Abstract
Chloroplast genomes in land plants harbor ∼20 group II introns. Genetic approaches have identified proteins involved in the splicing of many of these introns, but the proteins identified to date cannot account for the large size of intron ribonucleoprotein complexes and are not sufficient to reconstitute splicing in vitro. Here, we describe an additional protein that promotes chloroplast group II intron splicing in vivo. This protein, RNC1, was identified by mass spectrometry analysis of maize (Zea mays) proteins that coimmunoprecipitate with two previously identified chloroplast splicing factors, CAF1 and CAF2. RNC1 is a plant-specific protein that contains two ribonuclease III (RNase III) domains, the domain that harbors the active site of RNase III and Dicer enzymes. However, several amino acids that are essential for catalysis by RNase III and Dicer are missing from the RNase III domains in RNC1. RNC1 is found in complexes with a subset of chloroplast group II introns that includes but is not limited to CAF1- and CAF2-dependent introns. The splicing of many of the introns with which it associates is disrupted in maize rnc1 insertion mutants, indicating that RNC1 facilitates splicing in vivo. Recombinant RNC1 binds both single-stranded and double-stranded RNA with no discernible sequence specificity and lacks endonuclease activity. These results suggest that RNC1 is recruited to specific introns via protein–protein interactions and that its role in splicing involves RNA binding but not RNA cleavage activity.
INTRODUCTION
Group II introns are large ribozymes whose splicing proceeds via the same chemical steps as nuclear pre-mRNA splicing (reviewed in Michel et al., 1989; Bonen and Vogel, 2001; Pyle and Lambowitz, 2006). Group II introns are broadly distributed in bacteria and bacteria-derived organelles, but they are particularly prevalent in chloroplasts and in plant and fungal mitochondria. Despite their designation as ribozymes, few group II introns have been demonstrated to harbor catalytic activity, and genetic approaches have shown that proteins are required for the efficient splicing of many group II introns in vivo. However, few group II intron splicing factors have been identified, and even fewer have been analyzed at the biochemical level to elucidate the mechanisms by which they promote splicing.
Previously, we used genetic approaches to identify several nucleus-encoded proteins that are required for the splicing of group II introns in maize (Zea mays) chloroplasts: a CAF1/CRS2 complex, a CAF2/CRS2 complex, CRS1, and PPR4. Each of these is required for the splicing of distinct, but overlapping, subsets of the 17 group II introns in maize chloroplasts, and each is associated in vivo with those introns whose splicing it facilitates (Jenkins et al., 1997; Till et al., 2001; Ostheimer et al., 2003; Schmitz-Linneweber et al., 2005, 2006). CAF1, CAF2, and CRS1 are plant-specific proteins harboring a novel RNA binding domain called the CRM domain (Till et al., 2001; Ostheimer et al., 2003; Barkan et al., 2007), and the splicing functions of these proteins are conserved in their Arabidopsis thaliana orthologs (Asakura and Barkan, 2006). PPR4 is a member of the pentatricopeptide repeat (PPR) family (Small and Peeters, 2000; Schmitz-Linneweber et al., 2006), whereas CRS2 is related to peptidyl-tRNA hydrolase enzymes (Jenkins and Barkan, 2001; Ostheimer et al., 2005). Purified recombinant CAF1/CRS2, CAF2/CRS2, and CRS1 bind intron RNAs in vitro (Ostersetzer et al., 2005; our unpublished results). However, we have been unable to reconstitute splicing in vitro with these proteins alone, suggesting that additional proteins are required. Indeed, these proteins and their cognate introns are found in ribonucleoprotein (RNP) particles of ∼600 to 700 kD in chloroplast extract (Jenkins and Barkan, 2001; Till et al., 2001; Ostheimer et al., 2003), considerably larger than the sum of their known protein and RNA components.
To identify additional proteins involved in the splicing of chloroplast group II introns, we immunopurified CAF1, CAF2, and CRS1 RNPs from chloroplast extract and identified coimmunoprecipitated proteins by mass spectrometry. Here, we describe RNC1, a protein that was recovered in both CAF1 and CAF2 coimmunoprecipitates. RNC1 is a plant-specific protein that has two ribonuclease III (RNase III) domains. RNC1 is found in complexes containing a subset of group II introns in the chloroplast that includes, but is not limited to, CAF1- and CAF2-dependent introns. Many of the introns with which RNC1 is associated splice inefficiently in rnc1 mutants, showing that RNC1 promotes splicing in vivo. Despite its two RNase III domains, phylogenetic considerations and biochemical assays indicate that RNC1 lacks endonucleolytic activity. These and other results suggest that RNC1 promotes splicing via its RNA binding activity and that it is recruited to specific plastid introns via protein–protein interactions.
RESULTS
Identification of RNC1 in CAF1 and CAF2 Coimmunoprecipitates
To seek additional proteins involved in the splicing of chloroplast group II introns, we used mass spectrometry to identify proteins that coimmunoprecipitate with the splicing factors CRS1, CAF1, or CAF2. These proteins were shown previously to reside in intron-containing complexes of ∼600 to 700 kD in the chloroplast stroma (Till et al., 2001; Ostheimer et al., 2003; Schmitz-Linneweber et al., 2005). To reduce contamination of the immunoprecipitates by abundant stromal proteins, chloroplast stroma was first fractionated on sucrose gradients, and gradient fractions containing particles of ∼600 to 700 kD were then subjected to immunoprecipitation (equivalent to fractions 8 to 11 shown in Figure 3C below). The bulk of stromal proteins are found in either smaller (e.g., ribulose-1,5-bis-phosphate carboxylase/oxygenase [Rubisco]) or larger (e.g., ribosome) particles (see Figure 3C below), so this size selection provided substantial enrichment for intron RNPs.
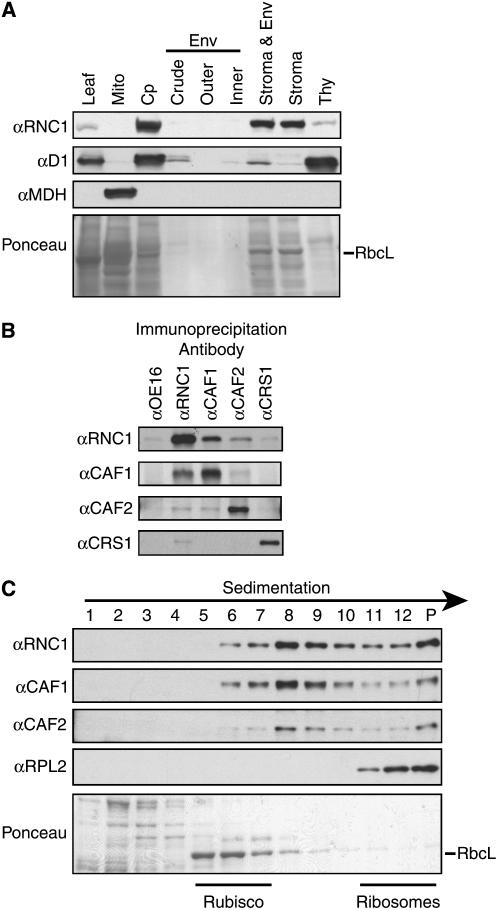
Figure 3.
RNC1 Is Associated with CAF1 and CAF2 in the Chloroplast Stroma.
(A) Immunoblots of leaf and subcellular fractions. Chloroplasts (Cp) and chloroplast subfractions were from the fractionated chloroplast preparation described and verified previously (Williams and Barkan, 2003); the samples in these lanes are derived from the same initial quantity of chloroplasts. The blot was reprobed to detect a marker for thylakoid membranes (D1) and mitochondria (MDH). Env, envelope; Mito, leaf mitochondria; Thy, thylakoid membranes.
(B) Coimmunoprecipitation of RNC1 with CAF1 and CAF2. Stroma was subjected to immunoprecipitation with the antibodies named at top. The presence of specific proteins in the immunoprecipitation pellets was tested by immunoblot analysis with the antibodies listed at left. Immunoprecipitations with OE16 antibody served as a negative control.
(C) Cosedimentation of RNC1 with intron ribonucleoprotein particles. Stromal extract was sedimented in sucrose gradients under conditions in which particles of greater than ∼70S pellet (P). An equal volume of each gradient fraction was analyzed by probing immunoblots with the antibody indicated at left. RPL2 is a protein in the large ribosomal subunit and marks the position of ribosomes. The RbcL band in the Ponceau S–stained blot at bottom marks the position of Rubisco. The peaks of CAF1, CAF2, and RNC1 between Rubisco and ribosomes coincide with the positions of several group II intron RNAs (Jenkins and Barkan, 2001; Till et al., 2001).
Immunoprecipitation pellets were separated by SDS-PAGE, and contiguous gel slices containing proteins between ∼15 and 120 kD were excised. Proteins were subjected to in-gel digestion with trypsin and analyzed by tandem mass spectrometry (MS/MS). Consistent with earlier data demonstrating the independence of the CAF1/CRS2, CAF2/CRS2, and CRS1 splicing complexes, CAF1 and CRS2 were identified in the CAF1 immunoprecipitate, CAF2 and CRS2 were identified in the CAF2 immunoprecipitate, and none of these proteins was identified in the CRS1 immunoprecipitate (data not shown). Several abundant proteins were identified in all three immunopurified samples (e.g., Rubisco, chaperonin-60, GAPDH, and RNA polymerase subunits) and were presumed to be contaminants. Of more interest were several uncharacterized proteins that were identified in the CAF1, CAF2, and/or CRS1 immunoprecipitates. Among these was a protein harboring two RNase III domains (InterPro identifier IPR000999), which was identified in CAF1 and CAF2 immunoprecipitates but not in the CRS1 immunoprecipitate. This protein was an attractive candidate to be an authentic partner of CAF1 and CAF2 because its identification was robust (11 and 8 peptides were identified in the CAF1 and CAF2 immunoprecipitates, respectively) (Figure 1A; see Supplemental Table 1 online) and it harbors a motif known to influence RNA metabolism. Furthermore, its Arabidopsis and rice (Oryza sativa) orthologs have not been reported in plastid proteome studies, suggesting that it is a low-abundance protein and thus is unlikely to be a contaminant. We named this protein RNC1, after the bacterial gene rnc that encodes RNase III.
Figure 1.
The RNC1 Protein.
(A) Multiple sequence alignment of maize RNC1 (Zm RNC1) with its rice and Arabidopsis orthologs (Os RNC1 and At RNC1, respectively). The domain organization of RNC1 is shown diagrammatically at top. Identical residues are shaded in black, and similar residues are shaded in gray. Gray bars beneath the alignment mark the two RNase III domains. Lines above the alignment mark the tryptic peptides identified by mass spectrometry in both the CAF1 and CAF2 coimmunoprecipitates; additional peptides were identified in the CAF1 but not the CAF2 immunoprecipitation and vice versa (see Supplemental Table 1 online). At RNC1 corresponds to At4g37510 and Os RNC1 corresponds to Os01g59510. The transit peptide cleavage sites predicted by ChloroP (Emanuelsson and von Heijne, 2001) are after amino acids 28, 30, and 51 for Zm RNC1, Os RNC1, and At RNC1, respectively.
(B) Multiple sequence alignment of the two RNase III domains from Zm RNC1, Os RNC1, and At RNC1 with the single RNase III domain from E. coli RNase III. Residues that coordinate a catalytic Mg2+ ion in bacterial RNase III are circled and are numbered according to the E. coli protein. The RNase III signature motif is underlined.
RNC1 is a member of an orthologous group that includes one protein in Arabidopsis (At4g37510.1) and one in rice (Os01g59510.1) (see POGs/PlantRBP at http://plantrpb.uoregon.edu). An alignment between the maize, rice, and Arabidopsis RNC1 orthologs is shown in Figure 1A. These three proteins are highly similar except at their N termini, which are predicted by the TargetP algorithm (Emanuelsson and von Heijne, 2001) to be chloroplast transit peptides. The RNC1 orthologs are not closely related to any other predicted proteins in rice or Arabidopsis (BLASTP e-value < 1e-5). RNC1 and its orthologs were examined for functional motifs at InterPro (Quevillon et al., 2005). Two RNase III domains were identified in maize and rice RNC1 (Os RNC1) and one in Arabidopsis RNC1 (At RNC1). However, the high sequence similarity among the three orthologs in regions corresponding to both RNase III domains (Figure 1A) strongly suggests that At RNC1 also has two RNase III domains. RNase III domains harbor the active site of RNase III–like enzymes, which cleave double-stranded RNA substrates (reviewed in MacRae and Doudna, 2007). Proteins with RNase III domains have been divided into three classes based upon their domain organization, typified by bacterial RNase III, eukaryotic Dicer, and eukaryotic Drosha. Proteins in each class include one or more of the following accessory domains: a double-stranded RNA binding domain (dsRBD), a DEAD box domain, and a PAZ domain. RNC1 does not fit into any of these classes, as it lacks conserved domains other than its two RNase III domains.
