Abstract
The growth of perennial plants in the temperate zone alternates with periods of dormancy that are typically initiated during bud development in autumn. In a systems biology approach to unravel the underlying molecular program of apical bud development in poplar (Populus tremula × Populus alba), combined transcript and metabolite profiling were applied to a high-resolution time course from short-day induction to complete dormancy. Metabolite and gene expression dynamics were used to reconstruct the temporal sequence of events during bud development. Importantly, bud development could be dissected into bud formation, acclimation to dehydration and cold, and dormancy. To each of these processes, specific sets of regulatory and marker genes and metabolites are associated and provide a reference frame for future functional studies. Light, ethylene, and abscisic acid signal transduction pathways consecutively control bud development by setting, modifying, or terminating these processes. Ethylene signal transduction is positioned temporally between light and abscisic acid signals and is putatively activated by transiently low hexose pools. The timing and place of cell proliferation arrest (related to dormancy) and of the accumulation of storage compounds (related to acclimation processes) were established within the bud by electron microscopy. Finally, the identification of a large set of genes commonly expressed during the growth-to-dormancy transitions in poplar apical buds, cambium, or Arabidopsis thaliana seeds suggests parallels in the underlying molecular mechanisms in different plant organs.
INTRODUCTION
The growth habit of perennial plants in the temperate zone is characterized by alternating periods of growth and dormancy. The changes in daylength, light quality, and temperature provide the most important environmental cues for synchronizing growth with seasonality. In autumn, photoperiodic tree species perceive dormancy-inducing short days (SDs), which are mediated through phytochrome and the CONSTANS (CO)/FLOWERING TIME LOCUS T (FT) regulon (Olsen et al., 1997; Böhlenius et al., 2006). SDs trigger the cessation of internode elongation and the transition from a growing apex to a bud. The morphology of the apex is transformed to a bud through sequential changes in the differentiation of young primordia: first, toward enveloping bud scales, and then to embryonic leaves (Rohde and Boerjan, 2001).
Autumnal bud development is a composite of bud formation, simultaneous acclimation to dehydration and cold, and acquisition of dormancy. These three processes are confounded and interconnected with each other but happen at different levels: cells throughout the plant will acclimate to dehydration and cold for survival, whereas only the youngest derivatives of the meristem participate in bud formation, and yet only the meristem (and those parts of the bud that will resume growth in spring) become dormant. Expression of a dominant-negative version of ETHYLENE TRIPLE RESPONSE1 (ETR1) or overexpression of ABSCISIC ACID-INSENSITIVE3 (ABI3) prevents the formation of closed apical buds upon SD induction (Rohde et al., 2002; Ruonala et al., 2006). In both cases, at least some degree of dormancy is established, indicating that bud formation and dormancy per se are distinct, partially independent aspects of bud development.
The environmental and hormonal factors controlling the induction of bud development, such as photoperiod, low temperature, and abscisic acid (ABA), have been known for years (Sylven, 1940; Nitsch, 1957; Eagles and Wareing, 1964; Weiser, 1970). However, their temporal integration, their mutual interactions, and particularly their molecular targets remained largely unknown (Horvath et al., 2003; Tanino, 2004; Rohde and Bhalerao, 2007). Moreover, the simultaneous activity of various closely intertwined cellular, physiological, and morphological processes confounds the dissection of the underlying developmental programs and their respective signals. Only in a few cases could processes be separated and assigned to the action of a particular signaling route (Welling et al., 2002; Mølmann et al., 2005).
To bridge the gap in our understanding of the molecular regulation of bud development and dormancy, transcript and metabolite profiling were applied to a complete time course of apical bud development in poplar (Populus tremula × Populus alba) from SD induction to complete dormancy. The gene expression and metabolite profiles established the temporal sequence of events and processes during bud formation and dormancy induction. Through a directed search for marker genes, bud formation, dormancy, and acclimation to dehydration and cold are dissected and described by specific sets of genes. Metabolism is inactivated and reconfigured toward the acquisition of cold tolerance and the accumulation of storage compounds. Light, ethylene, and ABA are the major signals that act consecutively to control bud development by setting, modifying, or terminating these processes. Finally, genes differentially expressed (DE) during apical bud development were compared with those during dormancy induction in poplar cambium or Arabidopsis thaliana seeds. A large set of genes commonly related to the growth-to-dormancy transitions suggests parallels in the underlying molecular mechanisms in different plant organs.
RESULTS
Transcript and Metabolite Profiling Reveal the Molecular Players throughout Bud Development
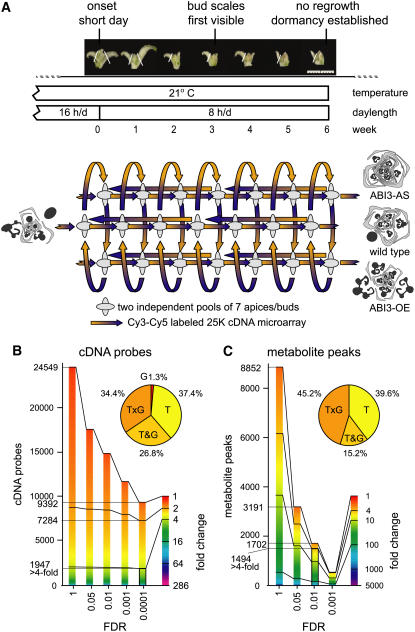
To identify the genes and metabolites that control bud development, developing buds of wild-type poplar were analyzed at weekly intervals during 6 weeks of SD treatment with microarray transcript profiling and gas chromatography–mass spectrometry (GC-MS) metabolite profiling (Figure 1A). Transgenic poplars that upregulate or downregulate the ABI3 transcription factor (TF) (hereafter designated ABI3-OE and ABI3-AS, respectively) were included to gain deeper insight into bud formation as well as into ABA/ABI3-dependent signal transduction during bud development. Wild-type ABI3 activity enables the maturation of young leaf organs inside the bud prior to dormancy (Rohde et al., 2002). ABI3-OE plants fail to develop a closed bud under SD conditions, lack encasing bud scales, and develop larger embryonic leaves than wild-type plants. By contrast, ABI3-AS plants have less developed embryonic leaves and more pronounced bud scales (Figure 1A). Both ABI3-AS and ABI3-OE plants become dormant. Regrowth tests with trees from the same experiment indicated that dormancy was established after 6 weeks of SDs in all genotypes (data not shown).
Figure 1.
DE Genes and Metabolites during SD Induction of Bud Formation and Dormancy.
(A) Apical bud formation and dormancy induced in wild-type (P. tremula × P. alba) poplar and two transgenic lines with antisense downregulation or overexpression of ABI3 (ABI3-AS and ABI3-OE, respectively) during 6 weeks of SD treatment. Photoperiod and temperature regime, bud phenology stages, and sampling points are indicated on the time axis. Apices and buds were sampled at weekly intervals (LDs up to 6 weeks of SDs). The sampled material is depicted; white lines indicate positions where leaves were removed from the sample. At each sampling point, two independent pools of seven apices or buds (depicted by ellipses) were used for combined transcript and GC-MS metabolite profiling. An interconnected, fully balanced loop design consisted of 42 two-color microarrays (time on the horizontal axis, genotype on the vertical axis) and six extra microarrays comparing more distantly related samples within the time series. Schematic drawings of transversal sections of apices or buds show identical organ composition under LDs and contrasting bud morphology after SD treatment (black, embryonic leaves; gray, bud scales/stipules) (Rohde et al., 2002). Bar = 1 cm.
(B) and (C) Fractions of the sampled transcriptome (B) or metabolome (C) considered as DE in function of fold change and FDR. The fold change is depicted in false color, and cDNA probes or metabolite peaks are ordered by magnitude of fold change. Distribution of time (T), genotype (G), time and genotype (T&G), and time–genotype interaction (T×G) effects are given for 1947 DE cDNA probes (more than fourfold, FDR < 0.0001) and for 1494 DE metabolite peaks (more than fourfold, FDR < 0.01).
Temporal expression profiles for 24,549 spotted poplar cDNA probes were obtained with an interconnected loop design for microarray hybridizations (Figure 1A; see Supplemental Table 1 online). Among 9392 significantly DE cDNA probes (false discovery rate [FDR] < 0.0001), 7284 cDNA probes (29.7%) have a more than twofold change in expression, including 1947 cDNA probes (7.9%) with a more than fourfold change (Figure 1B). The fold change in expression can occur either between any two genotypes at a given time or between any two points in time for a given genotype. The majority of the significant probes have both time and genotype effects (Figure 1B). For genes represented by multiple independent cDNA probes on the microarray, a single expression profile per gene was calculated to remove this redundancy from the data set (see Methods and Supplemental Table 2 online). A set of 1091 unique genes with a more than fourfold change in expression (FDR < 0.0001) was selected for further analysis. This set includes 81 of the 918 poplar TFs (8.8%) represented on the microarray (Riaño-Pachón et al., 2007) (see Supplemental Table 3 online). Another 200 TFs (30.6%) are more than twofold DE during bud development.
GC-MS metabolite profiling identified 8852 metabolite peaks. Among 1702 significantly differentially accumulating peaks (FDR < 0.01), 1494 metabolite peaks have a more than fourfold change (16.9% of 8852 peaks) (Figure 1C; see Supplemental Table 4 online). Similar to the transcripts, the majority of the significant peaks have both genotype and time effects (Figure 1C). The 1702 peaks (FDR < 0.01) correspond to 176 compounds (see Supplemental Table 5 online), of which 162 with more than fourfold differential accumulation were selected for further analysis. These include 110 unidentified compounds (67.9%), 13 organic acids (8.0%), 16 amino acids (9.9%), and 14 sugars or sugar alcohols (8.6%).
Progressive Changes in Transcriptome and Metabolite Composition Characterize Bud Development and Parallel Aberrant Bud Development in ABI3-OE and ABI3-AS
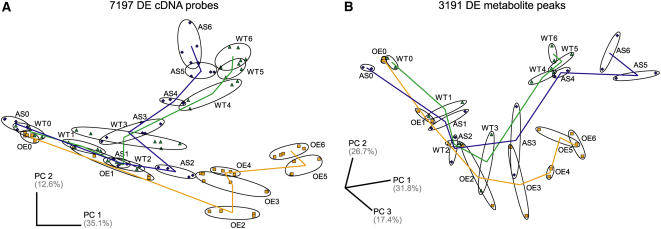
SD-induced bud development is associated with progressive changes in transcriptome and metabolite composition, as revealed by principal component analysis (PCA) (Figure 2). The transcriptomes of wild-type, ABI3-AS, and ABI3-OE poplars are highly similar under long days (LDs), consistent with the absence of a morphological phenotype in transgenic lines during active growth (Figure 2A) (Rohde et al., 2002). The first principal component primarily resolves samples in chronological order. Along the second principal component axis, ABI3-OE and ABI3-AS transcriptomes progressively deviate, in opposite manner, from the wild type, reflecting contrasting bud morphologies under SDs (Figures 1A and 2A). Gene expression patterns in ABI3-OE start to differ from those of wild type and ABI3-AS after 2 weeks of SDs (Figures 1A and 2A), preceding the transition of the apex to a bud structure, which is marked by the appearance of bud scales after 3 weeks of SDs in the wild type.
Figure 2.
Global Changes in Transcript and Metabolite Composition during SD-Induced Apical Bud Development.
(A) PCA plot of the transcriptome composition of 96 individually hybridized samples, represented each by the combined expression data of 7197 of all 9392 significantly DE cDNA probes (FDR < 0.0001) with expression data in all hybridizations. Ellipses group replicate hybridizations of all samples for a genotype at a given time, including the two independent biological replicates.
(B) PCA plot of metabolite composition of 42 individual samples, represented by the combined expression data of 3191 significantly DE metabolite peaks (FDR < 0.05). Ellipses group the two independent biological replicates for a genotype at a given time.
