Abstract
We describe a boron (B) transporter, Os BOR1, in rice (Oryza sativa). Os BOR1 is a plasma membrane–localized efflux transporter of B and is required for normal growth of rice plants under conditions of limited B supply (referred to as -B). Disruption of Os BOR1 reduced B uptake and xylem loading of B. The accumulation of Os BOR1 transcripts was higher in roots than that in shoots and was not affected by B deprivation; however, Os BOR1 was detected in the roots of wild-type plants under -B conditions, but not under normal conditions, suggesting regulation of protein accumulation in response to B nutrition. Interestingly, tissue specificity of Os BOR1 expression is affected by B treatment. Transgenic rice plants containing an Os BOR1 promoter–β-glucuronidase (GUS) fusion construct grown with a normal B supply showed the strongest GUS activity in the steles, whereas after 3 d of -B treatment, GUS activity was elevated in the exodermis. After 6 d of -B treatment, GUS activity was again strong in the stele. Our results demonstrate that Os BOR1 is required both for efficient B uptake and for xylem loading of B. Possible roles of the temporal changes in tissue-specific patterns of Os BOR1 expression in response to B condition are discussed.
INTRODUCTION
Boron (B) is an essential nutrient for plants, and B deficiency has been reported in 132 crops in >80 countries (Shorrocks, 1997). B deficiency generally affects the growing portion of the plant. B cross-links rhamnogalacturonan-II (RG-II) in the cell wall (Matoh et al., 1993), and borate-RG-II complexes have been detected in a wide range of plant species (Matoh et al., 1996; Matsunaga et al., 2004). The cross-linking of RG-II by B is required for the normal expansion of Arabidopsis thaliana rosette leaves (O'Neill et al., 2001). This requirement is probably one basis for the symptoms that appear in young portions of B-deficient plants. B deficiency also affects membrane functioning and metabolic activities (for review, see Bolanos et al., 2004), but it is likely that these effects are indirect consequences of the deficiency.
The B content in the cell walls of shoots of well-fertilized rice (Oryza sativa) plants is in the range of 5 mg B kg−1 of cell wall (Matoh et al., 1996). This value is 5- to 10-fold lower than the levels in dicotyledonous plants, the cell walls of which typically contain ∼25 to 45 mg B kg−1 of cell wall (Matoh et al., 1996). The B contents in leaf tissues of different plant species, grown in the same location, can differ considerably. Graminaceous species generally contain lower B content than dicotyledonous species (Marschner, 1995). The B requirements of graminaceous species are also lower than those of dicotyledonous species and correlate positively with the pectin contents of the cell walls (reviewed in Rerkasem and Jamjod, 2004). For example, symptoms of B deficiency appear in rice plants when the boric acid concentration in the hydroponic solution supporting these plants is <0.2 μM (Yu and Bell, 1998). In Arabidopsis, B deficiency symptoms are evident in wild-type plants grown in hydroponic solution containing 0.5 μM boric acid (Miwa et al., 2006). These differences between rice and dicotyledonous plants with respect to the B concentrations in the shoot tissues and the susceptibilities to B deficiency suggest differences in the B transport capacities of these plants.
It has long been believed that the passive transport of boric acid supplied sufficient B to the plant (Marschner, 1995). Recently, the active transport of B was reported in sunflower (Helianthus annuus) (Dannel et al., 1998). Soon thereafter, the Arabidopsis gene At BOR1 was identified as the first B transporter in a living system (Takano et al., 2002). At BOR1 is an efflux-type B transporter that functions in xylem loading and is essential for preventing B deficiency in shoots. Six BOR1-like genes are present in Arabidopsis. BOR1-like genes have also been found in a number of other plant species, including rice, suggesting the importance of BOR1 and similar genes in plants.
In excess, B is toxic. Therefore, it is important to regulate B transport in response to B conditions in the environment, as with other essential nutrients. For example, major transporters in plants, such as the ammonium transporters (AMTs) (Loque and von Wirén, 2004) and iron transporters (Ishimaru et al., 2006) are regulated at the transcriptional level and respond to the status of the corresponding nutrient. The iron transporter gene At IRT1 is regulated at both transcriptional and posttranscriptional levels (Connolly et al., 2002). Arabidopsis NIP5;1, a B channel crucial for B uptake under low B conditions, is regulated at the mRNA level by the B status (Takano et al., 2006). It has also been demonstrated that At BOR1 is regulated posttranscriptionally in response to B availability (Takano et al., 2005). This regulation is likely to be important in preventing the overaccumulation of B in shoots under high B conditions.
To understand the molecular mechanisms involved in B transport in rice, we focused on Os BOR1, the rice gene most similar to At BOR1. The roles of Os BOR1 in B transport and the regulation of Os BOR1 are described.
RESULTS
At BOR1-Like Genes in Rice
A database search of the rice genome for At BOR1-like genes identified four genes. We obtained cDNAs corresponding to these genes, determined their nucleotide sequences, and identified their intron-exon structures and open reading frames (ORFs). Based on the predicted amino acid sequences, we named these genes Os BOR1-4. The identifiers of these genes are shown in Table 1. The nucleotide sequences of the cDNAs corresponding to Os BOR1 and Os BOR3 are identical to sequences in the database (GenBank accession numbers AK070617 and AK072421, respectively). The nucleotide sequences of the Os BOR2 and Os BOR4 cDNAs were confirmed by direct sequencing of an RT-PCR product and three independently isolated RT-PCR products (accession numbers DQ421408 and DQ421409, respectively).
Table 1.
Nomenclature of At BOR1-Like Genes in Rice and Arabidopsis
| Rice
| ||||
|---|---|---|---|---|
| Gene name | Os BOR1 | Os BOR2 | Os BOR3 | Os BOR4 |
| GenBank accession number | AK070617 | DQ421408 | AK072421 | DQ421409 |
| Locus identifier | LOC_Os12g37840 | LOC_Os01g08040 | LOC_Os01g08020 | LOC_Os05g08430 |
|
Arabidopsis
| |||||||
|---|---|---|---|---|---|---|---|
| Gene name | At BOR1 | At BOR2 | At BOR3 | At BOR4 | At BOR5 | At BOR6 | At BOR7 |
| GenBank accession number | NC_003071 | NP_191786 | NP_187296 | NP_172999 | NP_177619 | NP_197925 | NP_194977 |
| NM_180138 | NM_116092 | NM_111520 | NM_101415 | NM_106139 | NM_122453 | NM_119403 | |
| Locus identifier | At2g47160 | At3g62270 | At3g06450 | At1g15460 | At1g74810 | At5g25430 | At4g32510 |
GenBank accession numbers and locus identifiers are shown. Locus identifiers are the current version, except for Os BOR2, which represents The Arabidopsis Information Resource version 3.
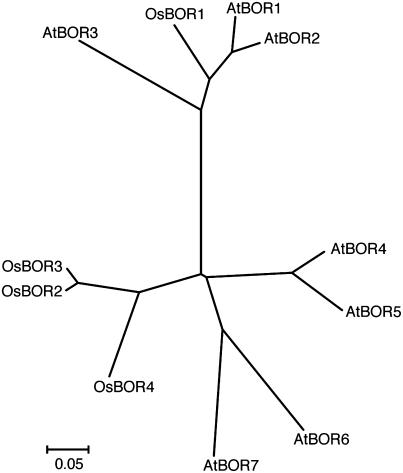
A phylogenetic analysis of the predicted amino acid sequences of the Arabidopsis and rice BOR1-like genes suggested that Os BOR1 is most similar to At BOR1, and Os BOR2, 3, and 4 are more distantly related to At BOR1 than Os BOR1 (Figure 1). Three of the four rice At BOR1-like genes, Os BOR2, 3, and 4, form a branch distinct from the Arabidopsis genes (Figure 1). Os BOR1 is predicted to encode a polypeptide of 711 amino acids. The Phobius program (Kall et al., 2004) predicted that Os BOR1 contains 10 transmembrane domains, as does At BOR1.