Figure 1B shows an alignment of the RNase III domain from Escherichia coli RNase III with the six RNase III domains in maize, rice, and Arabidopsis RNC1. Four residues that play important roles in RNase III catalysis are circled: E41, D45, D114, and E117 (E. coli numbering). These four residues coordinate a Mg2+ ion at the active site (Blaszczyk et al., 2001; Gan et al., 2006), and E41, D45, and E117 are strictly conserved among RNase III, Dicer, and Drosha enzymes (Blaszczyk et al., 2001; Han et al., 2004; Zhang et al., 2004). E117 and D45 are essential for catalysis, as E117A, E117Q, D45A, and D45N mutations inactivate E. coli RNase III (Sun and Nicholson, 2001; Sun et al., 2004; Zhang et al., 2004). These residues are not conserved in either of RNC1's two RNase III domains (Figure 1B): D45 corresponds to Gln in both domains, whereas E117 corresponds to Trp in the first domain and to Gln in the second. In E. coli RNase III, E41 and D114 act redundantly: mutation of either alone reduces metal ion affinity, but the double mutant is severely compromised (Sun et al., 2004). Neither of these residues is conserved in RNC1's first RNase III domain, and only E41 is conserved in the second domain (Figure 1B). Together, these amino acid substitutions strongly suggest that RNC1 lacks a functional endonuclease active site.
Recovery of rnc1 Insertion Mutants
To evaluate the physiological role of RNC1, we recovered mutant rnc1 alleles through a reverse-genetic screen of our collection of Mu transposon–induced nonphotosynthetic maize mutants (http://pml.uoregon.edu/) (Stern et al., 2004). The positions of three transposon insertions used for this study are diagrammed in Figure 2A. The rnc1-1, rnc1-2, and rnc1-3 alleles condition pale green, albino, and albino stunted seedling phenotypes, respectively (Figure 2B). The severity of these phenotypes correlate well with the positions of the transposon insertions: the insertion in rnc1-3 is within the open reading frame and is anticipated to be a null allele, whereas the insertion in rnc1-1 is 54 bp upstream of the start codon and is anticipated to be a weak allele. The rnc1-2 insertion is 11 bp upstream of the start codon, consistent with its intermediate phenotype. Plants that are homozygous for each of these alleles die after the development of three to four leaves, as is typical of nonphotosynthetic maize mutants. Complementation crosses were performed between plants heterozygous for each allele in all pairwise combinations; in each case, chlorophyll-deficient seedlings segregated in the F1 progeny, demonstrating that the pigment-deficient phenotypes result from the insertions in the rnc1 gene (see Methods for a summary of the complementation data). Furthermore, the phenotype conditioned by each heteroallelic combination is intermediate between those in the parental alleles (Figure 2B). Noncomplementing progeny of these crosses were used as the source of rnc1 mutant material in the experiments described below to ensure that the molecular defects observed result from disruption of the rnc1 locus.
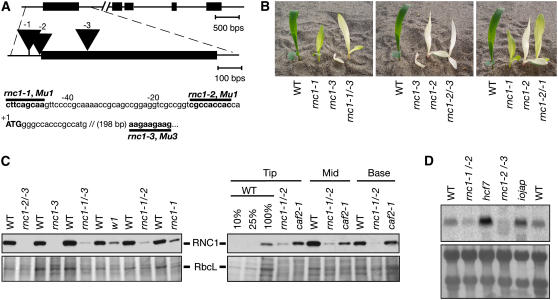
Figure 2.
Mutant Alleles of rnc1.
(A) Positions of Mu transposon insertions in the rnc1 gene. Protein-coding regions are indicated by rectangles, untranslated regions by lines, and insertions by triangles. The name of the mutant allele (rnc1-1, -2, or -3) is shown above each insertion. The positions of the insertions in the wild-type sequence are shown below, with the nine nucleotides that were duplicated during insertion and the name of the specific Mu element marked in boldface.
(B) Phenotypes of rnc1 mutant seedlings. The seedlings are homozygous for the rnc1-1, rnc1-2, or rnc1-3 allele or are the heteroallelic progeny of complementation crosses, as indicated. Plants were grown for 9 d.
(C) Loss of the RNC1 protein in rnc1 mutant leaf tissue. Immunoblots were probed with the RNC1 antibody (top panels); the same blots stained with Ponceau S are shown below, with the band corresponding to RbcL marked. The left panels show results for the first leaf harvested soon after it emerged from the coleoptile. The right panels show a developmental profile of the second leaf of 7-d-old seedlings; tissue was taken from the leaf base (youngest tissue), middle, and tip (oldest tissue). The albino mutants w1 and caf2 are shown to illustrate the effects on RNC1 protein level that result from the absence of plastid ribosomes.
(D) Loss of rnc1 mRNA in rnc1 mutants. An RNA gel blot of total seedling leaf RNA was probed with the rnc1 cDNA. RNA from hcf7 and iojap are shown to illustrate rnc1 RNA levels in mutants with mild (hcf7) or severe (iojap) plastid ribosome deficiencies. An image of the blot stained with methylene blue is shown below.
A polyclonal antibody was raised to a recombinant RNC1 fragment (amino acids 29 to 190). Immunoblot analysis of leaf extracts showed that RNC1 accumulates to reduced levels in each of the mutant lines and in the noncomplementing progeny of complementation crosses (Figure 2C). To determine whether RNC1 abundance changes during leaf development, we took advantage of the natural developmental gradient in the maize leaf: leaf cells and chloroplasts differentiate along an apical-to-basal gradient, with the more differentiated cells/chloroplasts found toward the tip. RNC1 accumulation in wild-type leaves peaks in the middle of the leaf and then declines in the older cells toward the tip. By contrast, the small amount of RNC1 generated in rnc1-1/rnc1-2 plants appears to build up during leaf development, such that it is at its highest level in the apical region (Figure 2C). As a consequence, the loss of RNC1 in the rnc1 mutants, compared with age-matched wild-type tissue, was most severe at early stages of leaf development (i.e., in the bases of seedling leaves and in the first leaf that was newly emerged from the coleoptile). RNA gel blot analysis showed that rnc1 mRNA levels are also reduced in the mutants (Figure 2D).
RNC1 Is Found in Complexes with CAF1 and CAF2 in the Chloroplast Stroma
RNC1 was initially identified from a chloroplast stromal preparation. To determine whether this is RNC1's sole intracellular location, immunoblots of leaf, chloroplast, mitochondria, and chloroplast subfractions were probed with the RNC1 antibody (Figure 3A). The results confirm that RNC1 is localized to the chloroplast stroma and show, in addition, that RNC1 has minimal association with the thylakoid membrane and that it is not found in mitochondria. CAF1 and CAF2 both coimmunoprecipitated with RNC1 from stromal extract (Figure 3B). Furthermore, immunoblot analysis of sucrose gradient–fractionated stroma showed that RNC1 cosediments with CAF1 and CAF2 in particles of ∼600 to 700 kD (Figure 3C). These results confirm that RNC1 is found in particles in the chloroplast stroma that include CAF1 and CAF2. RNC1 was not identified in the MS/MS analysis of the CRS1 coimmunoprecipitation. However, weak coimmunoprecipitation of CRS1 with RNC1 was observed in the directed tests shown in Figure 3B.
Coimmunoprecipitation Assays Identify Group II Introns Associated with RNC1
CAF1 and CAF2 are bound to group II intron RNAs in vivo (Ostheimer et al., 2003; Schmitz-Linneweber et al., 2005) in RNPs of ∼600 to 700 kD (Jenkins and Barkan, 2001). Given RNC1's coimmunoprecipitation and cosedimentation with CAF1 and CAF2 (Figure 3), it seemed likely that RNC1 is also associated with intron RNAs. In addition, RNC1's similarity to RNase III enzymes suggested that it might function in rRNA and/or mRNA processing. To gain insight into RNC1's physiological RNA ligands, we performed RNA immunoprecipitation-on-microarray (RIP-chip) assays as an initial screen (Schmitz-Linneweber et al., 2005): stromal extract was subjected to immunoprecipitation with the RNC1 antibody, and RNAs from the immunoprecipitation pellet and supernatant were labeled with red-fluorescing (F635) or green-fluorescing (F532) dyes, respectively, combined, and hybridized to a tiling microarray of the maize chloroplast genome. Three replicate immunoprecipitation reactions were performed and analyzed in this manner.
To highlight sequences whose enrichment occurs specifically when the RNC1 antibody is used for immunoprecipitation, the median enrichment ratio (F635:F532) for each array fragment was plotted according to chromosomal position, after subtracting the values reported previously for RIP-chip assays using antibody to PPR4, an rps12-int1 trans-splicing factor (Schmitz-Linneweber et al., 2006) (Figure 4A). Several RNC1-specific peaks stand out prominently, with two deep valleys resulting from PPR4's strong association with RNA from the two rps12 loci. As an additional negative control, two replicate assays were performed with stromal extract from rnc1-1/rnc1-2 mutant seedlings. This mutant stroma has reduced but still substantial levels of RNC1 protein because it was generated from complete seedling leaves rather than from the youngest leaf tissue, in which the RNC1 protein deficiency is most severe. Stronger mutant alleles or earlier developmental stages could not be used for this purpose because they yield insufficient stromal extract. These data are plotted in Supplemental Figure 1 online.
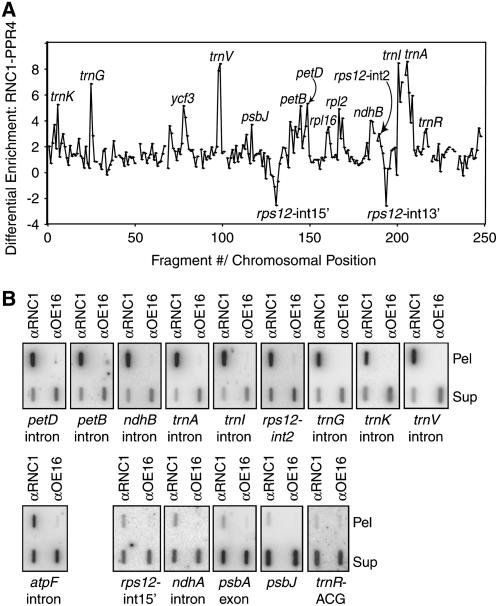
Figure 4.
Identification of RNA Ligands of RNC1 by Coimmunoprecipitation Analyses.
(A) RIP-chip data displayed to show differential enrichment of RNA sequences in RNC1 versus PPR4 immunoprecipitations. The median log2(F635:F532) for replicate spots across replicate immunoprecipitations with anti-PPR4 antibody (data from Schmitz-Linneweber et al., 2006) was subtracted from the median log2(F635:F532) for replicate immunoprecipitations with RNC1 antibody. The locus name is indicated for peaks that include fragments whose enrichment from wild-type stroma ranked in the top 15th percentile and if the sequences showed significant differential enrichment from either RNC1 versus PPR4 immunoprecipitations or from wild-type versus rnc1-1/rnc1-2 mutant stroma (see Supplemental Table 2 online).