Samples are labeled starting from LD (0) throughout 6 weeks SD treatment (1 to 6). ABI3-AS, blue diamonds; wild type, green triangles; ABI3-OE, orange squares. Percentage of variance explained is given for the respective principal component (PC) axes.
Metabolite composition changes are revealed during the first week of SDs, suggesting rapid adaptation to SDs (Figure 2B). Furthermore, the PCA discriminates between the metabolite compositions of wild-type and transgenic lines after 3 weeks of SDs, slightly later than the occurrence of transcriptome differences between respective lines (Figures 2A and 2B). Together, progressive temporal changes in transcripts and metabolites characterize bud development. ABI3, consistent with its molecular nature, profoundly affects gene expression in an SD-dependent manner, and this effect extends to primary metabolism.
Temporal Expression and Metabolite Clusters Identify Sequential Changes throughout Bud Development
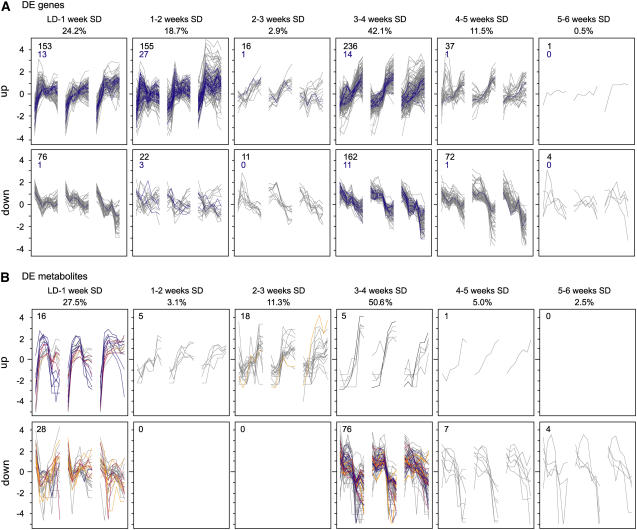
Bud development was further dissected using 945 genes and 160 metabolites of the respective fourfold sets that characterize wild-type development. Genes and metabolites were categorized in temporal clusters according to the time interval and directionality of their maximal change in expression to investigate whether particular pathways were sequentially activated (Figure 3). This global data analysis indicates two major phases of responses to SDs: an early adaptation to SDs and a late adaptation after a number of consecutive SDs, consistent with the PCA results (Figures 2 and 3). Globally, the level of most metabolites (71.6%) decreases, suggesting an overall inactivation of primary metabolism (Figure 3B).
Figure 3.
Temporal Categorization of Changes in Gene Expression and Metabolite Levels during Bud Development.
A total of 945 genes (A) and 160 compounds (B) of the respective fourfold set (with more than twofold change in expression in the wild type) were grouped by directionality and time interval of maximal change in expression in the wild type. Profiles are given as log2-transformed, mean-centered data. The top row indicates the time intervals and the percentage of this group within the respective fourfold set. In each graph, wild-type profiles are flanked by ABI3-AS (left) and ABI3-OE (right), with each consisting of seven sampling points.
(A) Within gene expression clusters, DE TFs are highlighted in blue. Numbers in the top left corner of each panel denote the total number of genes (black) and the number of TFs (blue) belonging to the cluster.
(B) Numbers in the top left corner of each panel denote the total number of compounds belonging to the cluster. Gray, unknown compounds; orange, sugars or sugar alcohols; blue, amino acids; red, organic acids; black, catechine.
A first profound change in gene expression occurs within the first 2 weeks after transfer to SD (Figure 3A). These early clusters contain light signal transduction components and, at 2 weeks of SDs, additionally ethylene signal transduction components. In parallel, the metabolite composition changes within the first week of SDs: two-thirds decrease, including 7 of the 14 sugars/sugar alcohols, and one-third increase, including several amino acids (Figure 3B). A second major transition takes place from 3 to 4 weeks of SDs. For 398 genes (42.1%) and 81 metabolites (50.6%), the most marked change in expression coincided with the transition to a bud structure. A substantial number of cell proliferation genes are downregulated at 3 to 4 weeks of SDs, while genes of the ABA pathway are upregulated. Five amino acids that initially increase after 1 week of SDs decrease, together with 10 other amino acids, after 3 weeks of SDs (Figure 3B). Interestingly, only five genes (0.5%) display their major change in expression from 5 to 6 weeks of SDs, the time that dormancy was established. Therefore, fixing the dormant state might rely on mechanisms other than gross transcriptional regulation.
In the following, the temporal clusters are systematically investigated for the presence of particular signal transduction or metabolic pathways. Comprehensive pathway information in poplar was inferred from the available literature for pathway components in Arabidopsis through linking poplar genes to their corresponding closest Arabidopsis homologs. For the sake of brevity, common gene names of the closest Arabidopsis genes are used to describe poplar genes.
SDs Activate Many Components of Light Signal Transduction
The change from LDs to SDs provoked expressional changes in many light signal transduction components. In addition to the detection and transduction of light signals, the ability to interpret photoperiod critically depends on the circadian clock (Eriksson and Millar, 2003). Both pathways converge at the CO/FT regulon that is required to transduce photoperiod to growth arrest and initiation of bud set (Böhlenius et al., 2006).
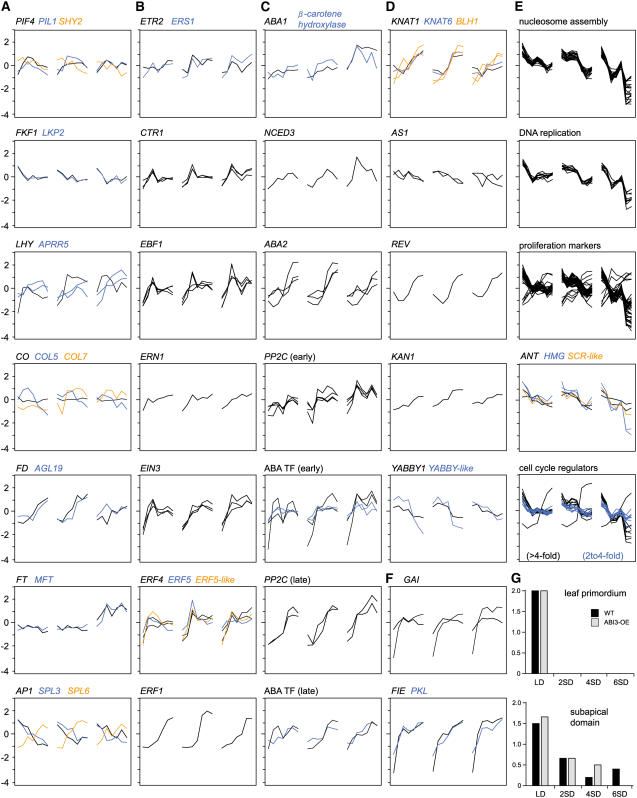
Of the 74 genes on the microarray involved in light signal transduction and the circadian clock, 38 genes (51.4%) are more than twofold DE, of which 9 genes (12.2%) are more than fourfold DE (Figure 4A; see Supplemental Table 2 online) (Wang and Deng, 2002). PHYTOCHROME A (PHYA) and PHYB are not regulated at the transcriptional level. However, nuclear phytochrome interactors, such as PHY-INTERACTING FACTOR4 (PIF4) and PIF3-LIKE1, are upregulated, and the AUX/IAA gene SUPPRESSOR OF PHYB2 (SHY2/IAA3) is downregulated early after the onset of SDs (Figure 4A). Downstream intermediates of PHYA signaling are also strongly DE at the transition from LDs to SDs (see Supplemental Table 2 online).
Figure 4.
Activation of Signaling and Developmental Pathways during SD-Induced Apical Bud Development.
(A) Light signaling, circadian clock, and CO/FT regulon.
(B) Ethylene signaling.
(C) ABA biosynthesis, signaling, and ABA-regulated TF.
(D) Meristem identity, meristem activity, organ development, and leaf patterning.
(E) Cell proliferation.
(F) Early SD-upregulated genes.
(G) Estimates of the frequency of mitotic figures in leaf primordium and the subapical domain during bud development in the wild type (black) and ABI3-OE (gray), given according to the frequency of occurrence of mitotic figures, with 0 (none), 1 (rare), or 2 (at least 2 to 4; frequent), observed in hundreds of cells in a transmission electron microscopic section. Obtained scores were averaged over three to five sections per sampling point and genotype. Typical mitotic figures are shown in Figures 5C and 5D.
In each graph, wild-type gene expression profiles are flanked by ABI3-AS (left) and ABI3-OE (right), with each consisting of seven sampling points. Expression profiles are given as log2-transformed, mean-centered data. For regulatory genes, more than twofold DE genes are shown; for genes encoding structural components, more than fourfold DE genes are shown. For full gene names, see text.
Other light signal transduction components involved in photoperiodic adjustment of the clock, such as FLAVIN BINDING, KELCH REPEAT, F-BOX PROTEIN1 (FKF1) and LOV DOMAIN KELCH REPEAT PROTEIN2 (LKP2), are downregulated after the onset of SDs (Figure 4A). FKF1 affects the circadian clock via TIMING OF CHLOROPHYLL a/b BINDING PROTEIN (TOC1) (Eriksson and Millar, 2003). While TOC1 and CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) are not on the microarray, LATE-ELONGATED HYPOCOTYL (LHY), constituting together with the former the central oscillator of the circadian clock, is upregulated after the onset of SDs (Figure 4A). Generally, the ARABIDOPSIS PSEUDORESPONSE REGULATOR (APRR) genes, which play essential roles close to the central oscillator, are upregulated. The most dynamic changes occurred early after the onset of SDs and in APRR5, the gene diurnally peaking closest to the time of harvest (Figure 4A). The fact that this gene is continually upregulated during 6 weeks of SDs suggests a continuous adjustment of clock genes over the whole period of consecutive SDs.
In Arabidopsis, seasonal events, such as the transition to flowering, also involve photoperiodic regulation. Recently, and particularly after the disclosure of a FT function in growth cessation (Böhlenius et al., 2006), pathways controlling the transition to flowering were hypothesized to control bud development (Horvath et al., 2003). Here, we had the opportunity to test whether these analogies extended beyond the photoperiodic pathway. Of 50 genes involved in the transition to flowering, 16 (32.0%) are more than twofold DE, of which 7 (14.0%) are more than fourfold DE (Boss et al., 2004) (Figure 4A; see Supplemental Table 2 online). CO expression, with samples taken in its diurnal expression minimum, does not change. However, seven CO-LIKE (COL) genes are more than twofold DE, and three of them are more than fourfold DE (Figure 4A). Four COL genes with strong genotype-dependent expression are either DE at 3 to 4 weeks of SDs or unregulated in ABI3-OE (Figure 4A; see below). FD, two APETALA1 genes, and two SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes are dramatically regulated in all genotypes (Figure 4A). Few other genes belonging to other flowering-time input pathways are DE (see Supplemental Table 2 online). Moderately DE genes include SUPPRESSOR OF CONSTANS1 (SOC1/AGL20), other SPL genes, and, interestingly, FCA, a gene encoding a protein with ABA binding ability in Arabidopsis (see Supplemental Table 2 online) (Razem et al., 2006). Together, these results suggest that the analogy between the transition to flowering and bud development is limited.