Figure 1.
Phylogenetic Analysis of At BOR1 and At BOR1-Like Genes in Arabidopsis and Rice.
A phylogenetic analysis was performed with MEGA 3.1 (http://www.megasoftware.net) using the neighbor-joining method (Saitou and Nei, 1987). Aligned sequences corresponding to residues 65 to 412 of Os BOR1 were used to generate the phylogenetic tree. The accession numbers and gene identifiers of each gene are shown in Table 1.
Os BOR1 Reduces the B Concentration in Yeast Cells
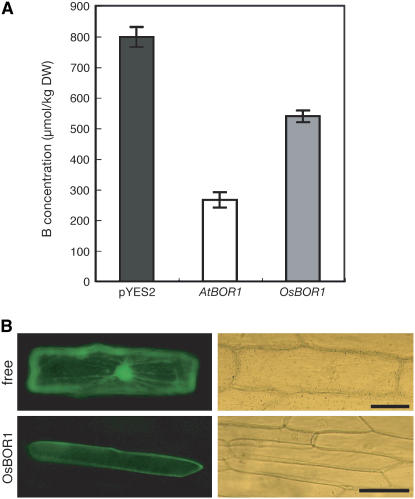
To examine the B efflux activity of Os BOR1, we expressed the gene in the Saccharomyces cerevisiae strain scbor1Δ (Takano et al., 2002; Nozawa et al., 2006). scbor1Δ cells lack an endogenous B efflux transporter. Cells in the mid-log phase cultured in liquid medium were exposed to 100 μM boric acid for 60 min, and the B concentration in the cells was determined. Transformants carrying the empty vector pYES2 and exposed to 100 μM B accumulated B to ∼800 μmol/kg dry weight (Figure 2A), whereas the B concentrations in cells expressing At BOR1 (Takano et al., 2002) or Os BOR1 were ∼270 and 540 μmol/kg dry weight, respectively (Figure 2A), reducing the B concentrations to 66 and 32% of the vector control, respectively. These results suggest that Os BOR1 is an efflux transporter of B, as is At BOR1.
Figure 2.
B Export Activity and Subcellular Localization of Os BOR1.
(A) B concentration in yeast cells expressing At BOR1 and Os BOR1. The B concentrations in scbor1Δ mutant cells carrying pYES2 (black bar) or pYES2 with the At BOR1 cDNA (white bar) or the Os BOR1 cDNA (gray bar) are shown. Cells were exposed for 60 min to medium containing 100 μM boric acid (n = 3). DW, dry weight.
(B) Subcellular localization of Os BOR1-sGFP in onion epidermal cells. Free and Os BOR1 indicate cells bombarded with the Pro35S:sGFP (pTH2; Chiu et al., 1996) or Pro35S:Os BOR1-sGFP constructs, respectively. Left and right panels represent fluorescence and light microscopy, respectively. Bars = 100 μm.
Os BOR1 Localizes to the Plasma Membrane
We investigated the subcellular localization of Os BOR1. A cauliflower mosaic virus 35S RNA (referred to as 35S) promoter:GFP-Os BOR1 cDNA fusion construct was introduced into onion (Allium cepa) epidermal cells by particle bombardment (Figure 2B). The sGFP-Os BOR1 fusion protein localized to the cell periphery, in contrast with the protein produced from Pro35S:sGFP (Figure 2B). We confirmed this plasma membrane localization of the sGFP-Os BOR1 fusion protein by plasmolyzing the cells with 1 M mannitol (Yamaguchi et al., 2005). The fluorescence signal was localized only at the thin layer of the cytosol and not at the cell wall (data not shown). Similar results were obtained with the bombardment of plasmids containing a Pro35S:Os BOR1-sGFP fusion gene (data not shown). These results suggest that Os BOR1 localizes to the plasma membrane.
Accumulation of Os BOR1 Transcripts in Rice and Their Detection in the Root Endodermis
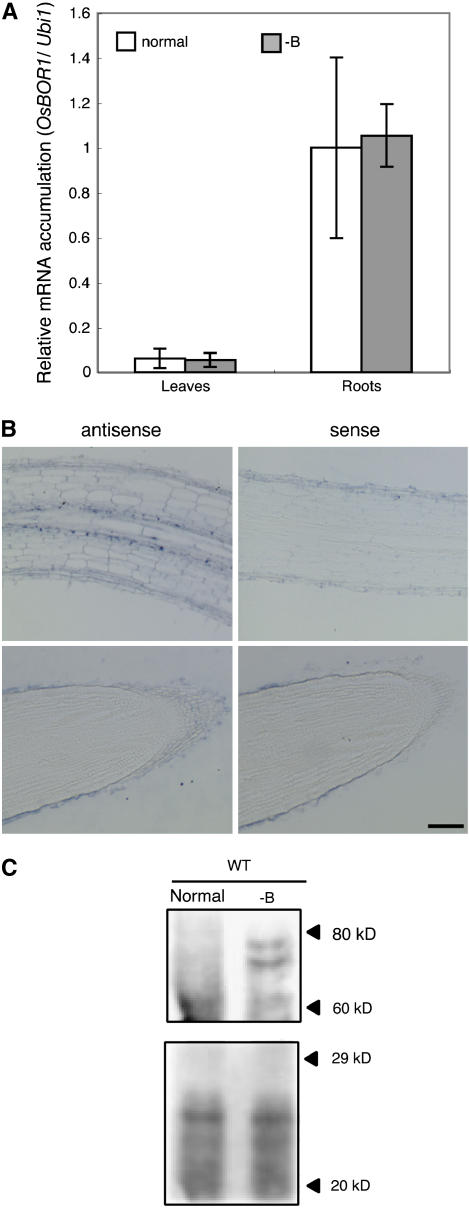
We examined the accumulation of Os BOR1 transcripts in shoots and roots in response to the B status. Plants were grown in medium (Fukuda et al., 2004) containing 18 μM boric acid (referred to as normal medium in this manuscript) for 10 d and then exposed to -B medium for 3 d. The accumulation of Os BOR1 transcripts relative to the Ubi1 transcript levels in leaves and roots was determined by RT-PCR. The relative accumulation of Os BOR1 transcripts to Ubi1 transcripts in roots was 10 times higher than the relative value in leaves (Figure 3A). The accumulation of Os BOR1 transcripts in leaves and roots was not affected by 3 d of B deprivation (Figure 3A).
Figure 3.
Accumulation of Os BOR1 Transcripts and Os BOR1 in Roots in Response to B Conditions, and Localization of Os BOR1 Transcripts.
(A) Quantitative RT-PCR determination of Os BOR1 mRNA accumulation in response to B conditions. Seventeen-day-old wild-type plants were grown in normal (white bars) or -B (gray bars) medium for 3 d. Averages ± sd of Os BOR1 mRNA accumulation relative to the Ubi1 transcript levels are shown for leaves and roots (n = 3).
(B) In situ hybridization analysis of Os BOR1 transcripts in rice roots. The top and bottom panels show longitudinal sections of the root elongation zone and root tips, respectively. The sections were hybridized with antisense or sense Os BOR1 probes. The magnifications of the four panels are identical. Bar = 10 μm.
(C) The roots of 17-d-old wild-type plants grown in normal or -B medium for 3 d were harvested. Five micrograms of microsomal proteins from roots were subjected to SDS-PAGE separation followed by immunoblot analysis using an anti-Os BOR1 antibody (top panel). The gel was stained with Coomassie blue (bottom panel). The numbers at right represent molecular masses (kilodaltons).
To analyze the cell-type specificity of Os BOR1 mRNA accumulation, we performed in situ hybridization using the Os BOR1 ORF region as a probe. In the roots of rice plants grown for 4 d in Murashige and Skoog medium, which contains 100 μM boric acid, Os BOR1 mRNA was detected in the endodermis and possibly the pericycle, but little Os BOR1 mRNA accumulated in root tips (Figure 3B).