(B) Verification of RIP-chip data by slot-blot hybridization. RNA purified from the pellets (Pel) and supernatants (Sup) of RNC1 immunoprecipitation reactions with wild-type stroma was applied to slot blots and hybridized with probes specific for the indicated sequence. Immunoprecipitations with an antibody to OE16 was used as a negative control. The total signal from the pellet plus supernatant is greater for sequences that are strongly enriched in RNC1 immunoprecipitations than in the control immunoprecipitation. This is due in part to the fact that one-sixth of the total pellet sample was analyzed in each slot, whereas only one-twelfth of the corresponding supernatant was analyzed. In addition, the signal is artifactually reduced in the supernatant samples due to saturation of the binding capacity of the membrane with the large amount of RNA.
t tests were performed to assess the significance of the difference in enrichment of each RNA sequence in (1) RNC1 immunoprecipitations from wild-type extract versus RNC1 immunoprecipitations from rnc1 mutant extract and (2) RNC1 immunoprecipitations versus PPR4 immunoprecipitations from wild-type extract. The results for the top-scoring fragments are summarized in Supplemental Table 2 online. These analyses provide strong support for an association between RNC1 and RNAs derived from trnA-UGC, trnI-GAU, trnV-UAC, ndhB, petD, and petB, because at least two fragments from each of these loci showed highly significant differential enrichment when analyzed by both methods. Support is also strong for an association with RNAs from trnG-UCC, trnK-UUU, rpl2, ycf3, the 3′ locus of rps12, and trnR-ACG, because either more than one fragment showed significant differential enrichment in one of the comparisons (e.g., 3′ locus of rps12) or one fragment showed highly significant differential enrichment in both comparisons (e.g., trnG-UCC). Results are also suggestive for rpl16 and psbJ, with borderline significance when analyzed both ways. Notably, the RIP-chip data did not provide even weak evidence for an association between RNC1 and plastid rRNAs, consistent with the hypothesis that this protein does not serve as a RNase III for rRNA processing.
All of the strongest candidates to emerge from the RIP-chip assay include group II intron sequences. The strong enrichment of intron RNAs derived from petD, petB, ndhB, rps12-int2, trnI-GAU, trnA-UGC, trnG-UCC, trnV-UAC, and trnK-UUU in the pellets of RNC1 immunoprecipitations was confirmed by slot-blot hybridization of RNAs that coimmunoprecipitate with RNC1 (Figure 4B, top). All of these sequences showed very strong enrichment in the RNC1 immunoprecipitation pellet and depletion from the supernatant, indicating that a substantial fraction of the molecules harboring these sequences are associated with RNC1. Three RNAs that did not exhibit strong enrichment in the RIP-chip assays (psbA mRNA, ndhA intron, and rps12-int1) were likewise found at only low levels in RNC1 immunoprecipitation pellets (Figure 4B, bottom right), albeit at higher levels than in the OE16 control. Finally, the two non-intron RNAs whose enrichment was suggested by the RIP-chip data, trnR-ACG and psbJ, did not reproduce in the slot-blot assays (Figure 4B, bottom right) and their metabolism is not disrupted in rnc1 mutants (see below), indicating that these were false-positives. Together, these results confirm that petD, petB, ndhB, rps12-int2, trnI, trnA, trnG, trnV, and trnK intron RNAs are associated with RNC1 in stromal extract. The results suggest further that RNC1 may associate to a small extent with other (perhaps many) chloroplast RNAs. Results of the genetic analyses below suggest, however, that the low-level interactions are not functionally relevant.
The atpF intron was unusual in that it did not score as a positive in the RIP-chip assays, but it was clearly enriched in RNC1 immunoprecipitations when assayed by slot-blot hybridization (Figure 4B, bottom left), albeit less strongly than those introns with RIP-chip support. We suspect that the RNC1–atpF intron interaction might be stabilized by magnesium and that it did not emerge from the original RIP-chip data because Mg2+ was not included in the immunoprecipitation buffer. The genetic data below suggest that RNC1 does, in fact, influence atpF intron metabolism.
All of the ligands with strong support from both the RIP-chip and slot-blot data are group II introns. These include known substrates of CAF1 (trnG-UCC and petD) and CAF2 (petB and ndhB) (Ostheimer et al., 2003), in accordance with the identification of RNC1 in coimmunoprecipitates with both CAF1 and CAF2. In addition, RNC1's ligands include RNAs from intron-containing genes for which splicing factors have not been described (trnI-GAU, trnA-UGC, rps12- int2, trnV-UAC, rpl2, and trnK-UUU).
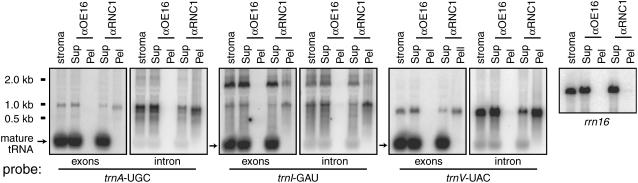
To determine whether RNC1 is bound solely to unspliced precursor RNAs or to both unspliced precursors and spliced products, coimmunoprecipitated RNAs were analyzed by RNA gel blot hybridization using intron-specific and exon-specific probes for the three best supported RNA ligands (trnI-GAU, trnA-UGC, and trnV-UAC) (Figure 5). Unspliced precursors (i.e., RNAs that hybridize to both exon and intron probes) were enriched in the immunoprecipitation pellet, but spliced tRNAs were not. These results support a role for RNC1 in tRNA splicing rather than in other aspects of tRNA metabolism. The trnI-GAU and trnA-UGC introns are encoded between the 16S and 23S rRNA genes and are cotranscribed with them. The RIP-chip data suggested, nonetheless, that the flanking rRNA sequences do not coimmunoprecipitate with RNC1. Lack of enrichment of 16S rRNA sequences was confirmed by RNA gel blot hybridization of RNC1 coimmunoprecipitations (Figure 5, right). These results provide additional evidence that RNC1 does not serve as a RNase III for chloroplast rRNA processing.
Figure 5.
Coimmunoprecipitation of Unspliced tRNA Precursors but Not Spliced tRNAs with RNC1.
RNA purified from an RNC1 immunoprecipitation supernatant (Sup) and pellet (Pel) was analyzed on duplicate RNA gel blots by hybridization to the indicated exon-specific and intron-specific probes. Equal fractions of the pellet and supernatant samples were analyzed. An immunoprecipitation with antibody to OE16 served as a negative control. RNA extracted from an equivalent amount of stroma was analyzed for comparison. Blots were initially probed with spliced cDNAs corresponding to the indicated mature tRNAs (exons) and were reprobed with intron-specific probes; residual signal at the positions of mature tRNAs remained after the intron probings. The right panel shows analogous samples probed to detect 16S rRNA. Pre-16S rRNAs were not detectable in any of the lanes.
rnc1 Mutants Are Deficient for Plastid Ribosomes
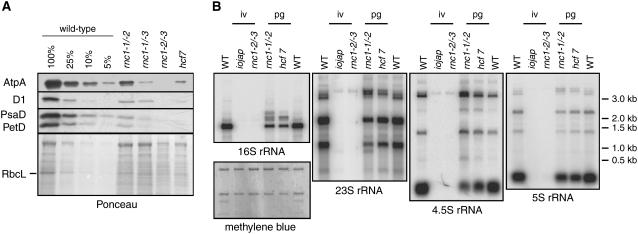
To gain insight into the role of RNC1 in the metabolism of its RNA ligands, chloroplast gene expression was characterized in the rnc1 insertion mutants. If RNC1 is required for the maturation of any of the tRNAs or ribosomal protein mRNAs with which it coimmunoprecipitates, then rnc1 mutants would be expected to have compromised chloroplast translation and, consequently, a reduced content of plastid ribosomes and of all plastid-encoded proteins. These predictions are supported by the data shown in Figure 6. Immunoblot analyses of leaf proteins show that core subunits of each photosynthetic enzyme complex that includes plastid-encoded subunits (photosystem II, cytochrome b6f, photosystem I, ATP synthase, and Rubisco) accumulate to reduced levels in rnc1 mutants (Figure 6A). Furthermore, the accumulation of all plastid rRNAs is reduced in rnc1 mutants, with the magnitude of the defect correlating with the magnitude of the RNC1 protein and chlorophyll deficiencies conditioned by each allele (Figure 6B). The plastid rRNA transcript populations are similar to those in previously described mutants with plastid ribosome deficiencies: pale green rnc1-1/rnc1-2 mutants have rRNAs similar to those in hcf7 mutants, which are similar in pigmentation and plastid protein content (Barkan, 1993); albino rnc1-2/rnc1-3 mutants lack detectable plastid rRNAs, as do albino iojap mutants, which lack plastid ribosomes (Walbot and Coe, 1979). As discussed previously, rRNA defects like these are pleiotropic effects of many or all mutations that disrupt the assembly of the ribosome and need not reflect a direct role for a gene in rRNA metabolism (Barkan, 1993; Williams and Barkan, 2003; Schmitz-Linneweber et al., 2006).
Figure 6.
Loss of Plastid Ribosomes and Plastid-Encoded Proteins in rnc1 Mutants.
(A) Reduced accumulation of photosynthetic enzyme complexes in rnc1 mutants. Immunoblots of total leaf extract (5 μg of protein or dilutions as indicated) were probed with antibodies to the protein labeled at left. AtpA, D1, PsaD, and PetD are subunits of ATP synthase, photosystem II, photosystem I, and the cytochrome b6f complex, respectively. The same blot stained with Ponceau S is shown below to illustrate the relative loading of the samples and the abundance of RbcL.
(B) Loss of plastid rRNAs in rnc1 mutants. Total seedling leaf RNA (0.5 μg) from the genotype indicated at top was analyzed by RNA gel blot hybridization using probes specific for the RNAs indicated at bottom. The pigmentation of the mutants is indicated at top: iv, ivory leaves; pg, pale green leaves. One of the blots stained with methylene blue is shown below to illustrate equal loading of the cytosolic rRNAs.
RNC1 Promotes the Splicing of Chloroplast Group II Introns in Vivo
To determine whether RNC1 facilitates the splicing of the introns with which it associates, the ratio of spliced-to-unspliced RNA from all plastid loci encoding group II introns was assayed in rnc1 mutants. To control for pleiotropic effects that might result from the ribosome deficiencies in rnc1 mutant plastids, RNA in pale green rnc1-1/rnc1-2 mutants was compared with that in pale green hcf7 mutants and RNA in albino rnc1-2/rnc1-3 mutants was compared with that in albino iojap mutants; the magnitude of the plastid rRNA deficiencies in these rnc1 alleles is well matched with that of the corresponding controls (Figure 6). The complete absence of chloroplast ribosomes in maize and barley (Hordeum vulgare) is accompanied by the failure to splice plastid introns in subgroup IIA (Hess et al., 1994; Jenkins et al., 1997; Vogel et al., 1999). Many subgroup IIA introns coimmunoprecipitate with RNC1 (trnK, trnI, trnA, rpl2, trnV, and rps12-int2); for these introns, splicing defects observed in the weak rnc1 alleles that retain chloroplast ribosomes (i.e., rnc1-1/rnc1-2) would support a role for RNC1 in splicing, whereas splicing defects observed in the strong rnc1 alleles would be uninformative, as they could result from the ribosome deficiency.
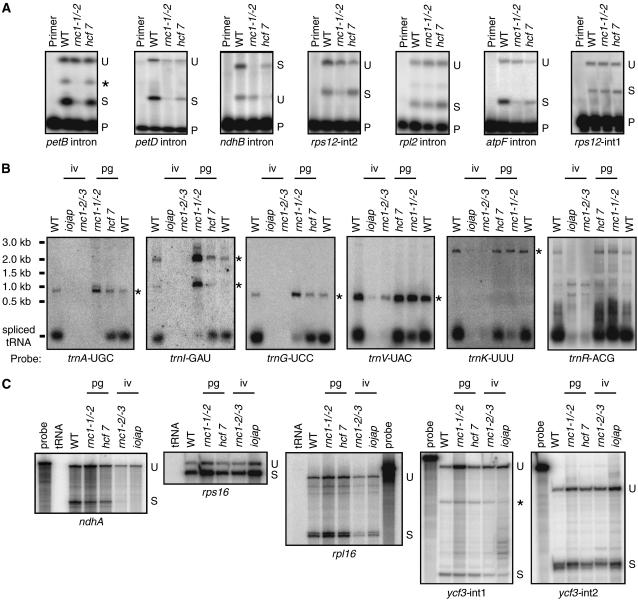
Splicing was assayed by poisoned-primer extension, RNase protection, and/or RNA gel blot hybridization, according to which assay yielded the clearest data for each intron (Figure 7; see Supplemental Figure 2 online). The results showed that RNC1 is required for the efficient splicing of most of the introns that emerged as RNC1 ligands in the coimmunoprecipitation assays. Poisoned-primer extension assays revealed a clear decrease in splicing for petB, petD, ndhB, and rps12-int2 in rnc1-1/rnc1-2 mutants compared with the hcf7 control (Figure 7A); RNase protection assays confirmed these results for petB, petD, and ndhB (see Supplemental Figure 2A online). RNA gel blot hybridizations (Figure 7B) demonstrated a clear splicing defect for trnA, trnI, trnG, trnV, and trnK in rnc1-1/rnc1-2 mutants compared with the hcf7 control; the loss of mature trnA, trnI, and trnG, and the concurrent overaccumulation of their unspliced precursors, was particularly striking. The rps12-int2, trnA, trnI, trnV, and trnK introns are subgroup IIA introns and are among the group of introns that fail to splice in the absence of plastid ribosomes (Vogel et al., 1999). However, the mild plastid ribosome deficiency in rnc1-1/rnc1-2 mutants cannot account for their reduced splicing, as they are spliced normally in hcf7 mutants (Figure 7B). A tRNA encoded by a locus lacking an intron, trnR-ACG, accumulated normally in the rnc1 mutants (compared with the appropriate control), demonstrating that tRNA metabolism is not globally disrupted in these mutants.