In addition to light signal transduction, and consistent with a rapid downregulation of gibberellin (GA) biosynthesis and cell elongation by SDs (Olsen et al., 1995), two genes encoding the GA response modulator GA-INSENSITIVE (GAI), considered to maintain a repressed state of GA signaling, were dramatically upregulated immediately after the onset of SDs (Figure 4F) (Fleet and Sun, 2005). Moreover, PICKLE (PKL) and FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), both involved in chromatin remodeling, are markedly upregulated at 1 week of SDs (Figure 4F). PKL functions as a general repressor of meristematic activity by negatively regulating meristematic genes such as AINTEGUMENTA (ANT) (Eshed et al., 1999). FIE is thought to be essential for the control of leaf and shoot development by regulating KNOTTED-like genes (Katz et al., 2004). Together, these regulatory genes in light signal transduction, GA signaling, and chromatin remodeling are part of the immediate molecular response to SDs and might control many downstream genes contained in the early temporal expression clusters.
Ethylene Biosynthesis and Signal Transduction Are Coordinately Triggered after 2 Weeks of SDs
Of a total of 53 genes on the microarray that are putatively involved in ethylene biosynthesis, perception, and downstream signaling, 21 genes (39.6%) were more than twofold DE, of which 9 genes (17.0%) were more than fourfold DE (Figure 4B) (Schaller and Kieber, 2002; Guo and Ecker, 2004). The temporal expression pattern of two ETHYLENE RESPONSE FACTOR4 (At ERF4) genes, At ERF5, and At ERF5-LIKE closely correlates with that for ethylene biosynthesis (ACC SYNTHASE6 [ACS6]), perception (ETR2 and ETHYLENE RESPONSE SENSOR1 [ERS1]), and signal transduction components (three CONSTITUTIVE TRIPLE RESPONSE1 [CTR1] genes, three ETHYLENE INSENSITIVE3 [EIN3] genes, four EIN3 BINDING F BOX1 [EBF1] genes, and ETHYLENE-REGULATED NUCLEAR-LOCALIZED PROTEIN [ERN1]) (Figure 4B), suggesting that these ERF genes are regulated in an ethylene-dependent manner. Thus, genes throughout the ethylene biosynthesis and signal transduction pathways are coordinately and transiently activated at 2 weeks of SDs but before the transition to the bud starting at 3 weeks of SDs. Only ERF1 expression is markedly upregulated at 3 to 4 weeks of SDs, implying regulation by another signal transduction pathway, possibly ABA (Figure 4B).
ABA Signal Transduction Peaks at 3 to 4 Weeks of SDs and Is Modified in ABI3-OE
Of 60 genes on the microarray that are putatively involved in ABA biosynthesis, seven genes encoding enzymes at four different steps are more than fourfold DE (Figure 4C; see Supplemental Table 2 online) (Finkelstein and Rock, 2002). Strikingly, genes encoding the rate-limiting (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 [NCED3]) and final (ABA2) steps in ABA biosynthesis are dramatically upregulated between 3 and 4 weeks of SDs in the wild type (Figure 4C) (Finkelstein and Rock, 2002). In addition, β-CAROTENE HYDROXYLASE and ABA1 are moderately upregulated in the wild type and ABI3-AS but more than fourfold induced in ABI3-OE by 2 weeks of SDs (Figure 4C). The upregulation of critical enzymes for ABA biosynthesis at 3 to 4 weeks of SDs is consistent with the previously reported transient peak in ABA content at 4 weeks of SDs (Rohde et al., 2002).
Of 64 genes on the microarray that are putatively involved in ABA signal transduction (Fedoroff, 2002; Finkelstein and Rock, 2002; Himmelbach et al., 2003), 20 genes are more than twofold DE (31.3%; 10 genes are more than fourfold DE), including homologs of MITOGEN-ACTIVATED PROTEIN KINASE3 (At MPK3), FAR-RED-INSENSITIVE219 (FIN219), HIGH RESPONSE TO OSMOTIC STRESS1 (HOS1), and ABI1, as well as five other PROTEIN PHOSPHATASE2C (PP2C) genes related to ABI1 and ABI2 (see Supplemental Table 2 online). Two of six PP2C homologs are upregulated between 3 and 4 weeks of SDs, coordinately with ABA2, and the other four are upregulated between 1 and 2 weeks of SDs (Figure 4C). Also, ABA-related TFs are upregulated at 3 to 4 weeks of SDs (ABA-RESPONSIVE ELEMENT BINDING PROTEIN3 [AREB3] and MYB62) or at 1 to 2 weeks of SDs (MYC2, ABA RESPONSE ELEMENT BINDING FACTOR2 [ABF2], G-BOX BINDING FACTOR3 [GBF3], and HB12). Together, ABA biosynthesis and part of the signal transduction pathway occur concomitantly with the transition of the apex to a closed bud structure but before the termination of meristematic activity.
Other hormonal pathways were either not represented in sufficient numbers of genes on the microarray (GA and cytokinin) or had no prominent or consistent changes in gene expression that would suggest pathway activation at a particular time (auxin; data not shown). In summary, SDs are the first and major trigger for bud development. Subsequently, ethylene and ABA signal transduction pathways are activated before and concomitant with the formation of the bud structure, respectively.
Factors Involved in Primordium Initiation and Differentiation to Bud Scales and Embryonic Leaves during Bud Formation
Although the bud structure becomes visible only after 3 weeks of SDs, the development of primordia is redirected from foliage leaves toward bud scales and embryonic leaves immediately after SD perception and continues until meristematic activity is terminated. Of 49 genes with various roles in meristem maintenance, primordium initiation, or leaf development, 18 genes (36.7%) are more than twofold DE, of which 10 genes (20.4%) are more than fourfold DE (Figure 4D; see Supplemental Table 2 online) (Bowman et al., 2002; Hake et al., 2004; Kepinski, 2006). In general, the major change in expression occurs at 3 to 4 weeks of SDs, concomitant with the transition to a bud structure.
Of the genes involved in maintaining the stem cell population in the central meristem, WUSCHEL and CLAVATA1 are not DE during bud formation (see Supplemental Table 2 online). Class I KNOX genes (BREVIPEDICELLUS/KNAT1 and KNAT6) and two members of the related BEL1-LIKE family (BEL1-LIKE HOMEODOMAIN1 [BLH1]) are coordinately upregulated during bud development (Figure 4D). In Arabidopsis, ASYMMETRIC LEAVES1 (AS1) is involved in the maintenance of class I KNOX repression after the displacement of cells from the stem cell population to founding organs (Byrne et al., 2000). Two AS1 genes are moderately downregulated during bud development, concomitant with the upregulation of KNAT1 and KNAT6 (Figure 4D).
Primordium differentiation and leaf development are further controlled by plant-specific TFs of the class III HD-ZIP, KANADI (KAN), and YABBY families (Bowman et al., 2002; Hake et al., 2004). REVOLUTA (REV), a member of the class III HD-ZIP family specifying adaxial cell fate, is upregulated after 3 to 4 weeks of SDs (Figure 4D). KAN and YABBY gene family members act partially redundantly in specifying abaxial cell fate (Bowman et al., 2002). KAN1 is upregulated, whereas YABBY1 and a YABBY-LIKE gene are downregulated, during bud development (Figure 4D). Together, the expression dynamics probably stem from the changing relative contribution of the meristem, young proliferative cells, and differentiating cells to the sampled organ. During bud formation, young embryonic leaves become densely packed and are not displaced from the apex by elongation, leading to a relatively higher number of meristematic cells (AS1, downregulated; KNAT1/KNAT6, upregulated) and an increase in young organs (REV/KAN, upregulated) that are halted in their differentiation (YABBY1 and YABBY-LIKE, downregulated).
To assign novel genes to the developmental program of bud scales, the expression profiles of ABI3-OE plants that essentially lack enveloping bud scales were explored. In total, 82 genes of the fourfold set were upregulated at 3 to 4 weeks of SDs in the wild type, the time that bud scales become visible, and were markedly less expressed in ABI3-OE, in agreement with a lack of bud scales (see Supplemental Table 2 online). Bud scales accumulate phenolic components, leading to their final red-brown coloration (Rohde et al., 2002). Consistently, catechine, a typical flavan-3-ol of Salicaceae and a precursor of condensed tannins, increases most markedly at 3 to 4 weeks of SDs in all genotypes but to a lesser extent in ABI3-OE (Figure 3B). The delineated set of 82 genes contains five genes encoding anthocyanin biosynthesis enzymes. Therefore, expression profiles of these genes are correlated with the time and extent of bud scale development and support the notion that the other 77 novel genes are likewise associated with bud scale differentiation and maturation. Although this gene set does not highlight a particular pathway, it contains five TFs that will be instrumental for targeted functional studies.
Meristem Inactivation Starts at 4 Weeks of SDs
The transition from an active to an inactive meristem proceeds sequentially, and the termination of meristematic activity is the hallmark of dormancy. The inactive meristem is characterized by nuclei with less developed nucleoli, indicating reduced synthesis of ribosomal structures (Figures 5A and 5B) (Arend and Fromm, 2003). Of 506 putative cell proliferation genes, 221 genes (43.7%) were more than twofold DE, of which 92 genes (18.2%) were more than fourfold DE, during bud development (see Supplemental Table 2 online) (Vandepoele et al., 2002; Wang et al., 2004; Beemster et al., 2005). The vast majority of these genes (86.9% of more than twofold DE and 92.4% of more than fourfold DE) are downregulated, mostly at 3 to 4 weeks of SDs (Figure 4E). Downregulation of cell proliferation genes was stronger and occurred slightly later in ABI3-OE than in the wild type (Figure 4E).
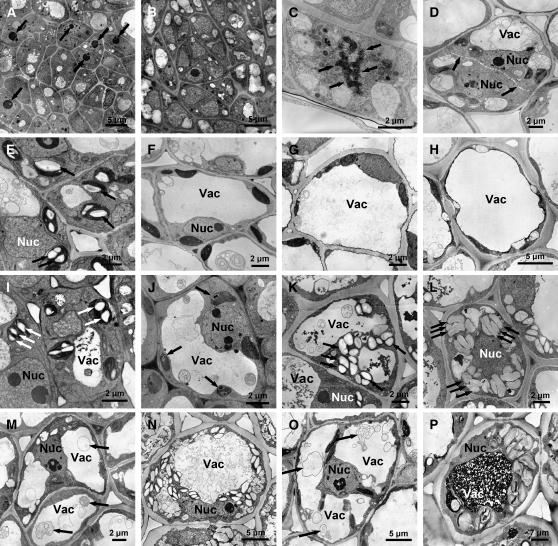
Figure 5.
Cellular Aspects of Bud Development in the Wild Type and ABI3-OE.
(A) Part of the apical meristem in the wild type under LD conditions. Arrows point to large nucleoli in meristematic cells.
(B) Part of the apical meristem in the wild type after 6 weeks of SDs. Most nuclei in meristematic cells lack visible nucleoli.
(C) Typical mitotic figure, cell in metaphase of cell division with separating chromosomes (arrows).
(D) Typical mitotic figure, cell in late telophase of cell division with a developing cell plate (arrows).
(E) to (H) Starch accumulation in wild-type leaf cells in LDs and after 2, 4, and 6 weeks of SDs. Starch granules are absent after 2, 4, and 6 weeks of SDs. Arrows in (E) point to starch granules.
(I) to (L) Starch accumulation in ABI3-OE leaf cells in LDs and after 2, 4, and 6 weeks of SDs. Starch gradually accumulates after 2 weeks of SDs. Arrows point to starch granules.
(M) and (N) Starch accumulation and vacuoles in wild-type cells of the subapical domain in LDs and after 6 weeks of SDs. Arrows in (M) point to vesicular material in lytic vacuoles.
(O) and (P) Starch accumulation and vacuoles in ABI3-OE cells of the subapical domain in LDs and after 6 weeks of SDs. Arrows in (O) point to vesicular material in lytic vacuoles.
Nuc, nucleus; Vac, vacuole.