Os BOR1 Accumulation Increases under -B Conditions
The accumulation of Os BOR1 was examined using immunoblot analysis with an antiserum raised against a partial Os BOR1 peptide. Microsomal fractions were prepared from the roots of wild-type plants grown in normal or -B medium. Two bands with predicted molecular masses of 82 and 76 kD were detected in samples prepared from -B plants. No corresponding bands were detected in the wild type under normal B conditions (Figure 3C). The predicted molecular mass of Os BOR1 is 79,336 D, similar to the positions of the bands. These results suggest that the accumulation of Os BOR1 is enhanced under B deprivation, similar to At BOR1 (Takano et al., 2005). Two bands were detected at this position. It is possible that the upper band represents an ubiquitinated form of Os BOR1. Identical band patterns were observed for Os BOR1 expressed in yeast (data not shown), suggesting that Os BOR1 is likely to be ubiquitinated in yeast.
Molecular Characterization of Tos17 Insertion Mutants in Os BOR1
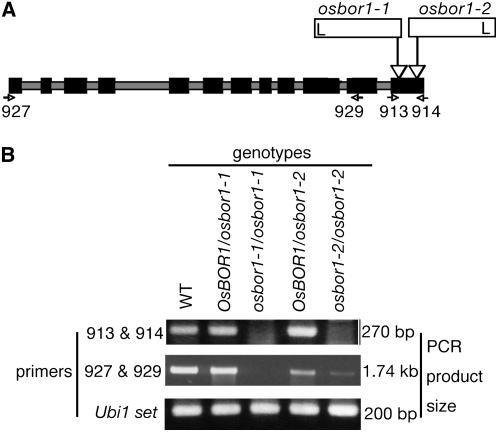
Two independent insertion lines of the retrotransposon Tos17 in the Os BOR1 gene were obtained from the Rice Genome Resource Center in Tsukuba, Japan (Miyao et al., 2003). DNA fragments of the sequences at both sides of the boundaries between Os BOR1 and the Tos17 insertions were amplified and the nucleotide sequences determined. In both lines, the Tos17 insertion had inserted in exon 12, but at different positions (Figure 4A). The Tos17 insertions are located 1941 and 2116 bp from the first ATG of the Os BOR1 cDNA, and these lines were named osbor1-1 and osbor1-2, respectively (Figure 4A).
Figure 4.
Schematic Presentation of Tos17 Insertion Sites in Os BOR1, and RT-PCR Detection of Os BOR1 Transcripts in the Mutants.
(A) Diagram of insertion positions of the retrotransposon Tos17 in Os BOR1. Black boxes and gray bars represent exons and introns, respectively. Open bars represent Tos17 insertions. The positions of the left border of the inverted repeat are shown (L). Arrowheads indicate the positions and orientations of the primers that were used for RT-PCR.
(B) RT-PCR analysis of Os BOR1 transcript accumulation in the leaves of 4-week-old mutant plants. The primers used are shown to the left, and the sizes of the PCR products are shown to the right. Os BOR1/osbor1-1, osbor1-1 heterozygote for the Tos17 insertion; osbor1-1/osbor1-1, osbor1-1 homozygote for the Tos17 insertion; Os BOR1/osbor1-2, osbor1-2 heterozygote for the Tos17 insertion; osbor1-2/osbor1-2, osbor1-2 homozygote for the Tos17 insertion.
The accumulation of Os BOR1 transcripts in these mutant lines was determined by RT-PCR using the primers 913 and 914 (Table 2), which anneal upstream and downstream of the Tos17 insertion sites in both osbor1-1 and osbor1-2 (Figure 4A). In both lines, a band of the expected size (270 bp) was detected in the wild type and the heterozygous mutants, but not in homozygous plants (Figure 4B). PCR using the primers 927 and 929, which anneal two positions upstream of the Tos17 insertions (Figure 4A), produced bands with the expected size (1.74 kb) from the wild type and osbor1-1 and osbor1-2 heterozygotes, but not from osbor1-1 homozygotes (Figure 4B). In osbor1-2 homozygotes, a weak band of the expected size was detected (Figure 4B). These results suggest that no detectable Os BOR1 transcript accumulates in osbor1-1, whereas in osbor1-2, a nearly full-length Os BOR1 transcript, likely to contain a portion of the Tos17 sequence, accumulates.
Table 2.
Primer Sequences
| Primer | Sequence |
|---|---|
| Ubi1-F | 5′-GACAAGGAGGGAATCCCG-3′ |
| Ubi1-R | 5′-GCATAGCATTTGCGGCA-3′ |
| 654 | 5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′ |
| 913 | 5′-CACTAGAAGCCGTGGTGAAA-3′ |
| 914 | 5′-CAGGTAGTTGCATAGCTCAT-3′ |
| 927 | 5′-ATGGAGGAGAGCTTCGTGCC-3′ |
| 928 | 5′-ATGGAGGAGAGCTTCGTGCC-3′ |
| 929 | 5′-ACCAGCAGCATGATCATGAG-3′ |
| 930 | 5′-GGTGCTCCAAATCAGGAAAT-3′ |
| 1019 | 5′-GAAAACCATGGCTCCTCCTCCTCCCTTTGGTGTTGATCCTT-3′ |
| 1096 | 5′-AAAAAGCAGGCTGGGTTGGCCAGTACAATTTTTC-3′ |
| 1097 | 5′-AGAAAGCTGGGTGTTTTCTCTTCTTGGACCCAAAAGAGG-3′ |
| 1136 | 5′-AAAAAGCAGGCTGGATGGAGGAGAGCTTCGTGCC-3′ |
| 1137 | 5′-AGAAAGCTGGGTGTCACTTTGGTGTTGATCCTT-3′ |
Disruption of Os BOR1 Causes Increased Sensitivity to B Deficiency
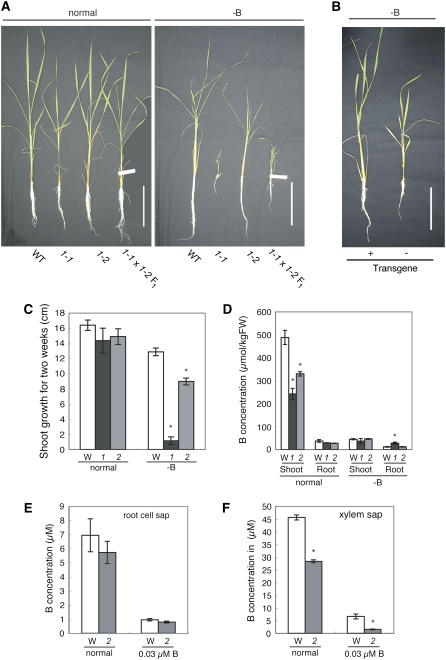
The growth of wild-type plants was slightly reduced under -B conditions (Figure 5A). Growth was obviously reduced in young leaves, similar to other plant species. The growth of osbor1-1 and osbor1-2 was indistinguishable from that of the wild type under conditions of normal B supply (Figure 5A), but under -B conditions, the growth of the mutants was reduced in both the aerial portions and the roots. The growth reduction in osbor1-1 mutant plants under -B conditions was more severe than that of osbor1-2 (Figure 5A). The reduced growth under -B conditions in two independent Tos17 insertion lines suggests that the defect was caused by the disruption of the Os BOR1 gene. osbor1-1 plants were smaller than wild-type plants at maturity, and most of the seeds of osbor1-1 plants did not mature.
Figure 5.
Physiological Analysis of osbor1 Mutant Plants and Complementation of osbor1 with the Pro35S:Os BOR1 cDNA Construct.
(A) One-week-old wild-type and mutant plants were grown for 4 weeks in normal or -B medium before the representative photos shown were taken. 1-1, osbor1-1; 1-2, osbor1-2; 1-1 × 1-2 F1, F1 progeny of a cross of osbor1-1 and osbor1-2. Bars = 10 cm.
