related proteins, ubiquitin-protein ligases, and protein kinase (GPK1) formed a major group. Genes encoding components of protein degradation pathways are repressed by glucose, whereas the expression of genes involved in protein biosynthesis is upregulated by glucose (Li et al., 2006). Consistent with the sugar-hypersensitive phenotypes seen in mur4-1, the expression of two ubiquitin-protein ligases was repressed in mur4-1 (see Supplemental Table 1 online). Their expression was restored to the wild-type level in mur4-1 prl1 (see Supplemental Table 3 online), suggesting that PRL1 mediates the expression of protein degradation pathways activated in mur4-1.
Figure 7.
Gene Expression Profiles in mur4-1, prl1, and mur4-1 prl1.
(A) Upregulated genes in mur4-1 were classified into six clusters according to their expression profiles in the wild type, mur4-1, prl1, and mur4-1 prl1. Cluster numbers and mutant alleles are indicated as follows: C, Col-0; m, mur4-1; p, prl1; mp, mur4-1 prl1.
(B) Downregulated genes in mur4-1 were classified into six clusters according to their expression profiles in the wild type, mur4-1, prl1, and mur4-1 prl1. Cluster numbers and mutant alleles are indicated as in (A).
(C) Model of cell wall signaling based on mutant and gene expression analysis.
DISCUSSION
Cell Division and Cell Elongation Responses to Sugars Are Altered in the hsr8-1 Mutant
Sugars modulate multiple plant growth, development, gene expression, and metabolic responses (Rolland et al., 2006). To identify genes involved in these responses, we have isolated hsr mutants exhibiting increased glucose-responsive expression of the ApL3 gene encoding a large subunit of AGPase, the first step of starch synthesis (Baier et al., 2004). In addition to increased ApL3 expression, the hsr8-1 mutant exhibited a range of hypersensitive sugar-responsive phenotypes, including increased starch and anthocyanin levels, reduced chlorophyll levels (Table 1), glucose-responsive hypocotyl elongation, and dark development, compared with the parental line (Figures 1 and 3). These phenotypes, which are similar to those found in other hsr mutants (Baier et al., 2004), revealed that mutations in the HSR8 gene enhance a wide range of responses to physiologically relevant sugar levels. During dark development, cell numbers in the shoot apex increase progressively in response to glucose (Figures 2G to 2L). The elevated cell division in response to low glucose levels seen in the hsr8-1 mutant suggests that this mutant affects links between sugar availability and cell proliferation. In cultured Arabidopsis cells, sucrose depletion arrests cells at the G1-to-S transition (Riou-Khamlichi et al., 2000) due to depleted CYCD3;1 levels (Menges et al., 2006), suggesting a potential mechanism to explain this phenotype. The increased hypocotyl cell elongation in response to low glucose concentrations and increased inhibition of hypocotyl elongation at higher glucose concentrations (Figure 3A) observed in the hsr8-1 mutant show that cell elongation regulation by sugars is also altered in the hsr8-1 mutant. These glucose-responsive cell division and cell elongation responses will be useful phenotypes in genetic screens for identifying mutants affecting sugar-dependent responses.
The hsr8 Mutation Establishes a Link between Cell Wall Changes and Sugar-Responsive Growth and Development
We demonstrated that HSR8 is At1g30620, previously identified as MUR4 encoding UDP-d-xylose-4-epimerase, which catalyzes the first step of arabinose synthesis in the Golgi (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003). Like hsr8-1, mur4 mutations do not exhibit light-grown phenotypes (Reiter et al., 1997; Burget and Reiter, 1999; Burget et al., 2003); consequently, their sugar-dependent growth and development phenotypes have not been recognized previously. The glucose-hypersensitive growth and developmental phenotypes caused by mutations in HSR8/MUR4 were abrogated by growth on low levels of arabinose, further confirming the identity of the HSR8 gene and revealing a link between arabinose biosynthesis and sugar-responsive growth and development. Among a variety of other mutants known to affect cell wall composition, the mur1-1 and mur3-1 mutations also exhibited hypersensitive sugar-responsive growth and development. mur1-1 is a mutation in a gene encoding GDP-d-mannose-4,6-dehydratase, which catalyses the first step in fucose synthesis (Bonin et al., 1997), and mur3-1, a mutation in a xyloglucan-specific galactosyltransferase, alters xyloglucan structure (Madson et al., 2003).