Figure 7.
Chloroplast Splicing Defects in rnc1 Mutants.
(A) Poisoned primer extension assays for mRNA splicing. Reverse transcriptase reactions were initiated with radiolabeled primers complementary to the exon downstream of the indicated intron; a dideoxy nucleotide that terminates reverse transcription after different distances on spliced and unspliced templates was included in the reactions. The splicing defects for the petB, petD, and ndhB introns were confirmed with RNase protection assays (see Supplemental Figure 2 online). The asterisk at the petB panel marks a product that terminates near the branch point adenosine; this likely originates from the lariat intermediate that is the product of the first step in splicing. Data are shown for the weak allele combination rnc1-1/rnc1-2 to minimize secondary effects due to the loss of plastid ribosomes. P, primer; S, spliced; U, unspliced.
(B) RNA gel blot analysis of tRNA splicing. Blots were probed with cDNAs derived from the indicated spliced tRNA. Asterisks mark the positions of unspliced precursors, as deduced from their size and by their hybridization to intron-specific probes (Figure 5; data not shown). The trnR-ACG gene does not contain an intron; it is shown because it emerged as a possible RNC1 ligand in the RIP-chip assays and to illustrate that tRNA metabolism is not globally disrupted in rnc1 mutants. iv, ivory leaves; pg, pale green leaves.
(C) RNase protection assays of mRNA splicing. Probes spanned either the 3′ splice junction (rps16, rpl16, and ycf3-int2) or the 5′ splice junction (ndhA and ycf3-int1). The asterisk marks a product anticipated to result from hybridization to the excised intron.
Three introns that did not emerge as RNC1 ligands from the immunoprecipitation assays, rps12-int1, ndhA, and rps16, are spliced similarly in rnc1 mutants and in the matched control mutants (Figures 7A and 7C). Thus, there is generally good correspondence between those introns that were strongly enriched in RNC1 coimmunoprecipitations and those whose splicing is disrupted in rnc1 mutants. There are a few exceptions, however. Two RNAs with strong support (rpl2 and ycf3) and one with weak support (rpl16) from the RIP-chip data exhibited little or no decrease in splicing in rnc1 mutants compared with their matched controls (Figures 7A and 7C).
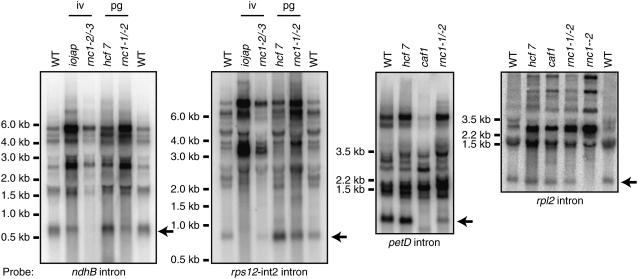
The rpl2 intron showed strong RNC1-dependent enrichment in the RIP-chip assays but its splicing was affected little, if at all, in rnc1-1/rnc1-2 mutants. These results suggest that RNC1 is bound to the rpl2 intron in vivo but is not required for optimal splicing. A firm conclusion cannot be made, however, because the residual RNC1 protein present in rnc1-1/rnc1-2 seedling leaves (Figure 2C) might be sufficient to promote the splicing of this intron. The rpl2 intron is in subgroup IIA and fails to splice in all mutants lacking plastid ribosomes, so its lack of splicing in plants with the stronger allele combination, rnc1-2/rnc1-3, is uninformative. To address the possibility that RNC1 may participate in some other aspect of rpl2 RNA metabolism, the rpl2 mRNA was analyzed by RNA gel blot hybridization using exon- and intron-specific probes (Figure 8; see Supplemental Figure 2B online), but no clear differences in the pattern of transcripts between hcf7 and rnc1-1/rnc1-2 plants emerged. Together, these results neither support nor refute a role for RNC1 in rpl2 splicing.
Figure 8.
RNA Gel Blots Showing the Levels of Excised Introns in rnc1 Mutants.
Total leaf RNA (5 μg per lane) was analyzed by RNA gel blot hybridization using probes specific for the indicated intron. RNA from a caf1 mutant, which has a strong defect in petD splicing, was included on the petD intron blot for comparison. Arrows point to presumed excised introns: their sizes match those of excised introns and they do not hybridize to exon probes (data not shown). The reduced splicing of the rpl2 and rps12-int2 introns in the strong rnc1 alleles (rnc1-2/rnc1-3 and rnc1-2 homozygotes) is not informative because these introns are in subgroup IIA and fail to splice in all maize mutants lacking plastid ribosomes. iv, ivory leaves; pg, pale green leaves.
The atpF intron is also an exceptional case in that its coimmunoprecipitation with RNC1 was detected by slot-blot hybridization but not in the RIP-chip assays. However, atpF splicing is reduced in rnc1-1/rnc1-2 mutants compared with the hcf7 control (Figure 7A), suggesting that RNC1 functions in concert with CRS1 (Jenkins et al., 1997; Till et al., 2001; Ostheimer et al., 2003) to promote atpF splicing. Although RNC1 was not originally detected as coimmunoprecipitating with CRS1 by mass spectrometry, immunoblot analysis (Figure 3B) detected weak RNC1/CRS1 coimmunoprecipitation.
A role for RNC1 in splicing rather than in stabilizing the spliced RNA is supported by several observations. First, intron sequences rather than flanking exon sequences were most strongly enriched in the immunoprecipitations, and where examined, intron-containing precursors but not spliced products coimmunoprecipitated with RNC1 (Figure 5); these results point to introns as RNC1's ligands. Second, a reduction in the level of spliced RNA was in some cases accompanied by an increase in the abundance of unspliced precursor (e.g., trnA, trnI, and trnG in Figure 7B). Third, a band that likely reflects the presence of the lariat intermediate in petB splicing (Figure 7A, asterisk) is reduced in rnc1-1/rnc1-2 mutants, suggesting that the first step in petB splicing is disrupted. Additional evidence that RNC1 functions in splicing is shown in Figure 8: RNA gel blot hybridizations with intron-specific probes for ndhB, rps12-int2, and petD showed that the ratio of excised intron to unspliced precursor RNAs is reduced in rnc1 mutants; this finding supports a role for RNC1 in intron excision rather than in intron turnover or some other aspect of RNA metabolism.
Together, these results provide strong evidence that RNC1 enhances the splicing of two CAF2-dependent introns (ndhB and petB) and two CAF1-dependent introns (petD and trnG). The results also provide strong evidence that RNC1 promotes the splicing of a set of plastid introns for which splicing factors have not been described previously: trnA, trnI, trnV, rps12-int2, and trnK. In addition, the data suggest a role for RNC1 in splicing the atpF intron, which also requires CRS1. That RNC1 coimmunoprecipitates with these introns and that its absence causes a decrease in their splicing together provide strong evidence that RNC1 plays a direct role in facilitating their splicing in vivo.
Recombinant RNC1 Binds RNA but Lacks Endonuclease Activity
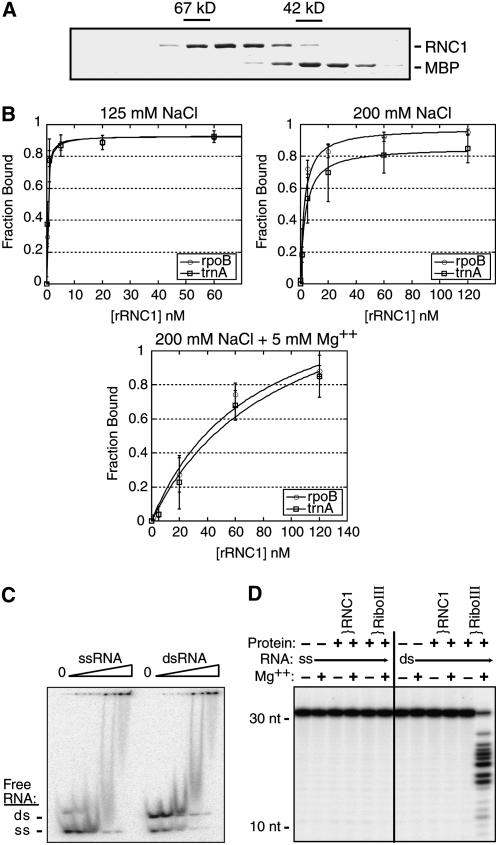
Untagged recombinant RNC1 (rRNC1) was generated by expression as a fusion with maltose binding protein (MBP), purification by amylose affinity chromatography, cleavage from the MBP moiety, and subsequent purification on a gel filtration column. The elution position of rRNC1 from the gel filtration column (Figure 9A) and analytical ultracentrifugation (data not shown) showed rRNC1 to be monomeric. The ability of rRNC1 to bind to one of its most strongly supported in vivo ligands, the trnA intron, and to a non-intron chloroplast RNA of similar size (rpoB-coding region) was tested with filter binding assays (Figure 9B). Because RNC1 could, in principle, recognize any of a variety of intron conformations, binding was tested to introns that had been heated and then treated in each of the following ways: (1) snap-cooled in a low concentration of monovalent salt and in the absence of Mg2+ to minimize RNA structure, (2) slow-cooled in the presence of 200 mM monovalent salt but no Mg2+ to promote the formation of secondary structures, and (3) slow-cooled in the presence of 200 mM salt and 5 mM Mg2+ to promote the formation of both secondary and tertiary contacts. rRNC1 bound RNA with high affinity (apparent Kd in the low nanomolar range in the absence of Mg2+) but did not discriminate between its specific and nonspecific substrates under any of these conditions. The addition of NaCl to 200 mM reduced the affinity of rRNC1 for both intron and non-intron RNAs, and the further addition of Mg2+ had an additional negative effect. Analogous results were obtained in assays using two other RNC1 ligands, the petB and trnV introns (data not shown). The inhibition of binding by monovalent and divalent salts suggests that binding is supported primarily by ionic interactions; the strong negative effect of Mg2+ suggests further that rRNC may not bind to the more collapsed conformers that form in the presence of Mg2+. This behavior is very different from that observed for the CRS1–atpF intron interaction, which is dependent on Mg2+ (Ostersetzer et al., 2005).
Figure 9.
In Vitro Assays with Recombinant RNC1.
(A) Elution of untagged rRNC1 from the gel filtration column. Fractions from the gel filtration column were analyzed by SDS-PAGE, and proteins were detected with Coomassie blue. Contiguous fractions from the relevant portion of the elution are shown. The elution of rRNC1 compared with BSA (67 kD) and MBP (42 kD) indicates that it is monomeric (predicted molecular mass, 59 kD). The peak rRNC1 fractions were pooled, and residual MBP was removed by passage over an amylose affinity column prior to use in in vitro assays.
(B) Filter binding assays showing RNA binding activity of rRNC1. Assays were performed with trace amounts of radiolabeled RNAs corresponding to an RNC1 ligand (trnA intron) and an RNA of similar size that did not show evidence of interacting with RNC1 in vivo (rpoB-coding region). RNA was heated in the absence of salts and then snap-cooled in the presence of 125 mM NaCl (top panel), slow-cooled in 200 mM NaCl (middle panel), or slow-cooled in 200 mM NaCl and 5 mM Mg2+ (bottom panel). Single-site binding isotherms were fit to the data using the following equation: fraction RNA bound = (maximum RNA bound × [protein])/(Kd + [protein]). Data points are averages from three assays, with error bars indicating 1 sd.
