Brad Day, Douglas Dahlbeck, and Brian J. Staskawicz (2006). NDR1 Interaction with RIN4 Mediates the Differential Activation of Multiple Disease Resistance Pathways in Arabidopsis. Plant Cell 18: 2782–2791.
In Figure 5A, sections of a protein gel image were inadvertently inserted that also had appeared in Figure 6. In addition, a single leaf panel was duplicated in Figure 7C. The experiments have been repeated as independent confirmation of the data, and the corrected images for Figures 5A and 7C appear below. Please note that all references to Figure 7 in the Results, Methods, and Discussion with regard to (previous) AvrB should now be replaced with AvrRpm1.
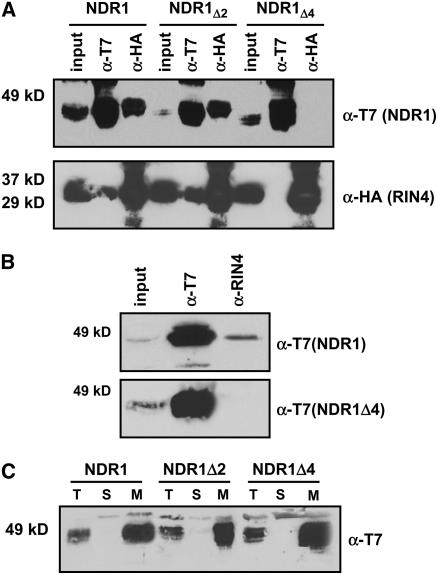
Figure 5.
The NDR1–RIN4 Interaction Occurs within the First Four Amino Acids of NDR1.
Stepwise deletions of two and four amino acids from the N terminus of NDR1 results in a differential pattern of association with RIN4.
(A) Coimmunoprecipitation and protein gel blot analysis of T7:NDR1 and HA:RIN4 from total protein extracts isolated from transiently expressed N. benthamiana. Loss of the NDR1–RIN4 interaction occurs following deletion of four amino acids from the N terminus of NDR1.
(B) Homologous expression of the NDR1Δ4 deletion mutant in Arabidopsis. Native promoter expression of T7:NDR1 and T7:NDR1Δ4 cDNAs in the Arabidopsis ndr1-1 mutant background confirms results of transient heterologous expression. Native RIN4 antibodies coimmunoprecipitate T7:NDR1 yet fail to coimmunoprecipitate T7:NDR1Δ4.
(C) Ultracentrifugation and localization of NDR1 and NDR1 deletion constructs confirm membrane localization. Total protein extracts from Arabidopsis plants (ndr1-1/HA:NDR1) expressing wild-type and deletion constructs were subjected to ultracentrifugation and separation by SDS-PAGE. Protein gel blot analysis (anti-T7) confirms plasma membrane localization for wild-type, Δ2, and Δ4 T7:NDR1 constructs.
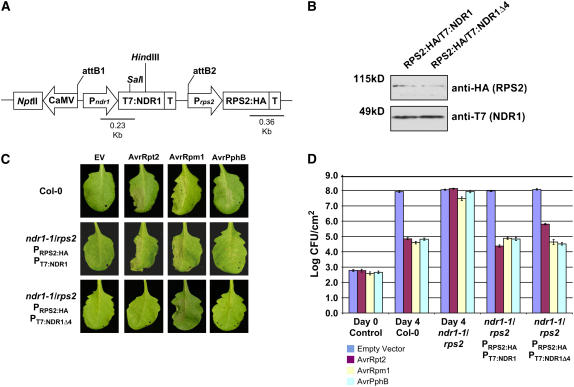
Figure 7.
Construction of a Homologous Expression System for Characterizing the RPS2–RIN4–NDR1 Interaction.
(A) rps2/ndr1-1 mutant Arabidopsis plants were transformed with a dual promoter construct expressing RPS2:HA and T7:NDR1/T7:NDR1Δ4 expressed under the control of their respective native promoters.
(B) Protein gel blot analysis of RPS2:HA/T7:NDR1 and RPS2:HA/T7:NDR1Δ4 transgenic plants. Total protein extracts were separated by SDS-PAGE and probed with anti-HA antibodies (RPS2) and anti-T7 (NDR1) to determine protein expression levels in complemented Arabidopsis lines.
(C) High-density (5 × 107 colony-forming units [cfu]/mL) inoculation of wild-type Col-0 (top panels), RPS2:HA/T7:NDR1 (middle panels), and RPS2:HA/T7:NDR1Δ4 (bottom panels) with P. syringae DC3000 expressing empty vector (EV; control), AvrRpt2, AvrRpm1, and AvrPphB effector proteins.
(D) Bacterial growth curve analysis of native promoter expression lines of Arabidopsis RPS2:HA/T7:NDR1- and RPS2:HA/T7:NDR1Δ4-complemented plants. Bacterial counts were determined at days 0 and 4 following inoculation with P. syringae strains (104 cfu/mL) expressing the indicated effector protein. Day 0 controls were plated 1 h after inoculation and represent the average bacterial growth count of all plant genotypes tested. The Arabidopsis ndr1-1/rps2 genotype corresponds to the parent genotype of the complemented expression system shown in (A). Data shown are the mean of four independent lines for both RPS2:HA/T7:NDR1 and RPS2:HA/T7:NDR1Δ4 constructs. Error bars indicate standard deviation calculated from the mean of replicate samples.