A wide range of cell wall polysaccharides are affected by the mur1, mur3, and mur4 mutations. It is conceivable that changes in several cell wall components, or a single component affected by all three mutants, may lead to altered sugar-responsive growth. Mutations in MUR4 cause decreased arabinose in pectins, xylans, xyloglucans, and AGPs (Burget and Reiter, 1999). Arabinosylated glycans such as RG-I are believed to play important roles in plant development (Burget and Reiter, 1999), and AGPs have been assigned various roles in plant development, including embryogenesis, cell–cell interactions, fertilization, cell proliferation, and cell expansion (Gaspar et al., 2001; Lee et al., 2005). Altered dark development responses were not seen in the arad1-2 mutation, which specifically decreases the levels of arabinan side chains of RG-I (Harholt et al., 2006). Furthermore, disruption of AGP function with βGlcY reagent also had no significant effect on dark development responses. This raises the possibility that changes in other arabinose-containing moieties may lead to altered sugar-responsive growth and developmental responses. It is also feasible that changes in specific arabinose-containing moieties are insufficient to activate sugar responses and that overall reduction of cell wall arabinose levels leads to altered sugar responses. This is consistent with the observation that only large changes in the cell wall lead to sugar hypersensitivity. For example, the strong alleles mur4-1, mur4-3, and mur4-4, which have less cell wall arabinose than the weaker alleles mur4-2, mur5-1, mur6-1, and mur7-1 (Reiter et al., 1997), had a greater extent of glucose-responsive dark development and hypocotyl elongation than the weaker alleles (Figures 5A and 5C; see Supplemental Figure 3 online).
The mur1 and mur3 mutations lead to reduced cell wall fucose levels (Reiter et al., 1997). In mur3-1 plants, a fucosylated side chain in xyloglucans is absent, implicating xyloglucans in sugar responses (Madson et al., 2003). However, mur2-1 plants, which are deficient in a xyloglucan-specific fucosyl transferase that also affects xyloglucans, did not exhibit altered sugar-responsive dark development and growth phenotypes, suggesting that only some types of xyloglucans may modulate sugar responses. The structure and function of RG-II are known to be altered by the mur1 mutation (O'Neill et al., 2001). Treatment of mur1-1 plants with low concentrations of borate rescued their growth defects and resulted in an increase in the extent of borate cross-linking of RG-II (O'Neill et al., 2001), indicating that the dwarf phenotype and brittle cell walls of mur1 plants were consequences of reduced cross-linking of RG-II by borate diesters. Treatment of hsr8-1, mur4-1, mur3-1, and mur1-1 seedlings with 1% glucose and 2 mM boric acid also reduced the extent of sugar-responsive dark development to that of wild-type seedlings, and the severely reduced hypocotyl elongation was also reversed to that seen in the wild type (Figures 5E and 5I). This observation suggested that RG-II may be a common factor in altered sugar responses. However, borate is known to cross-link a variety of cell wall components in addition to RG-II (Blevins and Lukaszewski, 1998), including pectic moieties that are known to be altered in mur4 (Burget et al., 2003) and xyloglucans known to be altered in mur3 (Blevins and Lukaszewski, 1998; Madson et al., 2003); therefore, borate responsiveness does not provide conclusive evidence for a sole role for RG-II in sugar responses. Furthermore, mur3 has specific effects on the structure of xyloglucans and is not likely to alter RG-II function directly (Madson et al., 2003). Thus, the present level of analysis indicates that alterations in several cell wall components, including RG-II, a specific class of xyloglucans, and either a subset of arabinose-containing cell wall moieties or large-scale reductions in cell wall arabinose, activate sugar responses.