(C) Gel mobility shift assay showing the relative affinity of rRNC1 for single-stranded and double-stranded RNA. A synthetic RNA oligonucleotide was radiolabeled and either heated and snap-cooled (ssRNA) or mixed with a twofold excess of its complement and slow-cooled in the presence of monovalent salts to promote duplex formation (dsRNA). The annealing was not complete, so the dsRNA substrate includes both dsRNA and ssRNA. RNA (40 pM) was incubated with increasing concentrations of rRNC1 (32, 80, 200, and 500 nM). The disappearance of unbound RNA is used to monitor protein binding.
(D) rRNC1 lacks RNase III–like endonuclease activity. RNA substrates were the same synthetic ssRNA and dsRNA 31-mers used in (C). Reactions contained rRNC1 at a concentration of 125 nM or E. coli RNase III (RiboIII) at 10 nM (dimer concentration).
A gel mobility shift assay was used with synthetic 31-mer oligonucleotides to determine whether rRNC1 has a preference for single-stranded or double-stranded RNA (Figure 9C). A discrete band reflecting an RNA/protein complex did not form in the presence of rRNC1, but protein binding could be monitored by the loss of free RNA as the protein concentration increased. rRNC1 bound with similar affinity to RNA in single-stranded and double-stranded forms; this is illustrated most clearly by its lack of preference for one form over the other when both were present in the same binding reaction (Figure 9C, right lanes). Together, the in vitro binding data suggest that RNC1 is a sequence-nonspecific RNA binding protein that can interact with both single-stranded and double-stranded RNAs.
The lack of conservation of critical catalytic residues in RNC1's RNase III domains (Figure 1B) strongly suggested that it lacks endonuclease activity. To test this prediction, rRNC1 and E. coli RNase III were incubated with single-stranded and double-stranded RNA 31-mers under conditions described for assaying the endonuclease activity of RNase III and Dicer enzymes, which require Mg2+ and a dsRNA substrate (Zhang et al., 2004). As expected, E. coli RNase III cleaved the double-stranded 31-mer in a Mg2+-dependent manner (Figure 9D). rRNC1, however, showed no evidence of endonuclease activity, even at high protein concentrations. Furthermore, incubation of RNAs harboring group II introns with rRNC1 under a variety of conditions detected neither intron-splicing nor intron-cleaving activity (data not shown). We cannot eliminate the possibility that rRNC1 was catalytically inactive due to protein misfolding or the lack of posttranslational modifications. Nonetheless, the failure to detect endonuclease activity in these assays, together with the lack of catalytic residues in RNC1's RNase III domains, provides strong evidence that RNC1 does not function as an endonuclease.
DISCUSSION
RNC1 Is a Catalytically Inactive Member of the RNase III Family That Promotes Group II Intron Splicing
We show here that RNC1 coimmunoprecipitates and cosediments with the chloroplast-splicing factors CAF1 and CAF2, that it is associated with both CAF-dependent and CAF-independent group II intron RNAs, that it is required for the efficient splicing of many chloroplast group II introns in vivo, and that it binds RNA with high affinity in vitro. These findings together provide strong evidence that RNC1 facilitates the splicing of a subset of group II introns in the chloroplast and that it does so via direct interaction with intron RNAs. The presence of two RNase III domains in RNC1 initially suggested that RNC1 might function as an endonuclease and that this activity could be relevant to its role in intron metabolism. However, several observations argue that RNC1 lacks endonucleolytic activity. First, several amino acids that are essential for catalysis by bacterial RNase III, Dicer, and Drosha enzymes are not conserved in RNC1's RNase III domains (Figure 1B). Second, RNC1's domain organization differs from that of previously characterized members of the RNase III domain family (MacRae and Doudna, 2007). Third, recombinant RNC1 lacks double-stranded RNA endonuclease activity in vitro. Finally, although an RNase III–like endonuclease could potentially be involved in intron turnover (Danin-Kreiselman et al., 2003), RNC1 is unlikely to play such a role because excised group II introns do not accumulate to increased levels in rnc1 mutants, as would result from a reduced rate of intron decay.
In bacteria, RNase III functions in the processing of a polycistronic rRNA precursor. The organization of rRNA operons is similar in chloroplasts and bacteria, so it is anticipated that a RNase III enzyme likewise participates in chloroplast rRNA processing. However, RNC1 is unlikely to serve this function because it appears to lack endonuclease activity. Furthermore, the coimmunoprecipitation data did not provide evidence for an association between RNC1 and rRNA sequences: the trnI and trnA introns, which are encoded between the 16S and 23S rRNA genes and cotranscribed with them, are strongly enriched in RNC1 coimmunoprecipitations, but the enrichment of the flanking rRNA sequences drops off sharply. Although rnc1 mutants do exhibit defects in rRNA metabolism, no rRNA processing defects were detected beyond those observed in other mutants with defects in plastid ribosome biogenesis (e.g., hcf7 and ppr4) (Barkan, 1993; Schmitz-Linneweber et al., 2006) and that result from disrupted ribosome assembly. The reduced splicing of several plastid tRNAs and one ribosomal protein mRNA in rnc1 mutants (trnI, trnA, trnG, trnK, trnV, and rps12-int2) can account for the loss of chloroplast translation and plastid ribosomes; the aberrant rRNA metabolism in rnc1 mutants is likely to be a consequence of the reduced availability of these essential components of the translation machinery.
If RNC1 does not function as a chloroplast RNase III, then what protein might do this? Bacterial RNase III enzymes harbor a dsRBD and a RNase III domain. A search of the POGs/PlantRBP database (Walker et al., 2007) (http://plantrbp.uoregon.edu/) for rice and Arabidopsis proteins with both of these domains identified orthologous proteins in rice (Os05g18850) and Arabidopsis (At3g20420) that are excellent candidates. All of the essential catalytic residues are conserved in their RNase III domains, they have predicted chloroplast-targeting sequences, and their domain organization mirrors that of bacterial RNase III enzymes (data not shown). No other proteins in rice or Arabidopsis meet all of these criteria.
RNC1 is unique among characterized proteins and among predicted plant proteins in that it has two RNase III domains and no other conserved domains. Eukaryotic Dicer resembles RNC1, however, in that it contains two RNase III domains and functions as a monomer (reviewed in MacRae and Doudna, 2007). Bacterial RNase III forms a homodimer, with dimerization mediated by interactions between the single RNase III domain in each monomer. In Dicer, a pseudodimeric active site is formed via interactions between its two RNase III domains. The paired RNase III domains in RNC1 monomers may likewise form a pseudodimer. The dsRBD in E. coli RNase III is not essential for RNA binding or catalysis (Sun et al., 2001). Given that RNC1 lacks a dsRBD, its lack of preference for binding double-stranded RNA over single-stranded RNA is not surprising.
RNC1 Functions Together with, and Independently of, Previously Described Chloroplast Splicing Factors
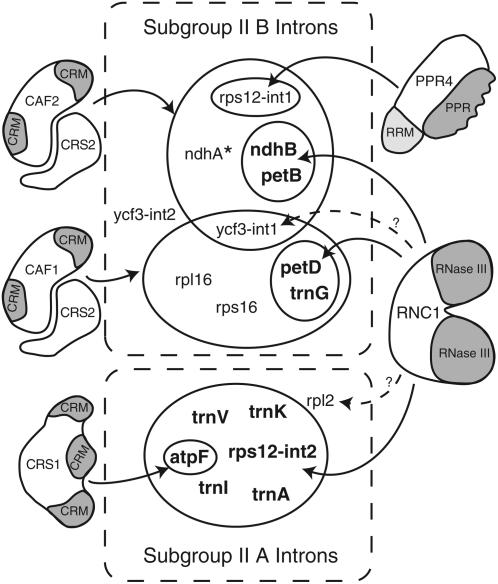
The intron specificities of RNC1 and previously described chloroplast splicing factors in maize are summarized in Figure 10. Of the 17 group II introns in maize chloroplasts, 10 had been shown previously to depend upon nucleus-encoded proteins for their splicing: CRS2/CAF1 and CRS2/CAF2 complexes are required for the splicing of overlapping subsets of nine introns in subgroup IIB (trnG, rps16, rpl16, petB, petD, rps12-int1, ndhB, ndhA, and ycf3-int1), PPR4 is required for the trans-splicing of rps12-int1, and CRS1 is required for the splicing of the atpF intron in subgroup IIA. RIP-chip data support the notion that these are the sole RNA ligands of CAF1 and PPR4 (Schmitz-Linneweber et al., 2005, 2006). The results presented here show that RNC1 associates with and promotes the splicing of at least four subgroup IIB introns that also require a CRS2/CAF complex: ndhB, trnG, petB, and petD. Thus, three nucleus-encoded proteins are now implicated in the splicing of each of these introns (RNC1, CRS2, and either CAF1 or CAF2).
Figure 10.
Nucleus-Encoded Chloroplast Splicing Factors in Maize and Their Intron Targets.
The 17 group II introns in the maize chloroplast genome are divided into subgroups IIA and IIB according to Michel et al. (1989). Nucleus-encoded splicing factors are shown, with identified domains shaded. The designated intron targets (solid arrows) have been shown to coimmunoprecipitate with the indicated proteins and to require those proteins for optimal splicing in vivo. Dashed arrows designate interactions detected by coimmunoprecipitation but for which a role in splicing has not been documented. Results are summarized from this work and from Jenkins et al. (1997), Vogel et al. (1999), Till et al. (2001), Ostheimer et al. (2003), and Schmitz-Linneweber et al. (2005, 2006). The Arabidopsis orthologs of CAF1, CAF2, and CRS1 have analogous functions (Asakura and Barkan, 2006). Arabidopsis HCF152 is the only other putative chloroplast splicing factor reported in land plants (Meierhoff et al., 2003); HCF152 is required for the accumulation of spliced petB RNA, but not for the accumulation of excised petB intron, so its role in splicing is not firmly established. In addition to these nucleus-encoded splicing factors, a role for plastid-encoded MatK in subgroup IIA splicing has been proposed (reviewed in Bonen and Vogel, 2001). * The ndhA intron shows a strong interaction with and dependence on CAF2 and a weak interaction with and dependence on CAF1.
RNC1 also associates with and promotes the splicing of several introns in subgroup IIA: the trnI, trnA, trnK, and trnV introns are strongly enriched in RNC1 coimmunoprecipitates and their splicing is strongly reduced even in weak rnc1 mutants. Two other subgroup IIA introns, rps12-int2 and atpF, also show an interaction with, and some dependence upon, RNC1. Aside from the CRS1–atpF intron interaction, nucleus-encoded proteins involved in the splicing of subgroup IIA introns in land plant chloroplasts have not been reported previously. However, a role for a plastid-encoded protein in their splicing has been proposed based upon their failure to splice in maize and barley mutants lacking plastid ribosomes (Hess et al., 1994; Jenkins et al., 1997; Vogel et al., 1997, 1999). A candidate for this plastid-encoded splicing factor is MatK, which is encoded within the trnK intron. MatK is related to the maturase proteins encoded within many bacterial and fungal group II introns that facilitate the splicing of their host intron (reviewed in Pyle and Lambowitz, 2006). Although a role for MatK in splicing has not been proven, it is possible that RNC1 functions in conjunction with MatK to facilitate the splicing of subgroup IIA introns in the chloroplast.