Besides genes involved in DNA replication and nucleosome assembly and a large group of marker genes preferentially expressed in proliferating cells, the set includes five regulatory genes (two HIGH-MOBILITY GROUP [HMG] genes, a SCARECROW-LIKE [SCL] gene, and two ANT genes) (Figure 4E). In Arabidopsis, ANT positively regulates cell proliferation by maintaining the meristematic competence of cells during organogenesis (Mizukami and Fischer, 2000). In agreement with this function, downregulation of ANT (and the other TFs) at 3 to 4 weeks of SDs might be critical for the arrest of primordium initiation and cell proliferation.
Furthermore, the more than fourfold downregulated genes include core cell cycle regulators, such as CYCLIN A1 (CYCA1), CYCA3, CYCB2, CYCD3, and CYCLIN-DEPENDENT KINASE B1 (CDKB1) and CDKB2 (Figure 4E) (Vandepoele et al., 2002; Wang et al., 2004). CDKA1, CYCA2, and CYCH1 as well as RETINOBLASTOMA PROTEIN, DP-E2F-LIKE PROTEIN3, and CKS1 are twofold to fourfold downregulated (see Supplemental Table 2 online). Notably, only a few genes are upregulated: CYCD1, CYCD2, and a class I TCP gene moderately, and CYCP3;2 strongly (Figure 4E). In Arabidopsis, P-type cyclins bind CDKA, whose expression marks cells that retain the competence to divide (Torres Acosta et al., 2004). Although CDKA is moderately downregulated during bud development, a fraction of cells within the bud might retain division competence during dormancy and additionally express CYCP3;2.
Together, these gene expression data suggest that cell division and proliferation are generally discontinued after 4 weeks of SDs. A systematic analysis of mitotic figures revealed that cell division ceased at different times in different parts of the bud. Under SD conditions, no cell divisions were observed in embryonic leaves shortly after their initiation, in contrast with the young leaves initiated in LDs. In the subapical domain, cell division ceased only after 4 weeks of SDs (Figures 4G, 5C, and 5D). Thus, the downregulation of cell cycle regulators and molecular markers of cell proliferation at 3 to 4 weeks of SDs primarily marks the termination of cell division and proliferation in the central meristem and youngest (latest initiated) leaf primordia after 4 weeks of SDs (after the first visible signs of bud formation).
Carbohydrate Metabolism and Energy Generation Are Reconfigured in Two Phases of Molecular Adaptation to SDs
Bud development involves an extensive reconfiguration of metabolism (Figure 6; see Supplemental Table 2 online) (Lange and Ghassemian, 2005). Metabolism shifted in two markedly separate phases: an early adaptation to SDs and a late adaptation after ∼3 weeks of consecutive SDs, which was already reflected in the global changes in gene expression and metabolite composition (Figure 2).
Figure 6.
Metabolic Rearrangements in Glyoxylate/Citric Acid Cycle, Carbohydrate Metabolism, and Starch Biosynthesis and Degradation during Bud Development.
(A) Metabolite abundance profiles.
(B) Expression profiles of genes encoding enzymes of the glyoxylate cycle.
(C) Expression profiles of genes encoding enzymes involved in starch degradation.
(D) Expression profiles of genes encoding enzymes involved in sucrose, raffinose, and starch biosynthesis.
In each graph, wild-type profiles are flanked by ABI3-AS (left) and ABI3-OE (right), with each consisting of seven sampling points. Gene expression profiles are given as log2-transformed, mean-centered data. Genes encoding enzymes are named by their EC number; for full names, see text. For metabolites, regression curves for peak abundance (selected ion current/100 ng dry weight) are given, together with 95% confidence bands.
The first phase is dominated by a response to the sudden change from a 16-h LD to an 8-h SD. Under LD conditions, diurnally accumulating starch exceeds the energy demand during the short dark period. Plastids contain numerous starch granules shortly after the onset of the light period, mostly in the young leaves and, to a lesser extent, in the subapical domain (Figures 5E and 5I). Under SD conditions, however, starch is remobilized during the night, resulting in the disappearance of starch granules in the wild type (and a reduction in ABI3-OE) by 2 weeks of SDs (Figures 5F and 5J). This is paralleled by a transient decrease in the levels of maltose, glucose, glucose-6-phosphate, fructose, fructose-6-phosphate, sucrose, and xylose after 1 week of SDs in all genotypes (Figure 6A), suggesting a transient shortage in available sugars. Additionally, the expression of genes encoding components of the photosynthetic apparatus continuously decreases after the onset of SDs (see Supplemental Table 2 online), contributing to a lower energy supply.
Concomitantly, genes encoding enzymes of the glyoxylate cycle are upregulated after 1 week of SDs (ISOCITRATE LYASE [EC 4.1.3.1], MALATE SYNTHASE [EC 4.1.3.2], GLYCOLATE OXIDASE [EC 1.1.3.15], and MALATE DEHYDROGENASE [EC 1.1.1.40]) (Figure 6B), whereas none of the other enzymes of the citric acid cycle was DE. The increasing abundance of citrate and aconitate, together with decreasing levels of fumarate, argues additionally for an increasing contribution from the glyoxylate cycle compared with the citric acid cycle for energy generation during bud development (Figure 6A). Furthermore, a continued decrease in glucose-6-phosphate and fructose-6-phosphate at later stages suggests a reduced respiration through glycolysis (Figure 6A).
Carbohydrate metabolism is modified toward the accumulation of storage compounds and cryo-protecting or dehydration-protecting solutes when growth becomes increasingly arrested after 2 weeks of SDs. After the initial decrease, glucose, fructose, sucrose, and xylose gradually reaccumulate toward 6 weeks of SDs in the wild type and ABI3-AS (Figure 6A). By contrast, ABI3-OE maintains low glucose, fructose, maltose, and xylose levels, while it accumulates high levels of sucrose and raffinose during later stages of bud development. Correspondingly, enhanced SUCROSE SYNTHASE (EC 2.4.1.13) expression, together with ectopic expression of a GALACTINOL SYNTHASE (EC 2.4.1.123) in ABI3-OE, probably drives carbohydrate compound composition from hexoses toward oligosaccharides of the raffinose family (Figure 6D).
Genes encoding enzymes involved in starch degradation (α-GLUCAN/WATER DIKINASE [EC 2.7.9.4], ISOAMYLASE [EC 3.2.1.68], STARCH PHOSPHORYLASE [EC 2.4.1.1], α-AMYLASE [EC 3.2.1.1], and β-AMYLASE [EC 3.2.1.2]) start to be upregulated by 2 weeks of SDs (Figure 6C), whereas those involved in starch biosynthesis (ADP-GLUCOSE PYROPHOSPHORYLASE [EC 2.7.7.27], STARCH SYNTHASE [EC 2.4.1.21], and 1,4-α-GLUCAN-BRANCHING ENZYME [EC 2.4.1.18]) are most strongly upregulated at 3 to 4 weeks of SDs (Figure 6D). Activation of genes encoding starch biosynthetic enzymes is consistent with the appearance of amyloplasts after 4 to 6 weeks of SDs. Starch storage after 4 weeks of SDs occurred predominantly in the subapical domain, clearly differing from starch storage in young leaves during active growth (Figures 5E, 5N, and 5P). In ABI3-OE, the accumulation of storage starch is enhanced in the subapical domain (Figure 5P) but also occurs ectopically in embryonic leaves by 4 to 6 weeks of SDs (Figures 5K and 5L). These observations are consistent with an enhanced upregulation of a STARCH SYNTHASE gene (EC 2.4.1.21) in ABI3-OE (Figure 6D). In conclusion, starch accumulation occurs in response to SDs without additional cold signals.
The transition to storage mode is further demonstrated by the change from lytic vacuoles with numerous vesicles and membranous material under LDs to storage vacuoles containing dark-stained material by 6 weeks of SDs (Figures 5M to 5P). Storage compounds accumulate predominantly in the vacuoles of cells in the subapical domain and, to a lesser extent, in embryonic leaves. Storage vacuoles were much more pronounced in ABI3-OE in both leaves and the subapical domain (Figure 5P).
Two Waves of Adaptive Responses Occur during Bud Development
Similar to seed maturation, adaptation to dehydration and cold occurs during bud development under natural conditions. SDs alone are sufficient to induce genes known to be responsive to dehydration, cold, and ABA or are expressed during seed maturation, here collectively termed adaptive response genes (Fowler and Thomashow, 2002; Hoth et al., 2002; Ruuska et al., 2002; Seki et al., 2002; Suzuki et al., 2003; Vogel et al., 2005; Benedict et al., 2006) (Figure 7A; see Supplemental Table 2 online). Of 3346 adaptive response genes on the microarray, 1278 genes (38.2%) were more than twofold DE, and 389 genes (11.6%) were more than fourfold DE, during bud development (Figure 7A). Cold-induced (17.6%) and drought-induced (21.5%) genes are most represented within the fourfold DE gene set (Figure 7A). Notably, these genes are induced in response to SDs alone, in the absence of any exogenous cold or drought.
Figure 7.
Expression of Adaptive Response Genes and ABI3-Dependent Genes during Bud Development.
(A) Genes associated with responses to ABA, cold, drought, and seed maturation analyzed for expression during bud development. For each of the four responses, the total number of genes on the microarray is given, and the Venn diagram indicates the number of genes shared with one or more of the other groups for more than twofold (black) and more than fourfold (orange) DE gene sets. Genes shared between ABA response and seed maturation (AS) and cold and drought responses (CD) are given separately. The fourfold DE genes of the four groups are clustered according to the time of maximal upregulation or downregulation, occurring early (before 3 weeks of SDs) or late (after 3 weeks of SDs).
(B) and (C) Ectopic and putative ABI3 targets identified among the fourfold DE genes. The 146 ectopic targets (B) are characterized by ABI3-dependent expression profiles in the absence of DE in the wild type (less than twofold DE). The 324 putative ABI3 targets (C) have ABI3-dependent expression profiles and more than twofold DE in the wild type. The genes are categorized according to the requirement of SDs for the genotype effect to be recognized (more than twofold DE between the wild type and ABI3-OE in LDs) and according to temporal expression dynamics. Arrows indicate the extent of upregulation or downregulation in different genotypes. Six genes that are upregulated in ABI3-AS after exposure to SDs (B) are not shown.
In each graph, wild-type profiles are flanked by ABI3-AS (left) and ABI3-OE (right), with each consisting of seven sampling points. Gene expression profiles are given as log2-transformed, mean-centered data. In all clusters, fourfold DE adaptive response genes are depicted in orange, and TFs are depicted in blue. Other ABI3-dependent genes are depicted in gray. Numbers in the top left corner of each panel correspond to the total number of genes (black), number of adaptive response genes (orange), and number of TFs (blue).
Furthermore, the adaptive response set contains 123 genes that were previously identified as targets of C-repeat/dehydration–responsive element binding factors (CBFs) in Arabidopsis (Fowler and Thomashow, 2002; Maruyama et al., 2004; Vogel et al., 2005; Sakuma et al., 2006). Of these genes, 67 (54.5%) are more than twofold DE, with 38 (30.9%) more than fourfold DE, during bud development. However, none of the components with high hierarchical positions in the CBF and/or cold-response pathways, such as INDUCER OF CBF EXPRESSION1 (ICE1), CBF1, CBF3, ZAT12, and ZAT10, was DE. Therefore, SDs trigger the same target processes and components as cold or drought but probably use different signal transduction routes to activate them.