Other mutations that lead to cell wall changes also have major effects on growth and defense responses. The cev1 and eli1 mutations in Ce SA3 (encoding a cellulose synthase subunit) exhibit constitutive expression of ethylene- and jasmonate-responsive stress genes, enhanced resistance to fungal pathogens, and elevated lignin levels (Ellis et al., 2002; Cano-Delgado et al., 2003). Reduced callose synthase levels lead to enhanced salicylic acid–dependent disease resistance (Nishimura et al., 2003). Mutations in the RHM1 gene encoding a UDP-rhamnose synthase suppressed the root hair mutant lrx1, which encodes an extracellular Leu-rich repeat extensin protein (Diet et al., 2006). Collectively, those studies and this report illustrate that properties of the plant cell wall are actively monitored during growth and development and signaled to multiple cellular responses. Such mechanisms have been proposed to monitor the performance and alteration of the cell wall (Pilling and Hofte, 2003; Somerville et al., 2004). The recent discovery of a membrane-bound receptor kinase that mediates hypocotyl elongation in response to reduced cellulose synthesis reveals one mechanism linking cell wall changes to cellular functions (Hematy et al., 2007).
PRL1 Reveals a Potential Mechanism Linking the Cell Wall with Sugar-Responsive Phenotypes
Mutations in the PRL1 gene suppressed the glucose-hypersensitive growth and developmental phenotypes caused by mutations in the HSR8/MUR4 gene (Figure 6) without altering cell wall monosaccharide composition. This genetic analysis suggests that changes in the cell wall are signaled via the nucleus by a mechanism involving PRL1 in sugar-responsive processes (Nemeth et al., 1998).
The prl1 mutation causes a range of phenotypes, including hypersensitive responses to glucose and sucrose, cytokinins, auxin, ethylene, and abscisic acid (Nemeth et al., 1998). PRL1 encodes a nucleus-located WD repeat protein that interacts with and inhibits the activity of the Arabidopsis SnRK1 proteins AKIN10 and AKIN11, which are structurally and functionally related to yeast SNF1 and mammalian AMPK regulatory kinases (Bhalerao et al., 1999). This mechanism may alter carbohydrate-responsive growth and storage processes, consistent with their regulatory function in carbon metabolism in plants, animals, and yeast (Gancedo, 1998; Halford and Hardie, 1998; Carlson, 1999) and in glucose-dependent growth in the moss Physcomitrella patens (Thelander et al., 2004).
To further understand how prl1 suppresses hsr8/mur4-activated sugar responses, we performed whole genome array analysis using wild-type, mur4-1, prl1, and mur4-1 prl1 seedlings. Expression of light-activated and sugar-repressed genes was upregulated in prl1, supporting previous analyses of the expression of individual genes in prl1 seedlings (Nemeth et al., 1998). Furthermore, the expression of known sugar-inducible genes was also upregulated in prl1, consistent with sugar-hypersensitive phenotypes of this mutant (Nemeth et al., 1998).
The expression of a significant proportion of mur4-1–regulated genes was dependent upon PRL1. We classified mur4-1–regulated genes into different groups according to their expression profiles in the wild type, mur4-1, prl1, and mur4-1 prl1 to identify genes whose expression levels were restored to that of the wild type in the mur4-1 prl1 double mutant. The expression levels of three sets of genes whose functions are relevant to altered dark development and hypocotyl elongation phenotypes seen in mur4-1/hsr8-1 fell into this class. Expression of a large set of genes involved in cell wall metabolism and modification was restored to wild-type levels in the double mutant, suggesting that PRL1 is required for the regulation of mur4-activated transcriptional responses leading to compensatory cell wall changes. As proposed above, changes in the cell wall trigger these transcriptional responses. The increased expression of several growth regulatory genes such as GLB1 (Hunt et al., 2002) and At PSK5 (Yang et al., 2001) in mur4-1 was repressed to wild-type levels in mur4-1 plr1. The increased expression of these genes in mur4-1 may contribute to altered dark development and hypocotyl elongation. The expression of two ubiquitin ligase genes repressed in mur4-1 was restored to wild-type levels in the mur4-1 prl1 double mutant, suggesting that PRL1 regulates a protein degradation pathway involved in the regulation of mur4-1–activated responses. Finally, the expression of genes involved in stress and disease responses was restored to wild-type levels, suggesting that PRL1 is involved in the regulation of stress responses caused by cell wall defects in mur4-1.