There is good correspondence between the introns that strongly coimmunoprecipitate with RNC1 and those whose splicing is disrupted in rnc1 mutants, but there are several exceptions. The RIP-chip data suggested that RNC1 associates with RNAs from the intron-containing genes ycf3, rpl2, and possibly rpl16, but the splicing of these RNAs was not detectably reduced in rnc1 mutants compared with the appropriate controls. Because the rpl2 intron, a member of subgroup IIA, fails to splice in all mutants lacking plastid ribosomes, the strong rnc1 alleles were uninformative, and it remains possible that RNC1 does promote rpl2 splicing, with the residual RNC1 in plants harboring weak mutant alleles being sufficient to do the job. Alternatively, RNC1 may act redundantly with other proteins to promote the splicing of these introns, it may be involved in some other aspect of the metabolism of these RNAs, its association with these introns may not be functionally important, or its apparent association with these introns may be artifactual, due to postlysis binding to nonnative ligands. Postlysis exchange of RNA binding proteins from their native targets has been demonstrated (Mili and Steitz, 2004), although the generally excellent correspondence between our RIP-chip data and mutant phenotypes (Schmitz-Linneweber et al., 2005, 2006; Y. Asakura and A. Barkan, unpublished results) suggests that this is the exception rather than the rule. The coimmunoprecipitation data in Figure 4B suggest that RNC1 may interact to a small extent with many or most chloroplast RNAs. Whether these associations reflect a scanning activity of RNC1 as it explores for potential intron binding sites, or perhaps physiologically relevant activities of RNC1 that are independent of splicing, remains to be determined.
Role of RNC1 in Splicing
RNC1 associates with and facilitates the splicing of multiple introns in both subgroups IIA and IIB, giving it the broadest substrate specificity of any group II intron-splicing factor described to date. Nonetheless, several introns show little association with RNC1 and no apparent dependence on RNC1 for their splicing (e.g., ndhA and rps16). Among those introns whose splicing it does influence, RNC1 associates preferentially with and has a more profound effect on some than on others. For example, the splicing of the trnI and trnA introns is highly sensitive to a partial loss of rnc1 function, and these RNAs are very strongly enriched in RNC1 coimmunoprecipitations, whereas rps12-int2 exhibits a weak splicing defect in mild rnc1 alleles and is enriched to a lesser extent in RNC1 coimmunoprecipitations. In fact, the splicing of introns that depend upon both CAF/CRS2 complexes and RNC1 is disrupted less in strong rnc1 mutants than in strong crs2, caf1, or caf2 mutants, implicating the CAF/CRS2 complex as the major player, with RNC1 playing a supporting role. Thus, RNC1 seems to be required for the splicing of some of the introns with which it associates (e.g., trnI and trnA) and to merely enhance the splicing of others (e.g., petB and petD).
Proteins that promote the splicing of group II introns fall into two categories: conserved intron-encoded maturases that have an ancient relationship with their host intron, and proteins encoded by the host genome that were recruited to function in splicing (reviewed in Pyle and Lambowitz, 2006). RNC1 adds to the diversity of host-encoded proteins shown to promote the splicing of group II introns. In land plant chloroplasts, reported host-encoded splicing factors include three CRM domain proteins (CRS1, CAF1, and CAF2), the peptidyl-tRNA hydrolase homolog CRS2, and two members of the PPR family, maize PPR4 and Arabidopsis HCF152; HCF152, however, may stabilize spliced petB mRNA rather than promote splicing, as excised petB intron accumulates normally in hcf152 mutants (Meierhoff et al., 2003). In Chlamydomonas reinhardtii chloroplasts, Raa1, Raa2, and Raa3 promote the trans-splicing of one or both of the split group II introns in the psaA gene; Raa3 exhibits limited similarity to pyridoxamine 5′-phosphate oxidases (Rivier et al., 2001), Raa2 resembles pseudouridine synthase enzymes (Perron et al., 1999), and Raa1 encodes a protein that includes repeated motifs that are reminiscent of tetratricopeptide and PPR motifs (Merendino et al., 2006). In yeast mitochondria, the DEAD box protein MSS116 promotes the splicing of all group I and group II introns (Huang et al., 2005). Despite this diversity, a commonality among many host-derived group I and group II intron splicing factors is their derivation from RNA-interacting proteins that evolved in other contexts. For example, tRNA synthetases were recruited to serve as group I and group II intron splicing factors in fungal mitochondria (reviewed in Lambowitz et al., 1999), CRS2 was derived from a peptidyl-tRNA hydrolase (Jenkins and Barkan, 2001), CRS1, CAF1, and CAF2 were derived from a preribosome binding protein (Barkan et al., 2007), and Raa2 was derived from a pseudouridine synthase (Perron et al., 1999). RNC1 adds a further example of this phenomenon, as it is derived from a RNase III–like protein whose ancestral function presumably involved cleavage of double-stranded RNA. The results presented here show that RNC1 lacks this ancestral endonuclease activity. Analogously, CRS2 seems to lack its ancestral peptidyl-tRNA hydrolase activity (Jenkins and Barkan, 2001; Ostheimer et al., 2005) and Raa2's pseudouridine synthase activity is not necessary for its role in splicing (Perron et al., 1999).
Few group II intron splicing factors have been studied in sufficient detail to gain insight into mechanisms, but to date, all act by enhancing the productive folding of the intron into its catalytically active structure (reviewed in Pyle et al., 2007). Proteins could, in principle, facilitate intron folding by stabilizing an inherently weak native substructure, destabilizing aberrant structures, or precluding the formation of competing nonnative structures. The bacterial LtrA maturase (Matsuura et al., 2001; Noah and Lambowitz, 2003; Rambo and Doudna, 2004) and maize CRS1 (Ostersetzer et al., 2005) bind in vitro with high affinity and specificity to elements in domains 1 and 4 of their cognate introns; this binding is accompanied by the internalization of intron elements that are at the core of the folded ribozyme (reviewed in Pyle and Lambowitz, 2006; Pyle et al., 2007). By contrast, two fungal DEAD box proteins, CYT19 and Mss116p, promote the splicing of several group II introns via nonspecific RNA interactions and do so in a manner that requires ATP but not helicase activity (Mohr et al., 2006; Solem et al., 2006; Grohman et al., 2007; Halls et al., 2007). Given its RNA binding activity and lack of endonuclease activity, it seems very likely that RNC1 likewise promotes intron folding. rRNC1 lacks sequence specificity under the conditions tested, but it remains possible that RNC1 does have intrinsic specificity for its in vivo intron targets when presented to them under physiological conditions. Alternatively, RNC1 might be recruited to specific introns via protein–protein interactions. In fact, structural modeling of the first RNase III domain in RNC1 based on the structure of the endonuclease domain of Aquifex RNase III revealed a candidate protein interaction surface: a large hydrophobic surface in the RNC1 domain that corresponds to a hydrophilic surface in the bacterial protein (data not shown).
Whereas a single nucleus-encoded protein is sufficient to promote the splicing of several mitochondrial group I introns (reviewed in Weeks and Cech, 1995; Lambowitz et al., 1999; Solem et al., 2002) and an intron-encoded maturase is sufficient to promote the splicing of the bacterial LtrB group II intron (reviewed in Lambowitz and Zimmerly, 2004), it is becoming apparent that the machinery for group II intron splicing in chloroplasts is considerably more complex. At least 14 nuclear genes are implicated in the splicing of the two trans-spliced group II introns in the psaA gene in C. reinhardtii chloroplasts (Goldschmidt-Clermont et al., 1990). In maize chloroplasts, three nucleus-encoded proteins (PPR4, CRS2, and CAF2) have been shown to be required for the trans-splicing of rps12-int1 (Ostheimer et al., 2003; Schmitz-Linneweber et al., 2006), and at least four are required for the efficient cis-splicing of the petB intron: RNC1, CRS2, CAF2, and a genetically defined but as yet uncloned gene dubbed crs3 (our unpublished results). The group II introns in land plant chloroplasts appear to include the key intron elements required for group II intron catalysis, yet none of them has been observed to self-splice in vitro. Thus, these introns offer an opportunity not only to elucidate mechanisms for integrating the expression of nuclear and organellar genes but also for the discovery of novel mechanisms by which proteins collaborate with RNA within complex ribonucleoprotein particles.
METHODS
Immunoaffinity Purification of CAF1 and CAF2 RNPs
Generation of the CAF1 and CAF2 antibodies was described previously (Ostheimer et al., 2003). Protein A/G agarose beads (250 μL, 25% suspension; Santa Cruz Biotechnology) were washed three times with PBS plus 0.1% Tween 20 and incubated with ∼100 μg of affinity-purified anti-CAF1 or anti-CAF2 antibodies for 3 h at 4°C. Beads were washed three times in PBS plus 0.1% Tween 20 and three times with 0.2 M sodium borate, pH 9. Antibodies were cross-linked to protein A/G by suspending the beads in 500 μL of 0.2 M sodium borate containing 10 mg/mL dimethyl pimelimidate (Pierce) and rotating for 1 h at 25°C. To stop the reaction, the beads were washed three times in 0.2 M ethanolamine, pH 8, and then incubated in the same buffer for 1 h. After three washes with PBS, beads were stored at 4°C until use.
Chloroplast stroma was prepared from maize (Zea mays) seedling leaves as described previously (Ostheimer et al., 2003) and stored at −80°C at a concentration of 5 to 10 mg/mL protein. Approximately 6 mg of stromal protein was layered onto each of two 10 to 30% sucrose gradients in KEX buffer [30 mM HEPES-KOH, pH 8.0, 150 mM KOAc, 10 mM Mg(OAc)2, and 5 mM DTT] and centrifuged for 4 h at 50,000 rpm in a Beckman SW55Ti rotor at 4°C. Gradient fractions containing CAF1 and CAF2 RNPs (∼600 to 700 kD) were pooled and incubated for 45 min at 4°C with 0.2 mL of protein A/G beads that had been prewashed with RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, and 5 μg/mL aprotinin). Beads were removed by centrifugation, and the precleared lysate was split into two equal aliquots and incubated with either anti-CAF1 or anti-CAF2 coupled protein A/G beads. Beads were incubated with rotation for 90 min at 4°C and then washed five times with 1 mL of RIPA buffer. Proteins were eluted from the beads by two sequential incubations in 75 μL of elution buffer (50 mM sodium phosphate, pH 6.8, and 3% SDS). The eluates were pooled, concentrated using the methanol/chloroform method (Wessel and Flugge, 1984), suspended in SDS loading buffer, and fractionated by SDS-PAGE (4 to 15% polyacrylamide). The gel was stained with silver, and 18 (CAF1 immunoprecipitation) or 23 (CAF2 immunoprecipitation) contiguous gel slices containing proteins between ∼15 and 120 kD were excised and analyzed by mass spectrometry.
Mass Spectrometry
Gel slices were manually washed, reduced, alkylated, digested with trypsin, and extracted principally as described by Shevchenko et al. (1996). The resulting peptides were dried and suspended in 15 μL of 5% formic acid. After verification of digestion by matrix-assisted laser-desorption ionization time-of-flight mass spectrometry peptide mass fingerprinting, all peptide samples were analyzed by nano-liquid chromatography electrospray ionization MS/MS using a Q-TOF mass spectrometer (Waters) interphased with a CapLC (Waters) using the following conditions for liquid chromatography. Peptides were loaded on a guard column (LC Packings; MGU-30-C18PM), followed by separation on a PepMap C18 reverse-phase nano column (LC Packings; nan75-15-03-C18PM), using 90-min gradients with 95% water, 5% acetonitrile, and 0.1% formic acid (solvent A) and 95% acetonitrile, 5% water, and 0.1% formic acid (solvent B) at a flow rate of 200 nL/min. Spectral data were extracted using Mascot distiller and searched with Mascot (Matrix Science) against maize (The Institute for Genomic Research code ZmGI-v6) and rice (Oryza sativa) (The Institute for Genomic Research code OsGI-v4). Search parameters were as follows: only full tryptic peptides, carbamidomethylation as fixed modification; Met oxidation as variable modification; peptide mass tolerance, ±1.2 D; fragment mass tolerance, ±0.8 D; maximally one missed cleavage. The Mascot XML output was automatically processed and further filtered using in-house software (Q. Sun and K.J. Van Wijk, unpublished results), followed by manual verification.