Most of the fourfold DE adaptive response genes were induced in two separate waves (Figure 7A). The first burst occurs within the first 2 weeks of SDs and is characterized by the induction of VEGETATIVE STORAGE PROTEIN (VSP), BARK STORAGE PROTEIN (BSP), drought-responsive proteins, LATE EMBRYOGENESIS-ABUNDANT (LEA) class transcripts, including DEHYDRIN (DHN), RESPONSIVE TO ABA18 (RAB18), and LOW-TEMPERATURE-INDUCED (LTI), which might confer desiccation tolerance, and a number of TFs, such as MYC2, HB12, ABF2, GBF3, ATAF1, NAC072, DROUGHT RESPONSE ELEMENT BINDING1 of the TINY clade (DRTY) (Nakano et al., 2006), and RELATED TO ABI3/VP12 (RAV2) (Figure 7A). These TFs respond to diverse signals, such as dehydration, cold, salinity, glucose, or light (Lee et al., 2001; Fujita et al., 2004, 2005; Kim et al., 2004). A second set of adaptive response genes, containing similar molecular functions as the first set, is induced at 3 to 4 weeks of SDs, together with the TFs MYB62, AREB3, and BLH1 (Figure 7A).
In Arabidopsis, Pro and putrescine constitute components of low-temperature-induced cold acclimation (Cook et al., 2004). None of the genes encoding enzymes performing the biosynthesis from Glu to either Pro or putrescine is DE: Pro levels do not significantly change, and putrescine levels quickly decrease under SDs (Figure 6A; see Supplemental Tables 2, 4, and 5 online). Thus, Pro and putrescine might not contribute to cold acclimation triggered by SDs or do not play a role in poplar, as corroborated by the absence of Pro accumulation in poplar cambium in response to a combination of SDs and low autumn temperatures (Druart et al., 2007).
ABI3-Dependent Processes during Bud Development
Transcriptomic changes in the transgenic poplar with modified ABI3 expression allowed the investigation of putative targets and processes that are influenced by ABI3 during bud development. Within the 1091 genes of the fourfold set, 146 genes (13.4%) were classified as ectopic ABI3 targets, because these were more than fourfold DE in ABI3-OE but not in the wild type. Another 324 genes (29.7%) were classified as putative ABI3 targets, because these were more than twofold DE in the wild type and clearly affected in ABI3-OE. The latter participate in bud development and are putatively regulated by ABI3 (Figures 7B and 7C). Strikingly, 97.3% of the ectopic and 96.3% of the putative targets required the additional presence of SDs to be significantly DE between genotypes, corroborating the need for cooperating factors for ABI3 action. Moreover, of the 470 genes affected by ABI3, only 23 genes were DE in ABI3-AS, indicating that the absence of ABI3 is probably compensated for by other factors during bud development (Figures 7B and 7C).
In Arabidopsis seeds, ABI3 regulates pathways that control late embryogenesis and seed maturation, including processes such as ABA sensitivity, desiccation tolerance, and storage protein accumulation (Parcy et al., 1994). Genes assigned to the seed maturation set represent 9.4% (44 of 470 genes) of all ABI3-dependent genes. Among the upregulated ectopic ABI3 targets in developing apical buds, as many as 55.6% of the genes with an organ-specific expression in Arabidopsis are characterized by predominant expression in seeds (including expressed protein genes; Figure 8B). Five well-known ABI3/ABA targets (three LEA genes, LTI, and Em6) were among the only 16 genes that are more than twofold DE between the wild type and ABI3-OE already under LD conditions (Figures 7B and 7C).
Figure 8.
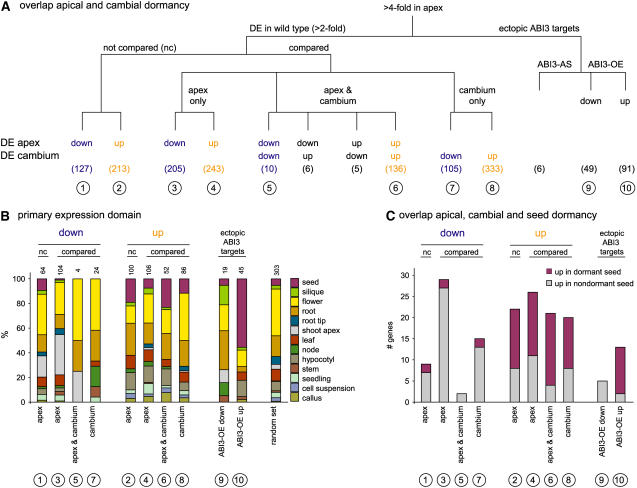
Common and Specific Changes in the Transcriptome during the Growth-to-Dormancy Transition in Poplar Apical Buds, Poplar Cambial Meristem, and Arabidopsis Seeds.
(A) Overlap of the apical and cambial growth-to-dormancy transition in poplar. Of the 945 genes classified as DE in the apex of the wild type, 340 cannot be compared with cambial expression data because the POP2 microarray is extended with an additional probe set compared with the POP1 microarray used by Schrader et al. (2004). The original cambium cDNA probe data are here given as genes. Gene sets found in only apex, only cambium, or both apex and cambium are delineated. The directionality of expressional change toward dormancy is specified for apex and cambium. Gene sets are numbered (in circles) for correspondence with (B) and (C).
(B) Relative distribution of organ-specific expression within gene sets defined in (A). Distributions are based on expression data of the closest Arabidopsis genes, retrieved from Genevestigator. Organ specificity was assumed when an organ contained >10% of the total signal of all 42 different organs represented in Genevestigator. The 42 different organs were subsequently grouped to the 13 major classes represented here. Gene sets from (A) show different organ-specific expression domains and also clearly deviate from those of 1000 random Arabidopsis genes given for reference. Numbers above the bars indicate the total number of genes in each gene set with organ-specific expression; genes without organ-specific expression are omitted.
(C) Overlap of the gene sets in (A) with those identified in dormant and nondormant Arabidopsis Cvi seeds (classification according to Cadman et al., 2006). Poplar genes are linked with the closest Arabidopsis homologs to the Arabidopsis expression data.
ABI3 influences as many as 36.5% of adaptive response genes (142 of 389 genes in the wild-type fourfold set), and 30.2% of all ABI3-dependent genes belong to the adaptive response gene set (142 of 470 genes) (Figures 7A to 7C). This clear overrepresentation of adaptive response genes among the ABI3-dependent genes emphasizes a role for ABI3 in differentiation and maturation prior to dormancy. Similar gene expression changes have been described in Arabidopsis that ectopically expresses ABI3 or its monocot ortholog VIVIPAROUS1 (VP1) (Parcy et al., 1994; Suzuki et al., 2003). The increased expression of adaptive response genes renders Arabidopsis ABI3 overexpressors more cold-tolerant (Tamminen et al., 2001).
ABI3-OE enhances the expression of 15 TFs, of which 6 are putatively involved in adaptive response: DREB5A, BIM2, ATAF1, NAC072, GBF3, and AUXIN REPONSE FACTOR3 (ARF3) (Figures 7A to 7C). All of these TFs, except ARF3, are upregulated early after the onset of SDs. Wild-type ABI3 is only expressed from 3 weeks of SDs onward, making it unlikely that they normally depend on ABI3. Because many of the TFs are known to respond to ABA, ABI3 overexpression, like that of VP1 (Suzuki et al., 2001), might cause a higher sensitivity to ABA. This suggestion is supported by the ectopic expression of ABA biosynthesis (ABA1) and signal transduction (PP2C) components in ABI3-OE, both of which are known to be inducible by ABA (Figure 7C) (Barrero et al., 2006). Thus, the role of ABI3 during bud development could consist of increasing ABA sensitivity at critical times, putatively at 3 to 4 weeks of SDs, when the ABA concentration peaks.
Another pathway that is influenced by ABI3 is light signal transduction (Kurup et al., 2000; Rohde et al., 2000; Mazzella et al., 2005). Among four ABI3-interacting proteins in Arabidopsis, a protein with a CO/COL/TOC1 domain was identified (Kurup et al., 2000). FT, being a target of CO in Arabidopsis and poplar, and MOTHER OF FT AND TFL1 (MFT), another FT family member, are strongly upregulated in ABI3-OE immediately after the onset of SDs but not in LDs. Putatively, ABI3 alters CO action in SDs, leading to the differential expression of FT and MFT. Notably, the ectopic expression of FT in ABI3-OE does not lead to prolonged growth at the apical meristem in SDs, as was observed in poplar that constitutively overexpressed FT (Böhlenius et al., 2006). This observation suggests that the prevention of growth cessation at the meristem requires high FT expression at the time of SD perception and that ectopic FT expression is not effective at later times. Furthermore, the increased transcript levels of FT and MFT are correlated with a decreased induction of FD and AGAMOUS-LIKE19 (AGL19) in ABI3-OE (Figure 4A).
In conclusion, SDs were required for >95% of the ABI3-dependent genes to be DE. In a straightforward scenario, photoperiod-derived or circadian clock–derived factors are needed as interactors for ABI3, but other more remote factors are equally good candidates. The large overlap with adaptive response genes associates ABI3 loosely to ABA-controlled adaptation processes and strengthens its implication in maturation and desiccation processes prior to dormancy itself. Consistent with ABI3 function prior to dormancy, ABI3 expression did not correlate with various dormancy states provoked in dormancy mutants or upon dormancy-modifying treatments (Baumbusch et al., 2004). Thus, ABI3 influences bud development prior to dormancy, possibly through modifying ABA sensitivity.
Common and Specific Components of Growth-to-Dormancy Transitions in Different Organs
Transcriptomic changes during bud development were compared with those associated with growth-to-dormancy transitions in other organs, such as the poplar cambial meristem and Arabidopsis seeds (Schrader et al., 2004; Cadman et al., 2006), to distinguish processes common to growth-to-dormancy transitions from processes specific for a respective organ and/or its developmental context. In contrast with the previous sections, here the establishment of dormancy covers the complete developmental transition to dormancy or out of dormancy.
In microdissected cambial cell layers, Schrader et al. (2004) identified 595 genes that are DE between an actively growing tree (July) and a dormant, cold-acclimated tree (October). On the basis of a direct gene comparison, 26.0% (157 of 605 genes) of the apical expression changes and 26.4% (157 of 595 genes) of the cambial expression changes are commonly associated with growth-to-dormancy transitions in apical bud and cambial meristem (Figure 8A; see Supplemental Table 2 online). Strikingly, of these 157 commonly DE genes, 146 genes (93.0%) exhibit consistent patterns of regulation (Figure 8A). A total of 448 genes are DE in apex, but not in cambium, and might be involved in morphological changes during apical bud development (Figure 8A). Conversely, the 438 genes that are DE in cambium, but not in apex, might be associated with processes that occur specifically in cambium, or at a later stage during dormancy establishment, or in response to cold exposure in the field-grown trees (Figure 8A).
The delineated gene sets were subsequently tested for the extent of specificity for a tissue rather than the transition to dormancy. In the absence of a large and robust quantitative expression database describing organ-specific expression for poplar genes, we used the organ-specific expression data of the respective Arabidopsis homologs (Genevestigator; www.genevestigator.ethz.ch). In general, 65% of the delineated genes have no organ-specific expression in Arabidopsis, whereas those with organ specificity are expressed in diverse organs (Figure 8B). However, among the genes that are downregulated in the apex during bud development, one-third (32.7%) of the organ-specific genes are predominantly expressed in the shoot apex. This fraction is highly enriched compared with the other gene sets (0 to 4%) and identifies genes putatively involved in apex-specific processes that are discontinued upon dormancy induction. Otherwise, genes with predominant expression in hypocotyl or seed are enriched in the gene sets that are upregulated during dormancy induction in apex and/or cambium (Figure 8B), including genes that contribute to desiccation tolerance, such as LEA/dehydrin proteins. Thus, the acquisition of desiccation tolerance is a common and integral component of bud development, which is similarly activated in cold-acclimated and dormant cambium.