(B) Growth of osbor1-2 containing (+) or lacking (−) the Pro35S:Os BOR1 transgene after 4 weeks of -B treatment. Bar = 10 cm.
(C) Shoot growth of the wild type (W) and osbor1-1 (1) and osbor1-2 (2) grown for 2 weeks in normal or -B medium. Averages and se (n = 3 to 5) are shown. An asterisk above a bar indicates a significant difference (P < 0.05) between the wild type and the mutant by the Student's t test.
(D) B concentration on a fresh weight basis in shoots and roots of wild type (W) and osbor1-1 (1) and osbor1-2 (2) grown in normal or -B medium for 2 weeks (the same materials as [C]). Averages and se (n = 3 to 5) are shown. An asterisk above a bar indicates a significant difference (P < 0.05) between the wild type and the mutant by the Student's t test.
(E) B concentrations in the cell sap of roots of wild type (W) and osbor1-2 (2) grown for 3 d in normal medium or medium containing 0.03 μM B. Seven-day-old plants germinated with deionized water were grown for 10 d in normal medium before the treatment. Averages and se (n = 3) are shown. The difference between the wild type and the mutant was not significant.
(F) Concentration of B in the xylem sap of wild-type (W) and osbor1-2 (2) plants grown for 3 d in normal medium or medium containing 0.03 μM B. The plants were precultured as in (E). Averages and se (n = 3) are shown. An asterisk above a bar indicates a significant difference (P < 0.05) between the wild type and the mutant by the Student's t test.
To confirm that the growth defects were caused by the insertion in Os BOR1, we conducted three experiments. First, we grew 10 progeny of osbor1-2 heterozygous plants under both normal and -B conditions. Under normal conditions, all of the plants were indistinguishable from wild-type plants. Under -B conditions, 3 of the 10 plants grew less well than the others (data not shown). The three poorly growing plants were found to be homozygous for the Tos17 insertion in Os BOR1, but none of the other seven plants were homozygous.
Second, we crossed a heterozygous osbor1-1 plant (genotype Os BOR1/osbor1-1) and a homozygous osbor1-2 plant (osbor1-2/osbor1-2) and observed the growth of the F1 progeny in -B medium. Seven of the 14 F1 progeny showed reduced growth under -B conditions. A typical F1 plant grown under these conditions is shown in Figure 5A. The growth of the other seven plants was similar to that of wild-type plants. All -B-sensitive plants were found to carry the osbor1-1/osbor1-2 alleles, and all of the other plants carried the Os BOR1/osbor1-2 alleles. F1 progeny were also grown under normal B conditions, and no growth defects were observed in plants carrying the osbor1-1/osbor1-2 alleles (Figure 5A).
Finally, we complemented osbor1 with the Os BOR1 cDNA. A full-length Os BOR1 cDNA under the control of the 35S promoter was introduced into homozygous osbor1-2 plants, and three independent transgenic lines were obtained. Hygromycin-resistant (Hygr) plants of the T1 generation of the transgenic lines were subjected to 4 weeks of -B treatment in hydroponic culture. In two of the three independent transgenic lines, all of the Hygr plants grew similarly to the wild type under -B conditions, whereas osbor1-2 mutant plants germinated in the absence of Hyg grew poorly (Figure 5B), establishing that the growth defects of osbor1-2 mutant plants under -B conditions were eliminated by the introduction of the Pro35S:Os BOR1 cDNA construct.
Taken together, these results show that the growth defects of osbor1-1 and osbor1-2 observed under -B conditions were caused by the disruption of Os BOR1. The results also demonstrate that osbor1-1 and osbor1-2 are recessive alleles.
Both of the osbor1 recessive alleles result in sensitivity to B deficiency, but the phenotype of osbor1-1 is more severe than that of osbor1-2 (Figure 5A). Two factors probably contribute to the different severities of the phenotypes. In osbor1-1, no Os BOR1 transcript was detected, whereas Os BOR1 transcript was detectable in osbor1-2 (Figure 4B). The Os BOR1 transcript in the osbor1-2 mutant contains the Tos17 sequence near the C terminus, and this transcript produces a functional B efflux transporter, as a reduction in the B concentration was observed in yeast cells expressing a cDNA corresponding to the osbor1-2 transcript (data not shown).
Detailed Growth Properties of osbor1 Mutants under -B Conditions
The osbor1-1 and osbor1-2 mutants were subjected to -B treatment, and both plant lines showed reduced growth in -B conditions in both shoots and roots, albeit with different severities. The growth of the shoots of the wild type and the osbor1-1 and osbor1-2 mutant plants were similar in normal medium (Figure 5C), but in the -B medium, the growth of osbor1-1 and osbor1-2 was ∼10 and 70% of that of the wild type, respectively. These results suggest that Os BOR1 is required for the maintenance of growth under -B conditions.
B Transport Properties of osbor1 Mutant Plants
Next, we measured the B concentrations in wild-type and osbor1-1 and osbor1-2 plants that had been grown in normal or -B medium for 2 weeks (Figure 5D). The B concentration in the shoots of osbor1-1 and osbor1-2 plants grown under the normal conditions was reduced to ∼50 and 70% that of wild-type plants, respectively (Figure 5D). In the roots of these plants, there was a trend that the concentration was less in the mutants than the wild-type plants. By contrast, in the shoots and roots of plants grown under the -B conditions, no significant reduction of B concentration between the two mutants and the wild type was observed. From these results, we concluded that Os BOR1 is likely to play a role in the uptake and xylem loading of B from roots to shoots.
To further characterize the transport properties in these mutants, we determined the B concentrations in root cell and xylem saps. osbor1-2 mutant plants were grown hydroponically for 18 d in normal B medium and then grown in normal B or -B (0.03 μM in this particular experiment) medium for 3 d before the collection of xylem and root cell saps. The B concentrations in root cell sap were similar in the wild type and the mutant after both normal and -B treatments (Figure 5E). In wild-type and osbor1-2 root cell saps, the B concentrations were in the range of 6 to 7 μM and ∼1 μM after 3 d of exposure to normal or 0.03 μM B medium, respectively. The B concentrations in root cell saps under normal conditions (6 to 7 μM) were less than half the B concentrations in standard nutrient solution (18 μM), suggesting that rice plants do not concentrate B upon its uptake from the medium. Under -B conditions, the B concentrations in root cell saps in both the wild type and the osbor1-2 mutant (∼1 μM) were >30-fold higher than those from plants grown in low B medium (0.03 μM), presumably representing the residual B from the preculture in normal medium.
By contrast, the B concentration in xylem sap of wild-type plants grown in normal medium containing 18 μM boric acid was ∼45 μM, suggesting that rice is able to concentrate B in the process of xylem loading. The B concentration in xylem sap collected from osbor1-2 plants grown in normal medium was 60% of that in the wild type (Figure 5F), suggesting that Os BOR1 is involved in the xylem loading of B. Moreover, in plants grown in -B medium for 3 d, the B concentration in the mutant was <20% that in wild-type plants (Figure 5F), suggesting a greater role for Os BOR1 under -B conditions than under normal conditions.
Tissue Specificity of Os BOR1 Promoter Activity in Roots Affected by B Status
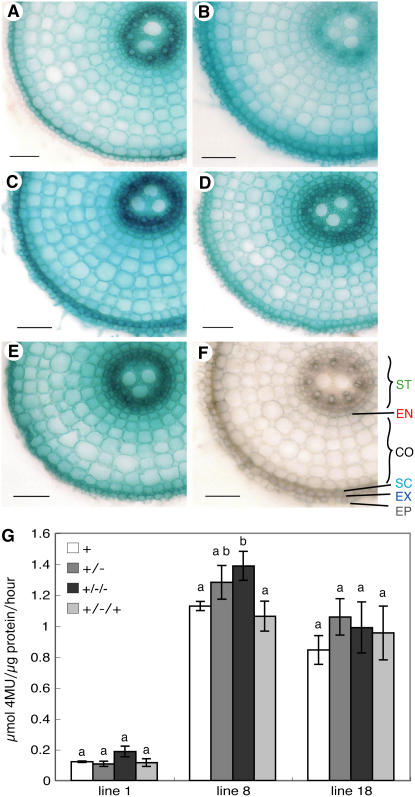
Twenty-nine independent transgenic rice lines carrying the ProOsBOR1:β-glucuronidase (GUS) construct were generated, and three lines (lines 1, 8, and 18) with relatively high GUS expression were selected for detailed analysis of GUS activities. After growth in normal medium, the plants were grown in -B medium for 3 or 6 d. Some of the -B-treated plants were then transferred back to normal medium for 3 d. We observed the cell-type specificities of the GUS activities in the roots ∼2 cm from the root tips, corresponding to the elongation zone. In this zone, root differentiation is completed and cell types can easily be recognized. We also used root tips containing meristematic tissue for GUS staining, but there was no clear tendency (data not shown).