Plant Material
Insertion alleles of rnc1 were identified in a reverse-genetic screen of a collection of ∼2300 Mu transposon–induced nonphotosynthetic maize mutants (http://chloroplast.uoregon.edu). DNA from pooled mutant seedlings was analyzed by PCR with an rnc1-specific primer (5′-TCGGTGGAGAGCTACTGCTC-3′ for rnc1-1 and rnc1-2; 5′-ATCGCAGCCATCGATCAGAA-3′ for rnc1-3) in conjunction with a Mu terminal inverted repeat primer (5′-GCCTCCATTTCGTCGAATCCCG-3′), using a method analogous to that described previously (Williams and Barkan, 2003). Sequences flanking both sides of each identified insertion were amplified by PCR and sequenced to locate the insertion sites. Mutations were tested for complementation by crossing plants heterozygous for each allele in all pairwise combinations. Twenty-four ears were recovered from rnc1-1/+ × rnc1-2/+ crosses, eight of which segregated chlorophyll-deficient mutants. Twenty-four ears were recovered from rnc1-2/+ × rnc1-3/+ crosses, 10 of which segregated chlorophyll-deficient mutants. Twenty-three ears were recovered from rnc1-1/+ × rnc1-3/+ crosses, one of which segregated chlorophyll-deficient mutants. Suppression of the mutant phenotype due to epigenetic silencing of Mu transposons may have caused the low recovery of chlorophyll-deficient mutants in this set of crosses, as described for other Mu-induced alleles with insertions upstream of the start codon (Martienssen et al., 1990).
Other maize mutants used in this work include hcf7 mutants, which are pale green and show reduced polysome assembly and aberrant metabolism of 16S rRNA (Barkan, 1993); fully albino iojap mutants, which lack plastid ribosomes (Walbot and Coe, 1979); w1 mutants, which are albino, lack plastid ribosomes, and do not have a primary defect in carotenoid biosynthesis (Han et al., 1993); and caf2 mutants, which are albino and have a severe plastid ribosome deficiency due to defects in the splicing of multiple group II introns (Ostheimer et al., 2003). The inbred line B73 (Pioneer HiBred) was used as the source of wild-type tissue for RIP-chip and chloroplast fractionation experiments. Seedlings were grown in soil in a growth chamber under a 16-h-light/8-h-dark cycle at 26°C and harvested between 7 and 9 d after planting. Plants harboring any of the mutant allele combinations analyzed here die after the development of approximately three leaves (∼3 weeks of growth) when grown in soil, as is typical of nonphotosynthetic maize mutants.
Analysis of RNA in Mutant Tissue
Seedling leaf RNA was extracted using Tri Reagent (Molecular Research Center). RNA gel blot hybridizations were performed as described previously (Barkan et al., 1994) using 5 μg of leaf RNA to detect chloroplast mRNAs, 0.5 μg of leaf RNA to detect chloroplast tRNAs and rRNAs, and 20 μg of leaf RNA to detect rnc1 mRNA. PCR fragments with the following residues were used as probes for RNA gel blots (residue numbers from GenBank accession number X86563): trnA-UGC intron, residues 98,079 to 98,704; trnI-GAU intron, residues 96,991 to 97,939; trnV-UAC intron, residues 53,156 to 53,834; trnR-ACG, residues 102,631 to 103,061; ndhB intron, residues 90,044 to 90,779; rps12-intron2, residues 92,281 to 92,861; petD intron, residues 74,777 to 75,423; rpl2 intron, residues 82,901 to 83,519; rrn16, residues 95,559 to 96,780; rrn23, residues 98,332 to 99,793; rrn5, residues 102,161 to 102,740; and rrn4.5, residues 101,951 to 102,270. RNase protection assays were performed with the method and probes described previously (Jenkins et al., 1997). Poisoned-primer extension assays to distinguish spliced from unspliced RNAs were performed as described previously (Asakura and Barkan, 2006) using the following primers and dideoxynucleotide: petB, 5′-GACGTTCTTAAAACCAATCATATAC-3′ (ddCTP); petD, 5′-CAGGATCATTTAAGTCAGGTTTC-3′ (ddCTP); atpF, 5′-GTTTTCGATTATCTAATAAATCTTT-3′ (ddCTP); ndhB, 5′-GCAACGACTGGAGTGGGGGA-3′ (ddATP); rps12-intron 1, 5′-GGTTTTTTGGGGTTGATAG-3′ (ddCTP); rps12-intron 2, 5′-TTGGCTTTTTGGCCCCATATT-3′ (ddCTP); and rpl2, 5′-GGCCGTGCCTAAGGGCATATC-3′ (ddCTP). Radioactive gels and blots were exposed to a PhosphorImager screen (Molecular Dynamics) and analyzed using ImageQuant software (GE Healthcare).
Antibody Production
A fragment of RNC1 (amino acids 29 to 190) with a C-terminal 6× His tag was expressed in Escherichia coli and used for the generation of antibodies in rabbits at the University of Oregon Antibody Facility. The expression clone pRNC1-C90 was produced by PCR amplification from DNA from the inbred line B73 (primers 5′-TATACCATGGTGCTTGCCGTTGCGGCCGAC-3′ and 5′-TACCACGTCGACAACGTACTGACCCAGGAACCA-3′). The PCR product was digested with NcoI and SalI and cloned into pet28b that had been digested with NcoI and XhoI. For antigen production, BL21 Star (DE3) cells (Invitrogen) were transformed with pRNC1-C90, and a 1-liter culture was grown with Luria-Bertani medium plus kanamycin (30 μg/L) to an OD600 of 0.6; protein expression was then induced by the addition of 0.8 mM isopropylthio-β-galactoside and cells were grown at 37°C for 120 min. After harvesting, cell pellets were suspended in 60 mL of buffer A (50 mM Na-phosphate, pH 8, 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol) and lysed by passage through a French press. Inclusion bodies were pelleted by centrifugation at 15,000g for 15 min and washed with buffer A containing 2 M urea. Antigen was solubilized in buffer A containing 8 M urea. The clarified antigen was purified by affinity chromatography on nickel-nitrilotriacetic acid agarose beads (Qiagen) that had been equilibrated in buffer A plus 8 M urea. Antigen was eluted in 2 mL of elution buffer (100 mM Na-phosphate, pH 4.5, and 8 M urea), precipitated with ethanol, suspended in PBS, and used for immunization. Antibodies were affinity-purified on a column in which the same RNC1 fragment had been covalently attached to cyanogen bromide–activated Sepharose.
Antisera to spinach (Spinacia oleracea) chloroplast RPL2 and MDH were generously provided by A. Subramanian (University of Arizona) and Kathy Newton (University of Missouri), respectively. Other antibodies used were generated by us and described previously (Voelker and Barkan, 1995).
Chloroplast Fractionation and Protein Analyses
Total leaf protein was extracted and analyzed by immunoblotting as described previously (Barkan, 1998). Chloroplast subfractions were those described by Williams and Barkan (2003). Stromal extracts (0.5 mg of stromal protein per experiment) were fractionated by sedimentation through sucrose gradients according to Jenkins and Barkan (2001).
Analysis of RNAs That Coimmunoprecipitate with RNC1
RNA coimmunoprecipitation assays via RIP-chip and slot-blot hybridization were performed as described previously (Schmitz-Linneweber et al., 2005).
Generation of Recombinant RNC1 for in Vitro Assays
A PCR product encoding RNC1 was generated by PCR amplification using B73 cDNA as a template with primers 5′-ATAAGATCTCGCCGCGTCCGCGTGCTT-3′ and 5′-TATGTCGACTCAGACTGCTTTAGGCTGGAC-3′. The PCR product was digested with BglII and SalI and cloned into BamHI/SalI-cut pMAL-TEV to generate plasmid pRNC1-C180. pMAL-TEV is pMAL-c2X (New England Biolabs) modified to include sequences encoding a TEV protease cleavage site at the EcoRI site. The protein produced after cleavage with TEV protease starts at RNC1 amino acid 24 (five amino acids before the transit peptide cleavage site predicted by ChloroP) and continues to the natural stop codon. The DNA sequence of the insert was verified prior to use for protein expression.
The MBP-RNC1 fusion protein was expressed by introducing pRNC1-C180 into Rosetta 2 cells (Novagen); cultures initiated from a single colony were grown at 37°C on Luria-Bertani medium with carbinicillin and chloramphenicol to an OD600 of 0.4, transferred to 20°C, and grown for another 1 h (OD600 ∼ 0.7). Protein expression was then induced by the addition of isopropylthio-β-galactoside to 0.4 mM, and incubation was continued for 6 h at 20°C. Harvested cells were suspended in 50 mL of ice-cold buffer A (40 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol) containing a Roche Complete (EDTA-free) protease inhibitor cocktail tablet. Cells were lysed at 4°C by sonication with a sonic dismembrator (Fisher 550) with three 1-min pulses (medium tip at setting 5), stirring on ice between each pulse. The lysate was then cleared by centrifugation at 13,000g for 15 min. Nucleic acids were precipitated by slowly adding polyethyleneimine to the lysate from a 10% stock, pH 7.5, to a final concentration of 0.2% and centrifugation at 13,000g for 15 min. The MBP-RNC1 fusion protein was purified from the supernatant by affinity chromatography on amylose-coupled agarose resin (New England Biolabs). The lysate was applied to a 2-mL column equilibrated in buffer A. The column was washed twice with 5 column volumes of buffer A containing protease inhibitors and twice more in buffer A alone. Bound protein was eluted in buffer A containing 15 mM maltose. The fusion protein was digested with 15 μg of TEV protease overnight on ice. TEV protease was purified from pRK793-transformed BL21-RIL cells as described (Kapust et al., 2001). Following filtration (0.45 μm; Amicon Ultrafree-MC), the protein was resolved by size-exclusion chromatography on a 1 × 30 cm Superdex 200 column (Pharmacia) in buffer A. RNC1 eluted as a monomeric species. The small amount of cleaved MBP in the RNC1 peak was removed by application to a 0.5-mL amylose column. The final preparation was dialyzed against buffer A plus 50% glycerol and stored at −20°C. The protein concentration was estimated spectrophotometrically at 280 nm assuming an extinction coefficient of 67,465 M−1 cm−1 (Pace et al., 1995).
RNA Binding Assays
Gel mobility shift assays used gel-purified synthetic RNA 31-mers derived from a randomly selected sequence: the 5′ untranslated region of the maize chloroplast petA mRNA (5′-TTCTAGTACAACTATTGCAGTAGAATGACAA-3′) in both the sense and antisense orientations. The sense strands were 5′ end labeled with T4 polynucleotide kinase and [γ-32P]ATP and purified by phenol chloroform extraction and ethanol precipitation. To generate double-stranded substrates, the labeled oligonucleotides (1 pmol) were annealed with the corresponding unlabeled antisense RNA oligonucleotide (2 pmol) in 24 μL of 40 mM Tris-HCl, pH 7.5, 180 mM NaCl, and 2 mM DTT by heating the samples to 95°C and cooling them slowly over 30 min to 37°C. Single-stranded substrates were heated in that same buffer and snap-cooled on ice. For binding reactions, the single- or double-stranded oligonucleotides were further diluted in annealing buffer and placed on ice. rRNC1 was diluted in its dialysis buffer containing 100 μg/mL BSA, and 5 μL was mixed with 25 μL of annealed nucleic acid and incubated at 25°C for 30 min. The final binding reactions contained 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2 mM DTT, 0.2 mM EDTA, 16.7 μg/mL BSA, 8.3% glycerol, 40 pM oligonucleotide, and the concentrations of protein indicated in the figure legends. Samples were applied to a running nondenaturing 5% polyacrylamide (29:1 acrylamide:bis-acrylamide) gel prepared in 1× THE buffer (Buck et al., 2005). Gels were run at 10 W (constant power) at 4°C with buffer recirculation until the bromphenol blue dye in an adjacent lane had migrated 10 cm. Gels were fixed in 30% methanol and 10% acetic acid, dried, and analyzed with a PhosphorImager.