Cadman et al. (2006) described gene sets that are DE between dormant and nondormant Arabidopsis seeds. Comparison of these gene sets with those of poplar should counterselect for genes involved in desiccation tolerance (Figure 8A), because desiccation tolerance has already been acquired during seed maturation prior to dormancy. For 64 of 447 genes that are downregulated upon dormancy induction in poplar apex and/or cambium, an Arabidopsis homolog is DE in dormant versus nondormant seeds (Figure 8C). The majority (49 genes, 76.6%) becomes upregulated in nondormant seeds and includes cell proliferation genes, consistent with preparation for growth resumption. Similarly, for 80 of 925 genes that are upregulated upon dormancy induction in apex and/or cambium, an Arabidopsis homolog is DE in dormant versus nondormant seeds, and of these, 58 genes (72.5%) are more highly expressed in dormant seeds than in nondormant seeds (Figure 8C). Together, these 107 consistently regulated genes are putatively associated with growth-to-dormancy transitions in seed and bud and/or cambium and include 12 TFs (see Supplemental Table 2 online). The 23 genes consistently expressed in all three organs include eight expressed genes and two as yet poorly characterized TFs in Arabidopsis: ATAF1 and SCL14. Therefore, comparing dormancy-related gene sets delineates components that are common to transitions to dormancy in various organs and differentiates between genes related to bud formation, cold acclimation and dehydration, and dormancy.
DISCUSSION
Extensive Transcriptional Regulation and Metabolic Reconfiguration Occur during Bud Development
The transition from active growth to dormancy is part of the adaptation to seasonal growth in perennials. To dissect the complex developmental program that underlies the transition from growth to dormancy, we reconstructed high-resolution temporal gene expression and metabolite profiles. These profiles revealed extensive molecular and biochemical changes during bud development. As much as 8% of the sampled transcriptome and 17% of the inspected GC-MS peaks changed profoundly (Figure 1). These dramatic changes are induced by SDs alone and yet identify only some of the pathways and downstream processes that are expected to occur in nature in response to gradually shortening days and declining temperatures. Importantly, transcriptional regulation and metabolic reconfiguration were found to be major components in many pathways that are involved in preparing for adverse winter conditions but not for the final installation of dormancy (Figure 3). Therefore, the fixation of dormancy might be controlled at a level other than gross transcriptional or metabolic regulation. Among the 1091 genes with more than fourfold DE, 244 genes annotated as expressed, hypothetical, or unknown proteins have now become associated with processes during bud development by their expression dynamics (see Supplemental Table 2 online). Altogether, besides its descriptive value, this data set opens ways for future targeted approaches, such as using groups of marker genes as a reference frame for studying genotypes with aberrant bud development or the downstream processes of other inducing factors. In addition, promoters with specific temporal patterns during bud development and regulatory genes are instrumental for targeted modifications of bud development.
A Timetable of Regulatory Events and Their Target Processes during Bud Development
Our results provide a comprehensive framework for the molecular processes that underlie bud development. Genes and metabolites have now become linked to consecutive events during bud development. The major regulatory genes and pathways as well as typical marker genes are presented in Figure 9 as a molecular timetable for bud development. Importantly, we attempted to dissect autumnal bud development into its three components (bud formation, the acclimation to dehydration and cold, and dormancy) and assigned specific groups of (marker) genes to each. The developmental progression is generally characterized by a consecutive activation of distinct signaling pathways with different consequences for each of the three parts of bud development (Figure 9). Bud formation begins immediately after the onset of SDs, although it is not readily visible from its initiation. The gradual imposition of dormancy starts later. Acclimation to dehydration and cold occurs concurrently with both. Furthermore, global analysis of transcript and metabolite profiles defined two major phases in bud development: an early response to SDs (first 2 weeks of SDs) and a second response after 3 weeks of consecutive SDs (Figures 2 and 9).
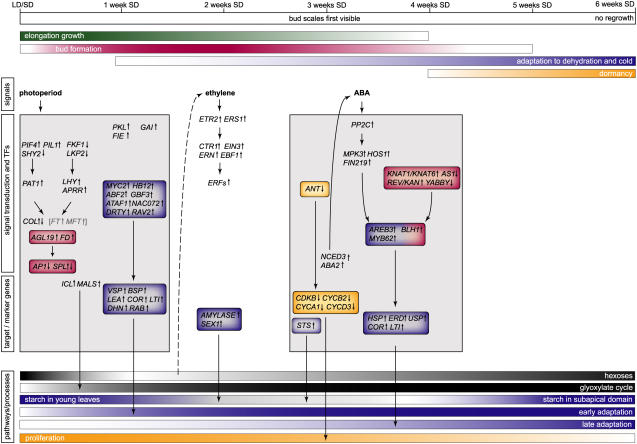
Figure 9.
Integrative Molecular Timetable of Bud Development in Poplar.
Autumnal bud development is a composite of bud formation (red), acclimation to dehydration and cold (blue), and dormancy (orange). Selected genes or processes that specifically belong to one of these aspects are highlighted accordingly. Simultaneous with bud development, elongation growth (green) gradually ceases in young derivatives that are displaced from the apex. Bud development is characterized by the sequential activation of light, ethylene, and ABA signal transduction pathways. The major transcriptional changes at the regulatory and target process levels are depicted at the time that the respective genes show their maximal change in expression. The two major phases of transcriptional and metabolic response are indicated by gray boxes. Below, tentative levels of cellular responses and/or the quantity of major metabolites are indicated with a graded scale. Arrows connect regulators and TFs to their putative downstream processes, without implying a genetic or direct molecular interaction. Because of its putative nature, the link between low sugar and ethylene signal transduction is shown with a dashed arrow. Genes shown in gray and within brackets are only found DE in ABI3-OE. ICL, ISOCITRATE LYASE; MALS, MALATE SYNTHASE; STS, STARCH SYNTHASE; SEX1, STARCH EXCESS1 (α-GLUCAN/WATER DIKINASE). For a full explanation and other names, see text.
The initial phase (first 2 weeks of SDs) covers the start of bud formation and a first wave of adaptive response genes (Figure 9). During that early phase, light signal transduction pathways are activated and carbohydrate metabolism is adjusted to the change in photoperiod. Transiently, the metabolic demand during long nights exceeds the amount of starch generated in SDs and leads to a sugar depletion at the end of the night (Figures 5, 6, and 9). Consistent with limiting carbohydrate levels, the glyoxylate cycle is upregulated immediately after SD perception, likely providing carbohydrate intermediates for gluconeogenesis or the generation of energy (Figures 6 and 9).
At the end of the first phase, ethylene biosynthesis and signal transduction pathways are transiently activated (Figures 4B and 9). This time precedes the final cessation of internode elongation and the first visible signs of bud formation, indicating that transient ethylene accumulation might be a crucial signal controlling developmental progression. In agreement with this hypothesis, ethylene-insensitive birch (Betula pendula) trees do not develop a closed bud (Ruonala et al., 2006). Transient activation of ethylene signaling lags the transfer to SDs by 2 weeks, excluding a direct regulation by SDs. Although a range of other environmental and developmental signals can induce ethylene biosynthesis and signaling, particularly the low sugar levels at 1 week of SDs could be the trigger during bud development (Figures 6 and 9). In Arabidopsis, acute low sugar levels resulting from an extended night transcriptionally activate ACS, ACC OXIDASE, ERS2, EIN3, and several ERF genes, including ERF1, At ERF2, At ERF4, and At ERF5 (Thimm et al., 2004), together closely resembling the gene set that has been identified during bud development (Figures 4B and 9). Thus, the low sugar levels downstream of a SD response might generate endogenous signals (such as ethylene) that, in turn, could contribute to progression to the next phase of bud development (Figure 9).
The second phase of bud development covers the morphological transition to a closed bud, the metabolic switch to storage mode after 3 to 4 weeks of SDs, and meristem inactivation. Most primary metabolites are downregulated at this time, indicating progressive inactivation of growth (Figure 3B). The only discernible hormonal signal at 3 to 4 weeks of SDs is ABA, as shown by a peak in ABA content in the apex and the induction of the rate-limiting ABA biosynthetic enzymes and ABA signal transduction (Figure 4C) (Rohde et al., 2002). ABA clearly contributes to a second wave of expression of adaptive response genes but probably also impinges on cell proliferation, which rapidly terminates after 4 weeks of SDs (Figure 9).
Strikingly, little transcriptional change is associated with the final stages of bud development, covering dormancy establishment (Figures 3 and 9). Thus, gross transcriptional regulation might not be important for imposing dormancy. The stable nature of dormancy could arise from long-term fixation of the expressional state of critical components, such as molecular regulators of cell division or growth, and might depend on chromatin remodeling. However, at least four different factors involved in chromatin remodeling (CDC48-LIKE, HISTONE1-3, FIE, and PKL) are strongly upregulated in apical buds immediately after SD perception rather than at the time of dormancy establishment (Figure 4F; see Supplemental Table 2 online).
Bud Formation
To generate a bud, primordium differentiation is directed, immediately after SD perception, to bud scales and embryonic leaves instead of foliage leaves (Rohde and Boerjan, 2001). This process is initiated from the first primordium originating in SDs onward and, therefore, must be initiated downstream of the photoperiodic signal. FIE and PKL, upregulated immediately after SD perception and repressing meristem identity (KNOTTED) and meristematic proliferation (such as ANT) genes in Arabidopsis, might participate in these early responses (Figures 4F and 9) (Eshed et al., 1999; Katz et al., 2004). The downregulation of GA biosynthesis and the marked upregulation of GAI that represses GA signaling within the first week of SDs, and the induction of ethylene biosynthesis and signal transduction at 2 weeks of SDs, might be necessary to prevent the elongation of young organs and internodes arising at the apex in SD conditions (Figures 4B and 4F).
Most changes in the expression of genes involved in organ development and in maintaining meristematic identity are pronounced at the time that bud scales become visible. During bud formation, young embryonic leaves get densely packed and are not displaced from the apex by elongation, leading to a relatively higher amount of meristematic cells and an increase in young organs that are halted in their differentiation (Figures 4D and 9). The contribution of ABA-triggered processes might consist mainly of allowing the young leaf organs at the apex to adapt to dehydration and cold.
Most interestingly, FD expression increased continually during bud development (Figure 4A). FD is expressed in the apical meristem and the young vegetative and floral primordia in Arabidopsis (Wigge et al., 2005). Together with FT, FD activates AP1 in Arabidopsis (Wigge et al., 2005). AP1 expression is also increased during bud development, although FT is expressed only in ABI3-OE (Figure 4A). It will be most interesting to explore the function of FD in bud formation, because it might represent an element of floral transition that has been recruited into the photoperiodic regulation of bud formation.
Bud Dormancy
Primordium initiation ceases when the meristem is finally inactivated, the latter being seen as a characteristic of dormancy induction (Rohde and Bhalerao, 2007). The timing of the differential expression of molecular markers and regulators of cell proliferation coincides with the transition to the bud and identifies 3 to 4 weeks of SDs as a critical time not only for bud structure development but also for dormancy initiation. ABA, peaking at 3 to 4 weeks of SDs, might contribute to the suppression of growth (Rohde et al., 2002). Microscopic analysis suggested that cell division ceases at different times within the bud: in embryonic leaves shortly after their initiation, and after 4 weeks of SDs in the subapical domain (Figure 4G). Therefore, downregulation of cell proliferation genes at 3 to 4 weeks of SDs primarily marks the progressive suppression of cell proliferation activity and primordium initiation from the meristem.
The proliferation genes, in turn, might be controlled by regulators of cell proliferation, such as ANT, that are coordinately downregulated at 3 to 4 weeks of SDs (Figures 4E and 9). CDKA and CDKB transcripts are downregulated, together with almost all other cell cycle–related genes, during bud development (Figure 4E). However, CDKA downregulation is milder than that of CDKB, suggesting that a subset of cells might retain CDKA expression. Because CYCD1 and CYCD2 are primarily expressed in G1, and CYCD3 is probably involved in the onset of S phase (Wang et al., 2004), upregulation of CYCD1 and CYCD2 and downregulation of CYCD3 are consistent with a relative increase in the G1-phase cell population. Accordingly, flow cytometric data have indicated that the major fraction of cells in dormant poplar buds is arrested in G1 (Rohde et al., 1997).