In the roots of transgenic plants grown in normal medium, strong GUS staining was observed in the stele (Figure 6A). GUS staining was also observed in the endodermis and exodermis (Figure 6A). After 3 d of exposure to -B medium, the staining in the exodermis became stronger than that in the stele (Figure 6B). Although GUS staining in the stele remained, signal intensities were much lower than those under the normal condition. After a further 3 d of exposure to -B medium, the staining in stele once again was stronger, and the staining in the exodermis remained strong (Figure 6C). In these plants, the staining in other types of cells was also stronger than in sections grown in -B medium for only 3 d (Figures 6B and 6C). When plants grown in -B medium for 3 d were transferred back into normal medium, the staining in the stele became stronger than that in the exodermis (Figure 6D). This pattern was similar to that of the plants grown under the normal condition (Figure 6A). Similar changes in the cell-type specificity of GUS activity in response to the B concentration in the medium were observed in two other independent transgenic lines (data not shown). We also used root tips containing meristematic tissue for GUS staining. The differential GUS patterns in response to B conditions were not evident in the root tips (data not shown).
Figure 6.
Histochemical Observation and Quantitative Analysis of GUS Activity in Roots of ProOsBOR1:GUS Transgenic Plants in Response to B Conditions.
(A) to (F) Transverse sections of roots at 1.5 to 2 cm from the tip treated with different B regimes. Sections were histochemically stained with X-Gluc to reveal GUS activity. Bars = 50 μm.
(A) Ten days of normal B treatment.
(B) Ten days of normal B treatment followed by 3 d of -B treatment.
(C) Ten days of normal B treatment followed by 6 d of -B treatment.
(D) Ten days of normal B treatment followed by 3 d of -B treatment and then 3 d of normal B treatment.
(E) Transgenic plant carrying Pro35S:GUS as a positive control for GUS staining.
(F) A nontransgenic plant. ST, stele; EN, endodermis; CO, cortex; SC, sclerenchyma cells; EX, exodermis; EP, epidermis.
(G) GUS activity of ProOsBOR1:GUS transgenic rice roots treated with different B regimes. +, 10 d of normal B treatment; +/−, 10 d of normal B treatment followed by 3 d of -B treatment; +/−/−, 10 d of normal B treatment followed by 6 d of -B treatment; +/−/+, 10 d of normal B treatment followed by 3 d of -B treatment and then 3 d of normal treatment. Averages and se (n = 3) are shown. Columns with different letters (a and b) indicate a significant difference (P < 0.05, Student's t test).
To ensure that these differences were not due to artifacts from the process of GUS staining, Pro35S:GUS transgenic rice plants were similarly stained. In these plants, GUS staining was observed throughout cross sections of roots (Figure 6E) and did not change with changing B conditions (data not shown). These results have established that the cell-type specificity of GUS activity in transgenic rice plants carrying the ProOsBOR1:GUS construct changes in response to the B content of the medium.
Os BOR1 Promoter Activity Does Not Strongly Respond to B Conditions
We also performed a fluorometric GUS assay using the same three transgenic lines (1, 8, and 18). The GUS activities in roots of the treated transgenic plants were not significantly altered, except for transgenic line 8 at 6 d of -B treatment (Figure 6G), in which it was upregulated by 20%. It is likely that the overall Os BOR1 promoter activity is regulated little, if at all, in response to the B conditions in the medium.
DISCUSSION
Os BOR1 Is an Ortholog of At BOR1
Four At BOR1-like genes exist in the rice genome and seven At BOR1-like genes, including At BOR1, are present in Arabidopsis (Figure 1). Among the four genes, Os BOR1 shows the strongest similarity to At BOR1 (Figure 1). A phylogenetic analysis suggested that three of the four genes are distinct from Arabidopsis genes. These differences in the number of genes and their phylogenetic relationships are likely to be a reflection of different B transport systems, requirements, and regulation in rice and Arabidopsis. The B requirement of graminaceous plants is generally lower than that of dicotyledonous plants, and the demand for B transport activity may also be lower in graminaceous plants than in dicotyledonous plants.
We previously demonstrated a role for At BOR1 in xylem loading (Takano et al., 2002) and its regulation by B nutrition (Takano et al., 2005). As reported here, the functions and regulation of Os BOR1 are generally similar to those of At BOR1: both are B efflux transporters (Figure 2A) that localize to the plasma membrane (Figure 2B), are expressed in pericycles (Figure 3B), are involved in the xylem loading of B (Figure 5F), and are required for normal growth especially under B-limited conditions (Figure 5A). The accumulation of both proteins is elevated under low B conditions (Takano et al., 2005; Figure 3C). Together with the sequence similarities, these functional similarities establish that Os BOR1 is an ortholog of At BOR1.
Os BOR1 Is Involved in the Entry of B into Roots
Os BOR1 has several unique features that distinguish it from At BOR1. One is its involvement in the uptake of B into roots. In plants grown for long periods under -B conditions, the B concentration in osbor1-1 roots was less than that in wild-type roots (data not shown). We have repeated similar experiments several times. Although in this particular experiment (short -B treatment) shown in Figure 5D, reduction in B concentration in roots of the mutants was not statistically significant, in several independently conducted experiments, and the reduction was evident and statistically significant after treatments with -B for a long period of time (data not shown). Based on the data obtained from similar experiments, we are confident that Os BOR1 is necessary for B uptake especially after exposure to -B for long periods. This is in clear contrast with At BOR1, which is involved in xylem loading but not in the uptake of B into roots (Takano et al., 2002).
This unique feature of Os BOR1 that distinguishes it from At BOR1 is likely to be due to the different cell-type specificities of expression in roots. In rice roots, Casparian strips, barriers that prevent ions and solutes from freely entering the stele, are found in both the exodermis and endodermis (Ma et al., 2006). The expression of Os BOR1 was observed in these tissues (Figures 6A to 6D). It is likely that the expression of Os BOR1 at these sites is important for both the uptake of boric acid into the roots and its loading into the xylem. In the case of Si, the polar localization of Lsi1 is likely to be important for the uptake of Si from the soil into the roots (Ma et al., 2006). Similarly, Os BOR1 may be critical for the transport of boric acid into the root stele. Determining the polar localization of Os BOR1 may clarify these issues.
Roles of Os BOR1 in the Long-Distance Transport of B
The B concentration in the xylem sap of osbor1-2 mutant plants was lower than that in the wild type after 3 d of exposure to either normal or low B conditions (Figure 5F), suggesting that Os BOR1 is involved in the loading of B into the xylem, irrespective of the B concentration in medium. This is in clear contrast with At BOR1, which is important in the efficient xylem loading of B, mostly under long-term -B conditions, but less important under conditions of normal B supply, presumably due the degradation of the At BOR1 protein (Takano et al., 2005). This is also apparently in contrast with our findings that no accumulation of Os BOR1 was detected after 3 d of exposure to normal medium (Figure 3C). It is likely that Os BOR1 accumulates to a level below the detection limit of our immunoblot analysis and that level may be sufficient for enhancing the xylem loading of B. As shown in Figure 5D, B concentrations in the aerial portion of the mutants were reduced under the normal condition, suggesting that Os BOR1 functions under normal B condition. It is possible that under the control condition, Os BOR1 accumulates only to a low level, which is likely to be below the detection limit of our immunoblot analysis.