Figure 7C shows a model of PRL1 function in the regulation of hsr8-1/mur4-1–activated sugar responses that is based on the sugar-hypersensitive growth and developmental phenotypes of hsr8-1/mur4-1 and an analysis of gene expression profiles. In this model, changes in the cell wall (in this case caused by mutants affecting polysaccharide synthesis) signal to PRL1 function in the nucleus, which then coordinates the expression of hormone- and stress-related genes (Nemeth et al., 1998) and genes related to cell wall modification and growth, leading to altered sugar-dependent growth and developmental responses. The regulatory pathway described in this report may contribute to the dynamic regulation of cell wall structure and function during growth and development and integrate metabolite regulation with the synthesis of new cell wall polysaccharides through PRL1 regulation of the SnRK1 proteins AKIN10 and AKIN11 (Bhalerao et al., 1999).
METHODS
Plant Material and Growth Conditions
All parental lines and mutants of Arabidopsis thaliana were in the Col-0 background, including A3L3, hsr8-1 (BC6 generation), hsr8-2 (SALK_010548), mur4-1(N8568), mur4-2 (N8569), mur4-3 (N8570), mur4-4 (N8571), mur1-1 (N6243), mur3-1 (N8566), prl1 (SALK_039427), and arad1-2 (SALK_029831). The Col-0 line is the parent of all of the mutants used. The A3L3 line is the parent of the hsr8-1 mutant and displayed identical responses to Col-0. The Wassilewskija line expressing the 35S:STP1 transgene was obtained from Steve Smith (University of Edinburgh). Seeds were surface-sterilized with 100% isopropanol and 20% (v/v) household bleach, washed at least five times with sterile water, stratified at 4°C for 6 d in the dark, dispersed on MS medium (Duchefa Biochemie) supplemented with 0.9% agar and 0.05, 0.5, 1, 2, and 3% glucose, exposed to light for 18 h, and then grown vertically in complete darkness at 23°C.
Identification of hsr8 Mutants
A3L3 (Col-0 background) seeds expressing a single copy of a T-DNA insert carrying an ApL3:LUC reporter gene were mutagenized and screened for increased luciferase activity as described (Baier et al., 2004). hsr8-1 seedlings showing higher luminescence than the parental line grown on the same plate were selected. hsr8-2 (SALK_010548) and mur4 were obtained from the Nottingham Arabidopsis Stock Centre. SALK_010548 T-DNA insertion in the HSR8 gene was confirmed by PCR and sequencing using primers SALK_010548LP (5′-TTTTCCCCAGATCAAAGGAAC-3′), SALK_010548RP (5′-TGGAATCAATTTGCCTTATGATC-3′), and LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′).
RNA Isolation, RT-PCR, and Quantitative Real-Time RT-PCR Analysis
Total RNA was extracted from Arabidopsis seedlings, roots, stems, leaves, and flowers using the RNeasy plant mini kit (Qiagen). RT-PCR analysis was performed as described (Li et al., 2006). cDNA samples were standardized on actin transcript amount using the primers AtactF and AtactR (Baier et al., 2004). The primers used for RT-PCR are as follows: AT5G09810-ACTIN (AtactF, 5′-GAGAAGATGACTCAGATC-3′, and AtactR, 5′-ATCCTTCCTGATATCGAC-3′); AT4G39210-ApL3 (ApL3F, 5′-CGTCTGAATCATGCAACC-3′, and ApL3R, 5′-GCATTTCCTGATCTTTGTATCCTCG-3′); AT4G15210-β-Amylase (β-AmyF, 5′-CGGAGAAGGGGAAGTTTTTC-3′, and β-AmyR, 5′-AATCTCATGCCCGTACTTCG-3′); and HSR8 (HSR8RTF, 5′-AACAACCTCATCGGTCTCGG-3′, and HSR8RTR, 5′-ATACTAATGAATGGCTGCTTCC-3′).
Quantitative real-time RT-PCR analysis was performed with an Opticon 2 DNA engine (MJ Research) using the SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich). TUB6 mRNA was used as an internal control, and relative amounts of mRNA were calculated using the comparative threshold cycle method. The primers used are described in Supplemental Table 5 online.