Filter binding assays were based on the procedure of Wong and Lohman (1993) with modifications as described by Ostersetzer et al. (2005). RNA substrates were prepared by transcription of PCR products containing a T7 RNA polymerase promoter at one end. Deoxyoligonucleotides used for PCR were as follows: rpoBT7for, 5′-TAATACGACTCACTATAGGGGATCCAGAGGGACCCC-3′ and rpoBrev, 5′-GGATCCTCGTGATTAGGC-3′; trnAfor, 5′-CTAATACGACTCACTATAGGGGGATATAGCTCAGTTGGTAGA-3′ and trnArev, 5′-TGGAGATAAGCGGACTCGAACCG-3′. The rpoB substrate is from the 3′ end of the rpoB mRNA and is 723 nucleotides long, while the trnA substrate includes the intron and both exons and is 879 nucleotides long. [α-32P]UTP was incorporated into the RNAs during in vitro transcription, the reactions were treated with DNase I, and RNAs were purified on denaturing polyacrylamide gels. Immediately prior to addition to the binding reactions, RNA was diluted in 4 mM Tris-HCl, pH 7.5, and 0.4 mM EDTA, heated at 95°C for 3 min, and then treated in one of three ways. For reaction set A, the RNA was immediately placed on ice for 3 min before the addition of an equal volume of 2× folding salts (20 mM Tris-HCl, pH 7.5, 180 mM NaCl, and 5 mM DTT). For reaction sets B and C, an equal volume of 2× folding salts that had been prewarmed to 65°C was added directly to the RNA at 95°C and the mixture was slowly cooled (∼20 min) to 35°C. Folding salts (2×) for reaction B contained 20 mM Tris-HCl, pH 7.5, 360 mM NaCl, and 5 mM DTT, while 2× salts for reaction C also included 12 mM MgCl2. Taking into account the salts from the protein dilution buffer, the final salt concentrations were 125 mM NaCl (reaction set A), 200 mM NaCl (reaction set B), and 200 mM NaCl and 5 mM MgCl2 (reaction set C). Binding reactions (30 μL) consisted of folded RNA substrates (10 pM, 25 μL) and 5 μL of diluted rRNC1 in RNC1 dialysis buffer containing 100 μg/mL BSA. After 30 min at 25°C, the reactions were filtered in a slot-blot manifold through sandwiched nitrocellulose and nylon membranes and washed with 125 μL of the respective binding buffer (4°C) lacking BSA and DTT. Binding reactions were quantified by phosphor imaging, and results were plotted using Microsoft Excel and Kaleidograph.
Endonuclease assays used the single-stranded and double-stranded RNA 31-mers prepared for the oligonucleotide binding assays described above. RNAs (4 nM) were incubated for 30 min at 25°C in a 10-μL reaction containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 0.5 mM EDTA, 1 mM DTT, and 5% glycerol plus or minus 10 mM MgCl2. E. coli RNase III (Epicentre) and rRNC1 were diluted in RNC1 dialysis buffer containing 100 μg/mL BSA. Reactions contained rRNC1 at a concentration of 125 nM or E. coli RNase III at 10 nM (dimer concentration). Reactions were stopped by the addition of 3 volumes of formamide dye mix (90% formamide, 20 mM Tris-HCl, pH 7.5, 20 mM EDTA, and 0.04% each xylene cyanol and bromphenol blue) and heating to 95°C. RNA was resolved on a 20% polyacrylamide (19:1 acrylamide:bis-acrylamide) gel containing 8 M urea and detected with a PhosphorImager.
Accession Number
Sequence data for rnc1 cDNA can be found in the GenBank data library under accession number EF650835.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RIP-Chip Data Showing Enrichment of RNA Sequences in RNC1 Immunoprecipitations Involving Wild-Type or rnc1-1/rnc1-2 Mutant Extract.
Supplemental Figure 2. Additional Analyses of Plastid RNA Metabolism in rnc1 Mutants.
Supplemental Table 1. RNC1 Peptides Identified by Mass Spectrometry of CAF1 and CAF2 Immunoprecipitation Reactions.
Supplemental Table 2. Statistical Analysis of RNC1 RIP-Chip Data.
Supplementary Material
Acknowledgments
We are grateful to Christian Schmitz-Linneweber for overseeing the sequencing of rnc1 cDNAs, to the Arizona full-length maize cDNA project for providing the cDNAs, and to Jana Prikryl for help plotting the RNA binding data. Antibodies to RPL2 and MDH were generously provided by A. Subramanian and K. Newton, respectively. Preliminary assays with recombinant RNC1 were performed by Aaron Brooks and Margarita Rojas. This work was supported by grants to A.B. (Grants MCB-0314597 and DBI-0421799) and to K.J.v.W. (Grant DBI-0211935) from the National Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Alice Barkan (abarkan@uoregon.edu).
Online version contains Web-only data.
References
- Asakura, Y., and Barkan, A. (2006). Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 142 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered RNA metabolism. Plant Cell 5 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297 38–57. [Google Scholar]
- Barkan, A., Klipcan, L., Ostersetzer, O., Kawamura, T., Asakura, Y., and Watkins, K. (2007). The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA 13 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 13 3170–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk, J., Tropea, J.E., Bubunenko, M., Routzahn, K.M., Waugh, D.S., Court, D.L., and Ji, X. (2001). Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 9 1225–1236. [DOI] [PubMed] [Google Scholar]
- Bonen, L., and Vogel, J. (2001). The ins and outs of group II introns. Trends Genet. 17 322–331. [DOI] [PubMed] [Google Scholar]
- Buck, A., Kazantsev, A., Dalby, A., and Pace, N. (2005). Structural perspective on the activation of RNase P RNA by protein. Nat. Struct. Mol. Biol. 12 958–964. [DOI] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., and Chanfreau, G. (2003). RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11 1279–1289. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., and von Heijne, G. (2001). Prediction of organellar targeting signals. Biochim. Biophys. Acta 1541 114–119. [DOI] [PubMed] [Google Scholar]
- Gan, J., Tropea, J.E., Austin, B.P., Court, D.L., Waugh, D.S., and Ji, X. (2006). Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 124 355–366. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M., Girard-Bascou, J., Choquet, Y., and Rochaix, J.-D. (1990). Trans-splicing mutants of Chlamydomonas reinhardtii. Mol. Gen. Genet. 223 417–425. [DOI] [PubMed] [Google Scholar]
- Grohman, J.K., Del Campo, M., Bhaskaran, H., Tijerina, P., Lambowitz, A.M., and Russell, R. (2007). Probing the mechanisms of DEAD-box proteins as general RNA chaperones: The C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry 46 3013–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls, C., Mohr, S., Del Campo, M., Yang, Q., Jankowsky, E., and Lambowitz, A.M. (2007). Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J. Mol. Biol. 365 835–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, C.-D., Patrie, W., Polacco, M., and Coe, E.H. (1993). Aberrations in plastid transcripts and deficiency of plastid DNA in striped and albino mutants in maize. Planta 191 552–563. [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Kim, Y.K., Jin, H., and Kim, V.N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, W.R., Hoch, B., Zeltz, P., Huebschmann, T., Koessel, H., and Boerner, T. (1994). Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.R., Rowe, C.E., Mohr, S., Jiang, Y., Lambowitz, A.M., and Perlman, P.S. (2005). The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. USA 102 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B., and Barkan, A. (2001). Recruitment of a peptidyl-tRNA hydrolase as a facilitator of group II intron splicing in chloroplasts. EMBO J. 20 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, B., Kulhanek, D., and Barkan, A. (1997). Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell 9 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust, R., Tözsér, K., Fox, J., Anderson, D., Cherry, S., Copeland, T., and Waugh, D. (2001). Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14 993–1000. [DOI] [PubMed] [Google Scholar]
- Lambowitz, A., Caprara, M., Zimmerly, S., and Perlman, P. (1999). Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In The RNA World, R. Gesteland, T. Cech, and J. Atkins, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 451–485.
- Lambowitz, A.M., and Zimmerly, S. (2004). Mobile group II introns. Annu. Rev. Genet. 38 1–35. [DOI] [PubMed] [Google Scholar]
- MacRae, I.J., and Doudna, J.A. (2007). Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 17 138–145. [DOI] [PubMed] [Google Scholar]
- Martienssen, R., Barkan, A., Taylor, W., and Freeling, M. (1990). Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes Dev. 4 331–343. [DOI] [PubMed] [Google Scholar]
- Matsuura, M., Noah, J., and Lambowitz, A. (2001). Mechanism of maturase-promoted group II intron splicing. EMBO J. 20 7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., and Schuster, G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino, L., Perron, K., Rahire, M., Howald, I., Rochaix, J.D., and Goldschmidt-Clermont, M. (2006). A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 34 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, F., Umesono, K., and Ozeki, H. (1989). Comparative and functional anatomy of group II catalytic introns—A review. Gene 82 5–30. [DOI] [PubMed] [Google Scholar]
- Mili, S., and Steitz, J.A. (2004). Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S., Matsuura, M., Perlman, P.S., and Lambowitz, A.M. (2006). A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc. Natl. Acad. Sci. USA 103 3569–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah, J., and Lambowitz, A. (2003). Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry 42 12466–12480. [DOI] [PubMed] [Google Scholar]
- Ostersetzer, O., Watkins, K., Cooke, A., and Barkan, A. (2005). CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., and Barkan, A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 22 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G.J., Hadjivassiliou, H., Kloer, D.P., Barkan, A., and Matthews, B.W. (2005). Structural analysis of the group II intron splicing factor CRS2 yields insights into its protein and RNA interaction surfaces. J. Mol. Biol. 345 51–68. [DOI] [PubMed] [Google Scholar]
- Pace, C., Vajdos, F., Fee, L., Grimsley, G., and Gray, T. (1995). How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 11 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K., Goldschmidt-Clermont, M., and Rochaix, J.-D. (1999). A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18 6481–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle, A., and Lambowitz, A. (2006). Group II introns: Ribozymes that splice RNA and invade DNA. In The RNA World, R. Gesteland, T. Cech, and J. Atkins, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 469–506.
- Pyle, A.M., Fedorova, O., and Waldsich, C. (2007). Folding of group II introns: A model system for large, multidomain RNAs? Trends Biochem. Sci. 32 138–145. [DOI] [PubMed] [Google Scholar]
- Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., and Lopez, R. (2005). InterProScan: Protein domains identifier. Nucleic Acids Res. 33 W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo, R.P., and Doudna, J.A. (2004). Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43 6486–6497. [DOI] [PubMed] [Google Scholar]
- Rivier, C., Goldschmidt-Clermont, M., and Rochaix, J. (2001). Identification of an RNA-protein complex involved in chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 20 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell 17 2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Williams-Carrier, R.E., Williams-Voelker, P.M., Kroeger, T.S., Vichas, A., and Barkan, A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained in polyacrylamide gels. Anal. Chem. 68 850–858. [DOI] [PubMed] [Google Scholar]
- Small, I., and Peeters, N. (2000). The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25 46–47. [DOI] [PubMed] [Google Scholar]
- Solem, A., Chatterjee, P., and Caprara, M. (2002). A novel mechanism for protein-assisted group I intron splicing. RNA 8 412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem, A., Zingler, N., and Pyle, A.M. (2006). A DEAD protein that activates intron self-splicing without unwinding RNA. Mol. Cell 24 611–617. [DOI] [PubMed] [Google Scholar]
- Stern, D., Hanson, M., and Barkan, A. (2004). Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci. 9 293–301. [DOI] [PubMed] [Google Scholar]
- Sun, W., Jun, E., and Nicholson, A.W. (2001). Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry 40 14976–14984. [DOI] [PubMed] [Google Scholar]
- Sun, W., Li, G., and Nicholson, A.W. (2004). Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry 43 13054–13062. [DOI] [PubMed] [Google Scholar]
- Sun, W., and Nicholson, A.W. (2001). Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant. Biochemistry 40 5102–5110. [DOI] [PubMed] [Google Scholar]
- Till, B., Schmitz-Linneweber, C., Williams-Carrier, R., and Barkan, A. (2001). CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA 7 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, R., and Barkan, A. (1995). Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 14 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Boerner, T., and Hess, W. (1999). Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 27 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., Hubschmann, T., Borner, T., and Hess, W. (1997). Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: Support for MatK as an essential splice factor. J. Mol. Biol. 270 179–187. [DOI] [PubMed] [Google Scholar]
- Walbot, V., and Coe, E.H. (1979). Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl. Acad. Sci. USA 76 2760–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, N.S., Stiffler, N., and Barkan, A. (2007). POGs/PlantRBP: A resource for comparative genomics in plants. Nucleic Acids Res. 35 D852–D856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, K.M., and Cech, T.R. (1995). Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry 34 7728–7738. [DOI] [PubMed] [Google Scholar]
- Wessel, D., and Flugge, U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138 141–143. [DOI] [PubMed] [Google Scholar]
- Williams, P., and Barkan, A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36 675–686. [DOI] [PubMed] [Google Scholar]
- Wong, I., and Lohman, T. (1993). A double-filter method for nitrocellulose-filter binding: Application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. USA 90 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Jaskiewicz, L., Westhof, E., and Filipowicz, W. (2004). Single processing center models for human Dicer and bacterial RNase III. Cell 118 57–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.