Bud Formation and Dormancy Are Confounded with Concomitant Adaptation to Dehydration and Cold
SDs alone are sufficient to induce genes related to adaptive responses in the absence of the respective abiotic stress factors, such as cold or drought. The adaptive response genes were divided into two temporally distinct subsets covering essentially similar molecular functions (Figures 7 and 9). This two-phased adaptive response suggests regulation by at least two distinct regulatory pathways. Both phases appear to be independent from the CBF/cold-responsive signal transduction pathway. Many of these genes, and many of the associated TFs, can be regulated by ABA, but activation of the first set of genes occurs well before the upregulation of critical ABA biosynthesis genes and the endogenous ABA accumulation at ∼4 weeks of SDs (Figures 4C and 7). Therefore, this part of the response probably depends on signal(s) other than ABA, possibly SD-derived factors. The second wave of expression of adaptive response genes occurred at 3 to 4 weeks of SDs, concomitant with upregulated NCED3 and ABA2 expression and a peak in endogenous ABA content (Rohde et al., 2002). Thus, the late adaptive response is probably ABA-dependent (Figure 9). Similar stress-related gene expression was promoted by dormancy-inducing conditions in Arabidopsis seeds (Cadman et al., 2006). Likewise, cold hardiness–related genes were upregulated in poplar cambium prior to the occurrence of low temperature in autumn (Druart et al., 2007).
Commonalities of Dormancy Regulation in Different Dormant Organs
Dormancy represents a survival strategy to persist during prolonged periods that are unfavorable for growth. Dormancy can be integrated at diverse periods of the plant life cycle and can become established in meristems of organs as different as bud and seed. It has been known for years that seed and bud dormancy share physiological characteristics (Wareing, 1956), but it has not previously been pinpointed at the molecular level.
A comparison of transcriptome data for growth-to-dormancy transitions identified common gene sets containing, among others, 21 genes and 2 TFs (ATAF1 and SCL14) that consistently change expression in seed, bud, and cambium (see Supplemental Table 2 online). Among poplar cambium and buds, 12 transcriptional regulators are shared. With the exception of WRKY75, all of them are upregulated within the first 2 weeks of bud development, preceding the actual transition to dormancy by several weeks, suggesting that they might not be involved in dormancy establishment itself. Moreover, 8 of the 12 TFs belong to the adaptive response sets (ATAF1, PLATZ1, BIM2, APRR5, HB12, WRKY33, WRKY75, and DRTY; Figure 7A), of which HB12 and DRTY additionally have very distinct autumnal expression dynamics in poplar cambium (Druart et al., 2007). Accordingly, a substantial fraction of transcriptome changes common to dormancy induction in cambium and apical buds is dedicated to molecular programs that control the accumulation of storage compounds and the acquisition of desiccation and cold tolerance (Figures 7 and 8). Additionally, GAI and three TFs that are poorly characterized in Arabidopsis (SCL14, a MYB of the CCA1 clan, and a CCAAT-HAP2) are contained within the shared set of 12 TFs but have not previously been associated with one of the adaptive responses, suggesting a role other than acclimation during the growth-to-dormancy transition. Together, the genes common to the growth-to-dormancy transition in different organs might be particularly valuable for future dormancy studies.
In conclusion, with this molecular timetable at hand, it will become possible to address the functional relevance of the marker and regulatory genes for the process of bud development. The specific assignment of genes to the various processes of bud development will be instrumental in further experimentally uncoupling bud formation and dormancy. Furthermore, the genes will be useful for association genetics approaches to investigate the strong latitudinal differentiation of bud set traits.
METHODS
Plant Material
Transgenic poplar (Populus tremula × Populus alba; Institut National de la Recherche Agronomique No. 717.1B4) lines with antisense downregulation (ABI3-AS) and overexpression of ABI3 (ABI3-OE) have been described previously (lines AS29 and S8, respectively) (Rohde et al., 2002). Rooted explants of wild-type and transgenic lines were transferred from tissue culture to soil, grown for 3 months (60 cm) in the greenhouse under a 16-h photoperiod (LDs), and transferred to a growth chamber with an 8-h photoperiod (SDs), 100 μmol·m−2·s−2 from incandescent light bulbs, 55% RH, and 21°C. All lines were harvested, in parallel, at weekly intervals starting from a LD control (week 0) and during 6 weeks of SD treatment, at 4 h (LDs) or 2 h (SDs) after dawn.
At each sampling point, two biological replicates of seven pooled apices or apical buds were harvested, immediately frozen in liquid nitrogen, and kept at −80°C. After grinding, samples were split for parallel transcript and metabolite profiling.
RNA Extraction and Expression Profiling
Total RNA was extracted according to a modified procedure of Chang et al. (1993). After LiCl precipitation, total RNA was purified with the RNeasy mini kit, including the on-column DNase treatment (Qiagen). Five micrograms of total RNA was amplified with the Amino Allyl MessageAmp antisense RNA (aRNA) procedure (Ambion). Concentrations and quality of aRNA were assessed with an mRNA Nano assay on an Agilent 2100 bioanalyzer. Five micrograms of aRNA was directly labeled by Cy3-AP3-dCTP or Cy5-AP3-dCTP (GE-Healthcare) incorporation during first-strand cDNA synthesis with random nonamer primers (Invitrogen) and Superscript II RT (Invitrogen). The glass-spotted poplar cDNA microarray (POP2) contains 24,735 cDNA probes that correspond to 16,494 genes, representing 34.6% of 45,550 predicted genes in the poplar genome. Array details, sequences, GenBank accession numbers, and closest Arabidopsis thaliana genes are available from the Umeå Plant Science Center public database (www.populus.db.umu.se). An automated slide processor (Lucidea ASP hybridization station; GE-Healthcare) was used for hybridizations. Microarray images were obtained using a ScanArrayLite 4000 microarray analysis system scanner (Packard Bell) and analyzed with Genepix Pro 5.1 (Axon Instruments). Adaptive circles were used to indicate spots (spot diameter resize feature, 80 to 125%; composite pixel intensity, 250). After manual elimination of bad spots (highly irregular spot shape, dust, or salt particles), the median background pixel intensity was subtracted from the mean spot pixel intensity applying the local method (default option).
Reconstruction of Expression Profiles and PCA
The data were log2-transformed and Lowess-normalized applying linear local regression (f value = 0.5; SAS). Data were analyzed with two interconnected analysis of variance models (Wolfinger et al., 2001). The normalization model applied was as follows: ycijklm = μ + Ai + (AD)ij + rcijklm, where ycijklm is the Lowess-normalized intensity measurement from cDNA probe c (c = 1, . . ., 24,556), microarray i (i = 1, . . ., 48), dye j (j = 1, 2), genotype k (k = 1, 2, 3), time l (l = 1, . . ., 7), and biological replicate m (m = 1, 2); μ represents an overall mean value, A is the main effect for microarrays, AD is the microarray–dye interaction effect, and r denotes the residual. We included no main effect for dyes because the design was balanced in the dyes (Figure 1). The gene model applied was as follows: rcijklm = Pc + (PA)ci + (PD)cj + (PG)ck + (PT)cl + (PGT)ckl + (PB)cm + εcijklm, where P is the mean value per cDNA probe, PA represents random spot effects, PD the probe-specific dye effects, PG the probe-specific genotype effects, PT the probe-specific time effects, PGT the probe-specific time–genotype interaction effects, PB the probe-specific replicate effects, and ε the random error. The biological replicate was considered as a block factor. Ai and (AD)ij in the normalization model and (PA)ci in the gene model were random effects, and the others were fixed. Variance components were estimated with the restricted maximum likelihood method. For the gene model, type3 F tests and P values were calculated for all fixed terms. Differences of the least-square means for the time, genotype, and time–genotype interaction effects along with associated t tests, P values, and FDR-adjusted P values were calculated (Benjamini and Hochberg, 1995). Expression values were given as the least-square means of all (PGT)ckl combinations (see Supplemental Table 1 online). For genes with multiple independent cDNA probes, the vast majority of probes showed highly correlated expression profiles, indicating high reproducibility of gene expression profiles. cDNA probes with conflicting expression profiles were excluded from further analysis to maintain a high-quality data set. A single representative mean expression profile was calculated per gene by averaging highly correlated expression profiles and was mean-centered for clustering purposes (see Supplemental Table 2 online, which includes a list of probe sets used per gene). All raw (Genepix output) and normalized data (see Supplemental Tables 1 and 2 online) are available at www.upscbase.db.umu.se under experiment UMA-0024.
Metabolite Profiling
Metabolites were extracted with hot methanol:chloroform:water (3:2:4, v/v/v) and derivatized as described previously (Desbrosses et al., 2005). Ribitol and isoascorbic acid were added as internal standards, and retention time indices were determined with a C12, C15, C19, C22, C28, C32, and C36 n-alkane mixture. Metabolite profiling was performed (Desbrosses et al., 2005) using a Hewlett-Packard HP6890 series (Agilent Technologies) gas chromatography instrument equipped with a FactorFour VF-5ms column (30 mL × 25 mm i.d., df = 0.25 mm, 10 m EZ-Guard; Varian) coupled to a Hewlett-Packard 5973 mass selective detector (Agilent Technologies) and an Agilent Technologies 7683B series autosampler. m/z peaks were integrated with xcms version 1.6.1 (Smith et al., 2006), available in R version 2.4.0 (max = 300; xcmsSet [fwhm = 7.5, snthresh = 2, mzdiff = 0.5]; group [bw = 10]; retcor(); group [bw = 7.5]). For 8852 m/z peaks, abundances were obtained (selected ion current per milligram of dry weight; see Supplemental Table 4 online).
With the quadrupole mass spectral and retention time index library, m/z peaks (FDR < 0.01) were annotated based on available mass spectra and assigned to compounds (AMDIS version 2.64) (Stein, 1999). Significant differences between sampling points or genotypes were analyzed by two-way analysis of variance, and the FDR was calculated in R (q value [λ = 0]). For metabolic compounds with multiple m/z peaks (FDR < 0.01), a single profile was calculated for clustering purposes (Figure 3B). Per m/z peak, data were first averaged over the two replicate pools at each condition and log2-transformed. Subsequently, per compound, expression profiles of corresponding m/z peaks were averaged over highly correlated profiles, in a procedure similar to that applied to the cDNA probes, and mean-centered (see Supplemental Table 5 online). For selected metabolites (Figure 6), regression was performed per genotype with representative m/z peaks (R; gls procedure). Starting from a piecewise linear model with knots at all sampling points, the mean model was subsequently reduced with the likelihood ratio test (α = 0.05).
Electron Microscopy
Apical buds were fixed with 5% (v/v) glutaraldehyde in 100 mM sodium cacodylate, pH 7.0. After fixation, embryonic leaves and small sections from the central part of the bud (including apical/subapical meristem, recently formed leaf primordia, and parts of parenchyma) were carefully cut and postfixed with 2% (w/v) osmium tetraoxide. Fixed tissue samples were stained with 1% (w/v) uranyl acetate in 20% ethanol, dehydrated in graded series of ethanol, and embedded in Spurr's epoxy resin. Ultrathin sections, cut with a diamond knife on an ultramicrotome (LKB), were transferred onto Formvar-coated copper grids and stained with lead citrate. Sections were examined with a Zeiss EM 10c transmission electron microscope operated at 80 kV.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Annotation, Fold Change in Expression, FDR Values, and Temporal Expression Profiles for 24,549 cDNA Probes.