Propidum Iodide Staining
Samples were submerged in 10 mL of 37% formalin, 5 mL of propionic acid, and 70 mL of 100% ethanol overnight, dehydrated in 85, 95, and 100% ethanol, stained in propidum iodide solution (5 μg/mL) for 1 d, rinsed with 0.1 M l-Arg, pH 8.0, dehydrated in an ethanol series (15, 30, 50, 70, 85, 95, and 100%), cleared through an ethanol:xylene series (75:25%, 50:50%, 25:75%, and 100% xylene), and then viewed.
Carbohydrate, Chlorophyll, Anthocyanin, and Cell Wall Analyses
Carbohydrate, chlorophyll, and anthocyanin analyses were performed as described (Arnon, 1949; Galtier et al., 1995; Baier et al., 2004). Glucose, fructose, and sucrose were measured sequentially in cleared supernatants of K2CO3-neutralized HClO4 extracts of ground, frozen plant material (Galtier et al., 1995). Glucose uptake measurement was performed as described (Baier et al., 2004). For cell wall analysis, plant material was harvested and ground into a fine powder with a combination of liquid N2 and dry ice. For monosaccharide analysis, ground material was resuspended in 0.1 M MOPS/NaOH, pH 7.0, and centrifuged for 10 min at 15,000 rpm at 4°C. The pellet was washed twice with 0.25× extraction buffer, incubated for 1 h at 40°C in phenol:acetic acid:water (2:1:1, w/v/v), and centrifuged. The supernatant was discarded, and the pellet was washed twice with water and then freeze-dried. The neutral sugar content was determined after dispersion in 72% (w/w) H2SO4 for 3 h at 20°C, dilution to 1 M, and hydrolysis at 100°C for 2.5 h (Selvendran et al., 1979). The alditol acetates were prepared and analyzed by gas chromatography (Blakeney et al., 1983).
Scanning Electron Microscopy
Seedlings grown for 4 d in the dark were frozen in nitrogen slush at −190°C. Ice was sublimed at −90°C, and the specimen was sputter-coated and examined on an XL 30 FEG (Philips) cryoscanning electron microscope fitted with a cold stage.
Mapping of HSR8 and Complementation Tests
The HSR8 gene was mapped in the F2 population of crosses to Arabidopsis ecotype Landsberg erecta using simple sequence length polymorphism (Bell and Ecker, 1994) and CAPS (Konieczny and Ausubel, 1993) markers. To fine-map the HSR8 locus, new molecular markers were developed according to public databases (see Supplemental Table 4 online). A genomic DNA fragment containing the entire HSR8 coding region, the 2.6-kb upstream sequence, and the 1.5-kb downstream sequence was inserted into the binary vector pGreen to generate the transformation plasmid pGHSR8com for complementation. The plasmid pGHSR8com was introduced into the hsr8-1, mur4-1, and mur4-3 mutants using Agrobacterium tumefaciens GV3101, and transformants were selected on dl-phosphinothricin medium. The HSR8 promoter:GUS construct (HSR8pro:GUS) was made using a PCR-based Gateway system. The promoter-specific primers for the HSR8 gene were TOPAT1G30620PROM-F (5′-CACCTGTCAGCACGACGGAGTTGG-3′) and TOPAT1G30620PROM-R (5′-TTTGATTCACTTCAGCTGGCG-3′). PCR products were subcloned into the pENTR/D-TOPO vector (Invitrogen) using TOPO enzyme and sequenced. The HSR8 promoter was then subcloned into Gateway binary vector (pGWB3) containing the GUS reporter gene.
RNA Preparation, cRNA Synthesis, and Microarray Hybridization
Total RNA was extracted from 6-d-old dark-grown seedlings of Col-0, mur4-1, prl1, and mur4-1 prl1 using the RNeasy plant mini kit (Qiagen). Affymetrix GeneChip array expression profiling was performed at the Nottingham Arabidopsis Stock Centre according to Affymetrix Expression Analysis Technical Manual II (http://www.affymetrix.com/support/technical/manuals.affx). Two independent biological replicates were conducted. Microarray data analysis and clustering were performed as described (Li et al., 2006). Rank products (Breitling et al., 2004) were compared with the rank products of 10,000 random permutations of the same data to assign E-values. To correct for the multiple testing problem, we used the false discovery rate (Storey, 2003), in which the E-value of each gene was divided by its position in the list of changed transcripts. A false discovery rate < 0.05 means that only 5% or fewer genes up to this position are expected to be observed by chance (false positive) and the remaining 95% are true positives.