Although the concentration of B in shoots under -B was not significantly different from the wild type (Figure 5D), the shoot growth of the mutants under -B was severely reduced (Figure 5C). Thus, the total amount of B transported to shoots was much less in the mutant than the wild type, especially in the case of osbor1-1. It also indicated that Os BOR1 is important for xylem B transport both under -B and normal conditions.
The Accumulation of B in the Root Cell Walls of Rice Plants
Arabidopsis has no apparent B-concentrating mechanism that acts following the entry of B into roots. However, in wild-type rice plants, the total B concentration in roots grown under -B conditions for 2 weeks was 14 μM (Figure 5D), much higher than the B concentration in the medium. It is likely that rice plants have a high capacity to concentrate B upon its entry into roots. This process does not seem to be mediated by Os BOR1 because osbor1-1 and osbor1-2 also showed the same or more concentrated B contents than that in the medium (Figure 5D). B concentrations in roots (Figure 5D) were much higher than that in root cell sap (Figure 5E), suggesting that rice plants have mechanisms to deliver and concentrate B to the insoluble fraction of root cells, which is most likely to be the cell wall. This is a distinct feature of rice plants that contrasts with Arabidopsis in terms of B distribution within the roots.
It is worth mentioning that in roots of osbor1-1 grown under -B, B concentration was higher than the wild type (Figure 5D). It is possible that the reduced transport of B to shoots resulted in high accumulation of B.
Changes in the Cell-Type Specificity of Os BOR1 Expression in Roots Are Affected by B Conditions
We found that the cell-type specificity of Os BOR1 expression is affected temporally by the B content of the medium (Figures 6A to 6D). This represents a major difference between Os BOR1 and At BOR1. There are several reports of nutrient transporter genes that are induced by the nutrient conditions, including the rice genes AMT1;2 (Sonoda et al., 2003) and IRT1 (Ishimaru et al., 2006) and the Arabidopsis genes YSL2 (Schaaf et al., 2005) and the AMTs (for review, see Loque and von Wirén, 2004), but in these cases, no changes in cell-type specific patterns of expression were reported.
As discussed above, Os BOR1 is required for both the efficient uptake of B into the roots and the xylem loading of B. The effect of long-term -B treatment is most likely to be represented by the experiment shown in Figure 6C, in which plants were exposed to -B for 6 d. Os BOR1 expression was observed in both the exodermis and the stele, suggesting that the Os BOR1 in these cell types is responsible for B uptake and xylem loading, respectively.
When roots were exposed to -B conditions for 3 d, Os BOR1 was strongly expressed in the exodermis (Figure 6B), suggesting that under such conditions Os BOR1 is more likely to be involved in B uptake from the medium into root steles. This cell type–specific pattern of expression is transient because after 6 d of -B treatment, the Os BOR1 expression in the stele returned back to high levels (Figure 6C). After several days of -B treatment, a steady flow of B from the soil to the stele may be established, which requires both the efficient uptake and the xylem loading of B. Although we did not examine the physiological consequences of the changes in the tissue specificity of Os BOR1 expression, these transient changes in the cell type–specific pattern of Os BOR1 expression are likely to reflect a transient shift in the B transport requirement of root tissues.
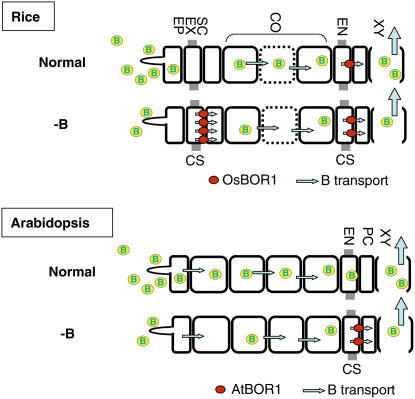
Based on these findings, we propose a model for the roles of Os BOR1 in B transport in rice, in contrast with those of At BOR1 in Arabidopsis (Figure 7). At BOR1 is expressed mainly in steles under -B conditions and is essential for the efficient xylem loading of B; under normal conditions, however, its accumulation is greatly reduced and xylem loading is likely to be a passive process. In rice, Os BOR1 expression was detected in both the exodermis and endodermis under -B conditions, and the gene may function in both B uptake and xylem loading. Expression in the exodermis may be important for the element to cross the outer Casparian strips, as rice submerged in water develops air spaces in cortical cells, which is likely to limit the symplastic transport of B to the steles. B is probably transported through the apoplasts in the cortical cells, in the roots of paddy rice. Then, B must cross the inner Casparian strip at the endodermis. Os BOR1 in the endodermis is likely to be important for the entry of B into the stele for xylem loading to take place. Changes in the cell-type specificity of the promoter activity are likely to reflect the roles of Os BOR1 under these conditions.
Figure 7.
Model of the Roles of Os BOR1 and At BOR1 in Roots under Normal and -B Conditions.
The diagram shows the cell layers in roots, with the epidermis at the left side and xylem vessels to the right. Gray boxes indicate the positions of the Casparian strips. In rice, Casparian strips are present in both the exodermis and the endodermis, whereas in Arabidopsis, the strips are found only in the endodermis. Under normal B conditions, Os BOR1 is not abundantly present, and considering the strong Os BOR1 promoter activity in the endodermis, it is likely to be present in endodermis at a high abundance. After 3 d of -B treatment, Os BOR1 accumulation is higher overall, particularly in the exodermis. The expression pattern in rice corresponds well with the rice root structure, given the presence of two Casparian strip layers and the cell death that occurs in the cortex under submerged conditions (represented in cells shown with dotted lines). In Arabidopsis, At BOR1 is responsible for xylem loading but not for uptake under -B conditions and is not present under normal conditions. CS, Casparian strip; EN, endodermis; CO, cortex; SC, sclerenchyma cells; EX, exodermis; EP, epidermis; PC, pericycle; XY, xylem.
In summary, we demonstrated that Os BOR1 is a plasma membrane–localized B transporter that plays a role in both the xylem loading of B and its uptake into roots. At BOR1 is required for the efficient xylem loading of B but has not been demonstrated to be involved in B uptake. The cell-type specificity of Os BOR1 expression changes with the B status of the medium, providing an example of a mineral-nutrient transporter whose tissue-specific expression pattern changes depending on the nutritional conditions.
METHODS
Plant Materials and Culture
Rice plants (Oryza sativa cv Nipponbare) were grown hydroponically in a temperature-controlled (30/25°C day/night) greenhouse under natural light. Seeds were germinated with ion-exchanged water and grown for a week before transfer to hydroponic culture (Fukuda et al., 2004). For B deficiency treatment, hydroponic solutions with no added boric acid, or containing 0.03 μM boric acid, were used. Fifteen to twenty plants were grown in 1.5-liter containers. The pH of the hydroponic solutions was adjusted to 5.4 to 5.6 every other day, and the nutrient solutions were renewed weekly.
Phylogenetic Analysis
A similarity search of the entire rice genome was performed with the At BOR1 amino acid and cDNA sequences using the BLASTP and TBLASTN programs at the National Institute of Agrobiological Sciences (http://riceblast.dna.affrc.go.jp/), the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/Genome/PlantBlast.shtml?4), and the Knowledge-Based Oryza Molecular Biological Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/). Locus numbers were obtained from The Institute for Genomic Research. Phylogenetic analysis was performed with MEGA 3.1 (http://www.megasoftware.net) using the neighbor-joining method (Saitou and Nei, 1987). Aligned sequences (residues 65 to 412 of Os BOR1) were used to generate the phylogenetic tree. The accession number and/or gene identifier of each gene is shown in Table 1.
Plasmid Construction
The sequences of all primers used in this study are shown in Table 2.