Supplemental Table 2. Annotation, Temporal Expression Profiles, Classifications, and Overlap with External Expression Data for 16,494 Poplar Genes.
Supplemental Table 3. Differential Expression within TF Families.
Supplemental Table 4. Annotation, Fold Change in Abundance, FDR Values, and Temporal Profiles for 8852 Metabolite Peaks.
Supplemental Table 5. Annotation, Temporal Profiles, and Classifications for 176 Metabolites.
Supplementary Material
Acknowledgments
We thank Bo Segerman (Umeå Plant Science Center) for support with microarray annotation issues, Tineke Casneuf for help with data analysis, Siegbert Melzer and Tom Beeckman for critical reading of the manuscript, and Martine De Cock for help in preparing it. This research was supported in part by the European Commission programs FORE (Grant HPRI-CT-2002-00198) and POPYOMICS (Grant QLK5-CT-2002-00953) and by grants from Vetenskapsrådet (to R.P.B). T.R. is indebted to the Institute for the Promotion and Innovation by Science and Technology in Flanders for a postdoctoral fellowship. A.R. is a Postdoctoral Researcher of the Research Foundation-Flanders.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Antje Rohde (antje.rohde@psb.ugent.be).
Online version contains Web-only data.
References
- Arend, M., and Fromm, J. (2003). Ultrastructural changes in cambial cell derivatives during xylem differentiation in poplar. Plant Biol. 5 255–264. [Google Scholar]
- Barrero, J.M., Rodríguez, P.L., Quesada, V., Piqueras, P., Ponce, M.R., and Micol, J.L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29 2000–2008. [DOI] [PubMed] [Google Scholar]
- Baumbusch, L.O., Hughes, D.W., Galau, G.A., and Jakobsen, K.S. (2004). LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J. Exp. Bot. 55 77–87. [DOI] [PubMed] [Google Scholar]
- Beemster, G.T.S., De Veylder, L., Vercruysse, S., West, G., Rombaut, D., Van Hummelen, P., Galichet, A., Gruissem, W., Inzé, D., and Vuylsteke, M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict, C., Skinner, J.S., Meng, R., Chang, Y., Bhalerao, R., Huner, N.P.A., Finn, C.E., Chen, T.H.H., and Hurry, V. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29 1259–1272. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57 289–300. [Google Scholar]
- Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A.M., Jansson, S., Strauss, S.H., and Nilsson, O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312 1040–1043. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 14 (suppl.): S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Ross, B., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Cadman, C.S.C., Toorop, P.E., Hilhorst, H.W.M., and Finch-Savage, W.E. (2006). Gene expression profile of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 46 805–822. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Cook, D., Fowler, S., Fiehn, O., and Thomashow, M.F. (2004). A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 101 15243–15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses, G.G., Kopka, J., and Udvardi, M.K. (2005). Lotus japonicus metabolic profiling. Development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol. 137 1302–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart, N., Johansson, A., Baba, K., Schrader, J., Sjödin, A., Bhalerao, R.R., Resman, L., Trygg, J., Moritz, T., and Bhalerao, R.P. (2007). Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage specific modulation of transcriptional and metabolic networks. Plant J. 50 557–573. [DOI] [PubMed] [Google Scholar]
- Eagles, C.F., and Wareing, P.F. (1964). The role of growth substances in the regulation of bud dormancy. Physiol. Plant. 17 697–709. [Google Scholar]
- Eriksson, M.E., and Millar, A.J. (2003). The circadian clock. A plant's best friend in a spinning world. Plant Physiol. 132 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V. (2002). Cross-talk in abscisic acid signaling. Sci. STKE 140 re10.1–re10.12, www.stke.org/cgi/content/full/sigtrans;2002/140/re10. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Rock, C.D. (September 30, 2002). Abscisic acid biosynthesis and response. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), 10.1199/tab.0058, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Fleet, C.M., and Sun, T.-p. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., Tran, L.-S.P., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39 863–876. [DOI] [PubMed] [Google Scholar]
- Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M.M., Seki, M., Hiratsu, K., Ohme-Takagi, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7 40–49. [DOI] [PubMed] [Google Scholar]
- Hake, S., Smith, H.M.S., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20 125–151. [DOI] [PubMed] [Google Scholar]
- Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479. [DOI] [PubMed] [Google Scholar]
- Horvath, D.P., Anderson, J.V., Chao, W.S., and Foley, M.E. (2003). Knowing when to grow: Signals regulating bud dormancy. Trends Plant Sci. 8 534–540. [DOI] [PubMed] [Google Scholar]
- Hoth, S., Morgante, M., Sanchez, J.-P., Hanafey, M.K., Tingey, S.V., and Chua, N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115 4891–4900. [DOI] [PubMed] [Google Scholar]
- Katz, A., Oliva, M., Mosquna, A., Hakim, O., and Ohad, N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37 707–719. [DOI] [PubMed] [Google Scholar]
- Kepinski, S. (2006). Integrating hormone signaling and patterning mechanisms in plant development. Curr. Opin. Plant Biol. 9 28–34. [DOI] [PubMed] [Google Scholar]
- Kim, S., Kang, J.-y., Cho, D.-I., Park, J.H., and Kim, S.Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40 75–87. [DOI] [PubMed] [Google Scholar]
- Kurup, S., Jones, H.D., and Holdsworth, M.J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21 143–155. [DOI] [PubMed] [Google Scholar]
- Lange, B.M., and Ghassemian, M. (2005). Comprehensive post-genomic data analysis approaches integrating biochemical pathway maps. Phytochemistry 66 413–451. [DOI] [PubMed] [Google Scholar]
- Lee, Y.-H., Oh, H.-S., Cheon, C.-I., Hwang, I.-T., Kim, Y.-J., and Chun, J.-Y. (2001). Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem. Biophys. Res. Commun. 284 133–141. [DOI] [PubMed] [Google Scholar]
- Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., Shimada, Y., Yoshida, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38 982–993. [DOI] [PubMed] [Google Scholar]
- Mazzella, M.A., Arana, M.V., Staneloni, R.J., Perelman, S., Rodriguez Batiller, M.J., Muschietti, J., Cerdán, P.D., Chen, K., Sánchez, R.A., Zhu, T., Chory, J., and Casal, J.J. (2005). Phytochrome control of the Arabidopsis transcriptome anticipates seedling exposure to light. Plant Cell 17 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mølmann, J.A., Asante, D.K.A., Jensen, J.B., Krane, M.N., Ernstsen, A., Junttila, O., and Olsen, J.E. (2005). Low night temperature and inhibition of gibberellin biosynthesis override phytochrome action and induce bud set and cold acclimation, but not dormancy in PHYA overexpressors and wild-type of hybrid aspen. Plant Cell Environ. 28 1579–1588. [Google Scholar]
- Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch, J.P. (1957). Growth responses of woody plants to photoperiodic stimuli. Proc. Am. Soc. Hortic. Sci. 70 512–525. [Google Scholar]
- Olsen, J.E., Junttila, O., and Moritz, T. (1995). A localised decrease of GA1 in shoot tips of Salix pentandra seedlings precedes cessation of shoot elongation under short photoperiod. Physiol. Plant. 95 627–632. [Google Scholar]
- Olsen, J.E., Junttila, O., Nilsen, J., Eriksson, M.E., Martinussen, I., Olsson, O., Sandberg, G., and Moritz, T. (1997). Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J. 12 1339–1350. [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem, F.A., El-Kereamy, A., Abrams, S.R., and Hill, R.D. (2006). The RNA-binding protein FCA is an abscisic acid receptor. Nature 439 290–294. [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón, D.M., Ruzicic, S., Dreyer, I., and Mueller-Roeber, B. (2007). PlnTFDB: An integrative plant transcription factor database. BMC Bioinformatics 8 42.1–42.10, http://www.biomedcentral.com/1471–2105/8/42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, A., and Bhalerao, R.P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12 217–223. [DOI] [PubMed] [Google Scholar]
- Rohde, A., and Boerjan, W. (2001). Insights into bud development and dormancy in poplar. In Trends in European Forest Tree Physiology Research, Cost Action A6: EUROSILVA, Tree Physiology, Vol. 2, S. Huttunen, H. Heikkilä, J. Bucher, B. Sundberg, P. Jarvis, and R. Matyssek, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 33–52.
- Rohde, A., De Rycke, R., Beeckman, T., Van Montagu, M., and Boerjan, W. (2000). ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, A., Prinsen, E., De Rycke, R., Engler, G., Van Montagu, M., and Boerjan, W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14 1885–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, A., Van Montagu, M., Inzé, D., and Boerjan, W. (1997). Factors regulating the expression of cell cycle genes in individual buds of Populus. Planta 201 43–52. [Google Scholar]
- Ruonala, R., Rinne, P.L.H., Baghour, M., Moritz, T., Tuominen, H., and Kangajärvi, J. (2006). Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J. 46 628–640. [DOI] [PubMed] [Google Scholar]
- Ruuska, S.A., Girke, T., Benning, C., and Ohlrogge, J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, G.E., and Kieber, J.J. (March 27, 2002). Ethylene. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society Plant Biologists), 10.1199/tab.0071, www.aspb.org/publications/arabidopsis/. [DOI]
- Schrader, J., Moyle, R., Bhalerao, R., Hertzberg, M., Lundeberg, J., Nilsson, P., and Bhalerao, R.P. (2004). Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 40 173–187. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 279–292. [DOI] [PubMed] [Google Scholar]
- Smith, C.A., Want, E.J., O'Maille, G., Abagyan, R., and Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78 779–787. [DOI] [PubMed] [Google Scholar]
- Stein, S.E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 10 770–781. [Google Scholar]
- Suzuki, M., Kao, C.-Y., Cocciolone, S., and McCarty, D.R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28 409–418. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Ketterling, M.G., Li, Q.-B., and McCarty, D.R. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylven, N. (1940). Lang-och kortkagstyper av de svenska skogstraden [Long day and short day types of Swedish forest trees]. Svensk Papperstidn. 43 317–324, 332–342, 350–354. [Google Scholar]
- Tamminen, I., Mäkelä, P., Heino, P., and Palva, E.T. (2001). Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana. Plant J. 25 1–8. [DOI] [PubMed] [Google Scholar]
- Tanino, K. (2004). Hormones and endodormancy induction in woody plants. J. Crop Improv. 10 157–199. [Google Scholar]
- Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., Selbig, J., Müller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Torres Acosta, J.A., de Almeida Engler, J., Raes, J., Magyar, Z., De Groodt, R., Inzé, D., and De Veylder, L. (2004). Molecular characterization of Arabidopsis PHO80-like-proteins, a novel class of CDKA;1-interacting cyclins. Cell. Mol. Life Sci. 61 1485–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouzé, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J.T., Zarka, D.G., Van Buskirk, H.A., Fowler, S.G., and Thomashow, M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41 195–211. [DOI] [PubMed] [Google Scholar]
- Wang, G., Kong, H., Sun, Y., Zhang, X., Zhang, W., Altman, N., dePamphilis, C.W., and Ma, H. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Deng, X.W. (April 4, 2002). Phytochrome signaling mechanism. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society Plant Biologists), 10.1199/tab.0074.1, www.aspb.org/publications/arabidopsis/. [DOI]
- Wareing, P.F. (1956). Photoperiodism in woody plants. Annu. Rev. Plant Physiol. 7 191–214. [Google Scholar]
- Weiser, C.J. (1970). Cold resistance and injury in woody plants. Science 169 1269–1278. [DOI] [PubMed] [Google Scholar]
- Welling, A., Moritz, T., Palva, E.T., and Junttila, O. (2002). Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol. 129 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- Wolfinger, R.D., Gibson, G., Wolfinger, E.D., Bennett, L., Hamadeh, H., Bushel, P., Afshari, C., and Paules, R.S. (2001). Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8 625–637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.