Accession Number
Microarray data from this article can be found in the ArrayExpress data library under accession number E-NASC-78.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Carbohydrate Levels and Uptake in the hsr8-1 Mutant.
Supplemental Figure 2. Dark Development and Hypocotyl Elongation of hsr8 Are Not Affected by βGlcY.
Supplemental Figure 3. Dark Development Phenotypes of the Wild Type and mur4-2.
Supplemental Figure 4. Quantitative Real-Time RT-PCR Analysis.
Supplemental Table 1. Analysis of Gene Expression Changes in mur4-1 using Affymetrix AtH1 Microarrays.
Supplemental Table 2. Analysis of Gene Expression Changes in prl1 using Affymetrix AtH1 Microarrays.
Supplemental Table 3. Cluster Analysis of Differentially Regulated Genes in mur4-1 and prl1.
Supplemental Table 4. PCR-Based Molecular Markers.
Supplemental Table 5. Primers Used in Quantitative Real-Time RT-PCR.
Supplementary Material
Acknowledgments
We thank Csaba Koncz for prl1 seeds, Steve Smith for 35S:STP1 transgenic lines, Georg Seifert and the Nottingham Arabidopsis Stock Centre for mur4 mutants, and Kim Findlay for assistance with scanning electron microscopy. This work was supported by Biotechnology and Biological Science Research Council Grant BB/C515620/1 and the Core Strategic Grant to the John Innes Centre.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael W. Bevan (michael.bevan@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. [DOI] [PubMed] [Google Scholar]
- Arnon, D.I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, M., Hemmann, G., Holman, R., Corke, F., Card, R., Smith, C., Rook, F., and Bevan, M.W. (2004). Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol. 134 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bhalerao, R.P., Salchert, K., Bako, L., Okresz, L., Szabados, L., Muranaka, T., Machida, Y., Schell, J., and Koncz, C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeney, A.B., Harris, P.J., Henry, R.J., and Stone, B.A. (1983). A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr. Res. 113 291–299. [Google Scholar]
- Blasing, O.E., Gibon, Y., Gunther, M., Hohne, M., Morcuende, R., Osuna, D., Thimm, O., Usadel, B., Scheible, W.R., and Stitt, M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins, D.G., and Lukaszewski, K.M. (1998). Boron in plant structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 481–500. [DOI] [PubMed] [Google Scholar]
- Bonin, C.P., Potter, I., Vanzin, G.F., and Reiter, W.D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. USA 94 2085–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Burget, E.G., and Reiter, W.D. (1999). The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphospho L-arabinose. Plant Physiol. 121 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget, E.G., Verma, R., Molhoj, M., and Reiter, W.D. (2003). The biosynthesis of L-arabinose in plants: Molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34 351–362. [DOI] [PubMed] [Google Scholar]
- Carlson, M. (1999). Glucose repression in yeast. Curr. Opin. Microbiol. 2 202–207. [DOI] [PubMed] [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3 1–30. [DOI] [PubMed] [Google Scholar]
- Cheng, W.H., Endo, A., Zhou, L., Penney, J., Chen, H.C., Arroyo, A., Leon, P., Nambara, E., Asami, T., Seo, M., Koshiba, T., and Sheen, J. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez, T., Vernhettes, S., Fagard, M., Refregier, G., Desnos, T., Aletti, E., Py, N., Pelletier, S., and Hofte, H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet, A., Link, B., Seifert, G.J., Schellenberg, B., Wagner, U., Pauly, M., Reiter, W.D., and Ringli, C. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian, K.D. (1980). Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catobolite repression in yeast. Mol. Gen. Genet. 178 633–637. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farras, R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier, N., Foyer, C.H., Murchie, E., Alred, R., Quick, P., Voelker, T.A., Thépenier, C., Lascève, G., and Betsche, T. (1995). Effects of light and atmospheric carbon dioxide enrichment on photosynthesis and carbon partitioning in the leaves of tomato (Lycopersicon esculentum L.) plants overexpressing sucrose phosphate synthase. J. Exp. Bot. 46 1335–1344. [Google Scholar]