To construct Pro35S:Os BOR1, the Os BOR1 ORF was amplified from the full-length cDNA clone J023062N03 (accession number AK070617), which was kindly provided by the Rice Genome Resource Center (http://www.nias.affrc.go.jp/). The specific primers 1136 and 1137 were used for the first round of PCR amplification. These primers contain the sequences of the attB adaptor for the Gateway BP reaction (Invitrogen). After a second round of PCR using the attB adapter primers, the PCR products were subcloned into pDONR-Zeo, an entry clone for the Gateway system (Invitrogen). After subcloning, the nucleotide sequences of the inserts were confirmed by DNA sequencing. This plasmid was named pYN11. pYN11 was subsequently used for an LR reaction, which allowed the Os BOR1 cDNA fragment to be fused into pMDC32 (Curtis and Grossniklaus, 2003), and the resulting plasmid, carrying Os BOR1 under the control of Pro35S, was named pYN22. The pMDC vectors were kindly provided by the University of Zurich (http://www.unizh.ch/botinst/Devo_website/curtisvector/) (Curtis and Grossniklaus, 2003).
To construct a plasmid for Os BOR1 cDNA expression in yeast, pYN11 was subjected to the LR reaction and the Os BOR1 cDNA fragments were transferred into pYES-DEST52 (Invitrogen). To generate GFP fusion constructs, the Os BOR1 cDNA fragment in pYN11 was transferred into pMDC43 after the LR reaction. This plasmid contains the Pro35S:sGFP-Os BOR1 fusion. Pro35S:Os BOR1-sGFP was constructed as follows. The Os BOR1 cDNA was amplified with the primers 951 and 1019, and the resulting PCR product was fused into pTF478 (Takano et al., 2002) between the KpnI and NcoI sites. This plasmid was used as a template for a subsequent PCR reaction with the primers 1136 and 1140, which contain the sequences of the attB adaptor for the Gateway BP reaction (Invitrogen). The resulting PCR product was subcloned into pDONR-Zeo. The insert was then transferred into pMDC32 via the LR reaction. The resulting plasmid containing the Pro35S:Os BOR1-sGFP insert was used in particle bombardment experiments to determine the subcellular localization of Os BOR1.
To generate the promoter-reporter construct, a genomic fragment containing the putative promoter region of Os BOR1 (positions −3188 to −1 bp from the initiation codon) was amplified by PCR from genomic DNA using the primers 1096 and 1097. These primers contain the sequences for the attB adaptor for Gateway cloning (Invitrogen). The fragments were subcloned into pDONR-Zeo. Next, the fragment was fused into pMDC162 (Curtis and Grossniklaus, 2003) using the LR reaction, and the resulting plasmid was named pYN35.
Determination of Os BOR1 Activity in Yeast
Experiments were performed as described by Takano et al. (2002), except that the Os BOR1 cDNA was used and the B concentration in the incubation medium was modified to 100 μM.
Particle Bombardment into Onion Epidermal Cells
Bombardment was performed as described by Takano et al. (2002), except that inner cells of white onion (Allium cepa) were used as the target. After the bombardment, the cells were held in darkness at room temperature (20 to 24°C) for 12 h, and GFP activity was observed using a fluorescence microscope.
Quantification of Transcripts by RT-PCR
After treatment of the plants, the roots were washed three times with ion-exchanged water and gently blotted with a paper towel to remove excess water. Total RNA was typically isolated from 0.1 g (fresh weight) of leaves or whole roots using an RNeasy plant mini kit (Qiagen). RT-PCR analysis was performed as described by Ohkama et al. (2002). Transcript accumulation was expressed relative to Ubi1 transcript accumulation (Christensen et al., 1992). Three real-time PCR analyses were performed for each RNA sample, and three RNA samples were independently prepared for each treatment. The average value of each RNA sample was calculated, and three average values were used to calculate the average and standard deviation for each treatment. The primers 913 and 914 were used as specific primers for Os BOR1, and the Ubi1-F and R primers were used as a control. All primer sequences are shown in Table 2.
In Situ Hybridization
Wild-type rice plants were germinated in liquid Murashige and Skoog medium, and 4 d later, roots were excised and fixed as described by Itoh et al. (2000). Microtome sections of 9 μm in thickness were placed on glass slides coated with Vectabond (Vector Laboratories). The Os BOR1 ORF was amplified by PCR with the primers 927 and 1019 (Table 2) and fused into pGEM-T Easy (Promega). After determination of the direction of the insert, digoxigenin-labeled antisense and sense probes were prepared using SP6 and T7 RNA polymerase (Roche), respectively. In situ hybridization and immunological detection of signals were performed using a TSA Plus DNP (AP) System (Perkin-Elmer). The detection was performed as described by Kouchi and Hata (1993).
Immunoblot Analysis
Preparation of microsomal proteins from bulk roots was performed as described by Takano et al. (2005). Samples containing 5 μg of protein, determined using the Bio-Rad Quick Start Bradford Dye Reagent, were mixed with an equal volume of 2× SDS-PAGE sample buffer (Takano et al., 2005) and incubated on ice for 10 to 20 min before loading. The rabbit anti-Os BOR1 antibody was raised against an Escherichia coli–expressed partial peptide of Os BOR1 (N355-E464) and used at 1000-fold dilution.
Establishment and Analysis of Transposon-Tagged Os BOR1 Lines
Two rice lines, NC0170 and NC0255, which carry insertions of the retrotransposon Tos17 in the Os BOR1 gene, were kindly provided by Hirohiko Hirochika and Akio Miyao (Rice Genome Resource Center; http://www.nias.affrc.go.jp/; Hirochika et al., 1996). NC0170 and NC0255 are referred to as osbor1-1 and osbor1-2, respectively. The positions of the Tos17 insertions in Os BOR1 were confirmed by PCR and sequencing. For PCR amplification of the boundary sequences between Tos17 and the rice genome, the Tos17 left border–specific primer 654 (Table 2) was used for both mutants. The Os BOR1-specific primers 928 and 930 (Table 2) were used for osbor1-1 and osbor1-2, respectively. The subsequent PCR products were sequenced and the positions of the insertions determined.
To detect the segregation of the Tos17 insertion in Os BOR1, the primers 654 and 928 were used for osbor1-1 and the primers 654 and 930 were used for osbor1-2. To detect the corresponding PCR product from Os BOR1, the primers 913 and 914 were used.
Rice Transformation
Rice transformation plasmids were introduced into Agrobacterium tumefaciens (EHA101 or GV3101). Agrobacterium-mediated transformation of rice plants was performed as described by Toki (1997), except that rice seeds were precultured for 5 d on medium containing 2 mg/L 2,4-D. Three independent transgenic plants that carried the osbor1-2 mutation were regenerated from the transformation with pYN22 (containing Pro35S:Os BOR1 cDNA), and 29 independent transgenic plants were regenerated from the transformation with pYN35 (containing ProOsBOR1:GUS).
Preparation of Plant Samples and Determination of B Concentrations
Appropriate portions of plants were harvested and their fresh weights determined. For sample collection, a region of ∼1 to 1.5 cm of the boundary between the aerial portions and roots was excluded. The B concentrations in the samples were determined as described by Takano et al. (2002).
To collect xylem fluid, plants were germinated and grown in normal medium for 18 d. When the plants were at the five-leaf stage, they were transferred to either normal medium or medium containing 0.03 μM B and grown for 3 d. Then, the shoots of the plants were removed with razor blades 1 to 2 cm above the junction between the shoot and the root. The xylem fluid (20 to 100 μL) that exuded through the cut ends of the stems and leaves was collected over several hours from the late afternoon into the evening.
To obtain root cell sap, roots of the same plants that were used for xylem fluid collection were harvested and stored at −80°C. After thawing at room temperature, fresh weights were determined and the samples were placed in 0.22-μm filter units (Ultrafree-MC; Millipore). The tissues were refrozen at −20°C and thawed at room temperature. Four to five freeze-thaw cycles were conducted, and the cell saps were collected by centrifugation at 1000g for 5 min at room temperature (Noguchi et al., 2000).
Histochemical Analysis of GUS Activities
Root sections of 1.5 to 2 cm from the root tips were washed with ion-exchanged water prior to harvest. Histochemical staining was performed according to Inoue et al. (2003), with the following modifications. The sections were incubated at 37°C for 60 min in GUS reaction buffer (Inoue et al., 2003). After staining, the sections were washed in 70% ethanol and held in 70% ethanol until observation. The sections were embedded in 4% agar and then cut into 130-μm sections using a DTK-100 microslicer (Dosaka EM). GUS staining was observed using an Axiophoto microscope (Carl Zeiss). For a positive control for GUS staining, transgenic rice plants containing the pIG121Hm vector were used (Hiei et al., 1994).
Determination of GUS Activities
Root tips (2 cm from the tip) of hydroponically grown plants were harvested and subjected to GUS assays (Jefferson, 1987). Protein concentrations were determined using Quick Start Bradford Dye Reagent (Bio-Rad).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK070617, DQ421408, AK072421, and DQ421409.
Acknowledgments
The full-length Os BOR1 cDNA clone and the mutant lines described in this article were obtained from the Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan (http://www.nias.affrc.go.jp/). The Os BOR2 and 3 clones were kindly provided by L. Monna. The transgenic seeds containing pIG121Hm were kindly provided by Y. Ito. Y. Nagato and H. Kitano provided advice on in situ hybridization. We thank N.K. Nishizawa and H. Inoue for assistance with GUS observation, E. Kato for antibody preparation, K. Aizawa for excellent technical assistance, and J. Takano for discussion. This work was supported in part by the Green Technology Project from the Ministry of Agriculture, Forestry, and Fisheries of Japan, by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a 21st Center of Excellence project (to T.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Toru Fujiwara (atorufu@mail.ecc.u-tokyo.ac.jp).
References
- Bolanos, L., Lukaszewski, K., Bonilla, I., and Blevins, D. (2004). Why boron? Plant Physiol. Biochem. 42 907–912. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Christensen, A.H., Sharrock, R.A., and Quail, P.H. (1992). Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18 675–689. [DOI] [PubMed] [Google Scholar]
- Connolly, E.L., Fett, J.P., and Guerinot, M.L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannel, F., Pfeffer, H., and Römheld, V. (1998). Compartmentation of boron in roots and leaves of sunflower as affected by boron supply. J. Plant Physiol. 153 615–622. [Google Scholar]
- Fukuda, A., Okada, Y., Suzui, N., Fujiwara, T., Yoneyama, T., and Hayashi, H. (2004). Cloning and characterization of the gene for a phloem-specific glutathione S-transferase from rice leaves. Physiol. Plant 120 595–602. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H., Higuchi, K., Takahashi, M., Nakanishi, H., Mori, S., and Nishizawa, N.K. (2003). Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 36 366–381. [DOI] [PubMed] [Google Scholar]
- Ishimaru, Y., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45 335–346. [DOI] [PubMed] [Google Scholar]
- Itoh, J., Kitano, H., Matsuoka, M., and Nagato, Y. (2000). SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 12 2161–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Kall, L., Krogh, A., and Sonnhammer, E.L.L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338 1027–1036. [DOI] [PubMed] [Google Scholar]
- Kouchi, H., and Hata, S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238 106–119. [DOI] [PubMed] [Google Scholar]
- Loque, D., and von Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55 1293–1305. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., Tamai, K., Yamaji, N., Mitani, N., Konishi, S., Katsuhara, M., Ishiguro, M., Murata, Y., and Yano, M. (2006). A silicon transporter in rice. Nature 440 688–691. [DOI] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants, 2nd ed. (San Diego, CA: Academic Press).
- Matoh, T., Ishigaki, K., Ohno, K., and Azuma, J. (1993). Isolation and characterization of a boron-polysaccharide complex from radish roots. Plant Cell Physiol. 34 639–642. [Google Scholar]
- Matoh, T., Kawaguchi, S., and Kobayashi, M. (1996). Ubiquity of a borate-rhamnogalacturonan II complex in the cell walls of higher plants. Plant Cell Physiol. 37 636–640. [Google Scholar]
- Matsunaga, T., Ishii, T., Matsumoto, S., Higuchi, M., Darvill, A., Albersheim, P., and O'Neill, M.A. (2004). Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes, and bryophytes. Implications for the evolution of vascular plants. Plant Physiol. 134 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, K., Takano, J., and Fujiwara, T. (2006). Improvement of seed yields under boron-limiting conditions through overexpression of BOR1, a boron transporter for xylem loading, in Arabidopsis thaliana. Plant J. 46 1084–1091. [DOI] [PubMed] [Google Scholar]
- Miyao, A., Tanaka, K., Murata, K., Sawaki, H., Takeda, S., Abe, K., Shinozaki, Y., Onosato, K., and Hirochika, H. (2003). Target site specificity of the Tos17 retrotransposon shows a preference for insertion in retrotransposon-rich regions of the genome. Plant Cell 15 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, K., Dannel, F., Pfeffer, H., Romheld, V., Hayashi, H., and Fujiwara, T. (2000). Defect in root-shoot translocation of boron in Arabidopsis thaliana mutant bor1-1. J. Plant Physiol. 156 751–755. [Google Scholar]
- Nozawa, A., Miwa, K., Kobayashi, M., and Fujiwara, T. (2006). Isolation of Arabidopsis thaliana cDNAs that confer yeast boric acid tolerance. Biosci. Biotechnol. Biochem. 70 1724–1730. [DOI] [PubMed] [Google Scholar]
- Ohkama, N., Takei, K., Sakakibara, H., Hayashi, H., Yoneyama, T., and Fujiwara, T. (2002). Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol. 43 1493–1501. [DOI] [PubMed] [Google Scholar]
- O'Neill, M.A., Eberhard, S., Albersheim, P., and Darvill, A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294 846–849. [DOI] [PubMed] [Google Scholar]
- Rerkasem, B., and Jamjod, S. (2004). Boron deficiency in wheat: A review. Field Crops Res. 89 173–186. [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method – A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schaaf, G., Schikora, A., Haberle, J., Vert, G., Ludewig, U., Briat, J.F., Curie, C., and von Wirén, N. (2005). A putative function for the Arabidopsis Fe-phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol. 46 762–774. [DOI] [PubMed] [Google Scholar]
- Shorrocks, V.M. (1997). The occurrence and correction of boron deficiency. Plant Soil 193 121–148. [Google Scholar]
- Sonoda, Y., Ikeda, A., Saiki, S., von Wirén, N., Yamaya, T., and Yamaguchi, J. (2003). Distinct expression and function of three ammonium transporter genes (OsAMT1;1 –1;3) in rice. Plant Cell Physiol. 44 726–734. [DOI] [PubMed] [Google Scholar]
- Takano, J., Miwa, K., Yuan, L., von Wirén, N., and Fujiwara, T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, J., Noguchi, K., Yasumori, M., Kobayashi, M., Gajdos, Z., Miwa, K., Hayashi, H., Yoneyama, T., and Fujiwara, T. (2002). Arabidopsis boron transporter for xylem loading. Nature 420 337–340. [DOI] [PubMed] [Google Scholar]
- Takano, J., Wada, M., Ludewig, U., Schaaf, G., von Wirén, N., and Fujiwara, T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18 1498–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki, S. (1997). Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 15 16–21. [Google Scholar]
- Yamaguchi, M., Sasaki, T., Sivaguru, M., Yamamoto, Y., Osawa, H., Ahn, S.J., and Matsumoto, H. (2005). Evidence for the plasma membrane localization of Al-activated malate transporter. Plant Cell Physiol. 46 812–816. [DOI] [PubMed] [Google Scholar]
- Yu, X., and Bell, P.F. (1998). Nutrient deficiency symptoms and boron uptake mechanisms of rice. J. Plant Nutr. 21 2077–2088. [Google Scholar]