- Gancedo, J.M. (1998). Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, Y., Johnson, K.L., McKenna, J.A., Bacic, A., and Schultz, C.J. (2001). The complex structures of arabinogalactan proteins and the journey towards understanding function. Plant Mol. Biol. 47 161–176. [PubMed] [Google Scholar]
- Gibson, S.I. (2005). Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 8 93–102. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., and Hardie, D.G. (1998). SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 37 735–748. [DOI] [PubMed] [Google Scholar]
- Harholt, J., Jensen, J.K., Sorensen, S.O., Orfila, C., Pauly, M., and Scheller, H.V. (2006). ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 140 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematy, K., Sado, P.E., Van Tuinen, A., Rochange, S., Desnos, T., Balzergue, S., Pelletier, S., Renou, J.P., and Hofte, H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17 922–931. [DOI] [PubMed] [Google Scholar]
- Hu, H., and Brown, P.H. (1994). Localization of boron in cell walls of squash and tobacco and its association with pectin (evidence for a structural role of boron in the cell wall). Plant Physiol. 105 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Taylor, J.P., Chen, J.G., Uhrig, J.F., Schnell, D.J., Nakagawa, T., Korth, K.L., and Jones, A.M. (2006). The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18 1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23 577–585. [DOI] [PubMed] [Google Scholar]
- Hunt, P.W., Klok, E.J., Trevaskis, B., Watts, R.A., Ellis, M.H., Peacock, W.J., and Dennis, E.S. (2002). Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99 17197–17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26 205–216. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 509–540. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Krapp, A., Hofmann, B., Schafer, C., and Stitt, M. (1993). Regulation of the expression of rbcS and other photogenic genes by carbohydrates: A mechanism for the “sink regulation” of photosynthesis. Plant J. 3 817–828. [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23 587–596. [DOI] [PubMed] [Google Scholar]
- Lee, K.J., Sakata, Y., Mau, S.L., Pettolino, F., Bacic, A., Quatrano, R.S., Knight, C.D., and Knox, J.P. (2005). Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17 3051–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, K., Van de Velde, S., Van Dijck, P., and Thevelein, J.M. (2004). Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16 293–299. [DOI] [PubMed] [Google Scholar]
- Leon, P., and Sheen, J. (2003). Sugar and hormone connections. Trends Plant Sci. 8 110–116. [DOI] [PubMed] [Google Scholar]
- Li, Y., Lee, K.K., Walsh, S., Smith, C., Hadingham, S., Sorefan, K., Cawley, G., and Bevan, M.W. (2006). Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Res. 16 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G.F., Caplan, J., Shoue, D.A., Carpita, N.C., and Reiter, W.D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, T., Oswald, O., and Graham, I.A. (2002). Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol. 128 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayordomo, I., and Sanz, P. (2001). Human pancreatic glucokinase (GlkB) complements the glucose signaling defect of Saccharomyces cerevisiae hxk2 mutants. Yeast 18 1309–1316. [DOI] [PubMed] [Google Scholar]
- Menges, M., Samland, A.K., Planchais, S., and Murray, J.A. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita, S., Suzuki-Fujii, K., and Nakamura, K. (1995). Sugar-inducible expression of a gene for beta-amylase in Arabidopsis thaliana. Plant Physiol. 107 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.H., Liu, Y.X., Hwang, I., Jones, T., and Sheen, J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336. [DOI] [PubMed] [Google Scholar]
- Nemeth, K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- O'Neill, M.A., Eberhard, S., Albersheim, P., and Darvill, A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294 846–849. [DOI] [PubMed] [Google Scholar]
- Pego, J.V., Kortstee, A.J., Huijser, C., and Smeekens, S.C. (2000). Photosynthesis, sugars and the regulation of gene expression. J. Exp. Bot. 51 (spec. No.): 407–416. [DOI] [PubMed]
- Percell, P.C., Smith, A.M., and Halford, N.G. (1998). Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase transcripts in leaves. Plant J. 14 195–202. [Google Scholar]
- Pilling, E., and Hofte, H. (2003). Feedback from the wall. Curr. Opin. Plant Biol. 6 611–616. [DOI] [PubMed] [Google Scholar]
- Price, J., Laxmi, A., St. Martin, S.K., and Jang, J.C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, W.D., Chapple, C., and Somerville, C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12 335–345. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Menges, M., Healy, J.M., and Murray, J.A. (2000). Sugar control of the plant cell cycle: Differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan, M., Gomez-Mena, C., Ruiz-Garcia, L., Salinas, J., and Martinez-Zapater, J.M. (1999). Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. 20 581–590. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57 675–709. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 (suppl.): S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., and Sheen, J. (2005). Sugar sensing and signaling networks in plants. Biochem. Soc. Trans. 33 269–271. [DOI] [PubMed] [Google Scholar]
- Rook, F., Corke, F., Baier, M., Holman, R., May, A.G., and Bevan, M.W. (2006). Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J. 46 1045–1058. [DOI] [PubMed] [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signaling. Plant J. 26 421–433. [DOI] [PubMed] [Google Scholar]
- Selvendran, R.R., March, J.F., and Ring, S.G. (1979). Determination of aldoses and uronic acid content of vegetable fiber. Anal. Biochem. 96 282–292. [DOI] [PubMed] [Google Scholar]
- Smeekens, S. (1998). Sugar regulation of gene expression in plants. Curr. Opin. Plant Biol. 1 230–234. [DOI] [PubMed] [Google Scholar]
- Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., Osborne, E., Paredez, A., Persson, S., Raab, T., Vorwerk, S., and Youngs, H. (2004). Toward a systems approach to understanding plant cell walls. Science 306 2206–2211. [DOI] [PubMed] [Google Scholar]
- Storey, J.D. (2003). The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 31 2013–2035. [Google Scholar]
- Takano, J., Noguchi, K., Yasumori, M., Kobayashi, M., Gajdos, Z., Miwa, K., Hayashi, H., Yoneyama, T., and Fujiwara, T. (2002). Arabidopsis boron transporter for xylem loading. Nature 420 337–340. [DOI] [PubMed] [Google Scholar]
- Thelander, M., Olsson, T., and Ronne, H. (2004). Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 23 1900–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Vanzin, G.F., Madson, M., Carpita, N.C., Raikhel, N.V., Keegstra, K., and Reiter, W.D. (2002). The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc. Natl. Acad. Sci. USA 99 3340–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadsen, D., and Smith, S.M. (2004). Identification of more than 200 glucose-responsive Arabidopsis genes none of which responds to 3-O-methylglucose or 6-deoxyglucose. Plant Mol. Biol. 55 467–477. [DOI] [PubMed] [Google Scholar]
- Willats, W.G., and Knox, J.P. (1996). A role for arabinogalactan proteins in plant cell expansion: Evidence from studies on the interaction of beta-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9 919–925. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S., Yoo, S.D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature 425 521–525. [DOI] [PubMed] [Google Scholar]
- Yang, H., Matsubayashi, Y., Nakamura, K., and Sakagami, Y. (2001). Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor. Plant Physiol. 127 842–851. [PMC free article] [PubMed] [Google Scholar]
- Yariv, J., Lis, H., and Katchalski, E. (1967). Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem. J. 105 1C–2C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv, J., Rapport, M.M., and Graf, L. (1962). The interaction of glycosides and saccharides with antibody to the corresponding phenylazo glycosides. Biochem. J. 85 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoine, M., Ohto, M.A., Onai, K., Mita, S., and Nakamura, K. (2006). The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signaling in Arabidopsis. Plant J. 47 49–62. [DOI] [PubMed] [Google Scholar]
- Zhou, L., Jang, J.C., Jones, T.L., and Sheen, J. (1998). Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc. Natl. Acad. Sci. USA 95 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.