Abstract
Glutathione biosynthesis is a key component in the network of plant stress responses that counteract oxidative damage and maintain intracellular redox environment. Using a combination of mass spectrometry and site-directed mutagenesis, we examined the response of Arabidopsis thaliana glutamate-cysteine ligase (GCL) to changes in redox environment. Mass spectrometry identified two disulfide bonds (Cys186-Cys406 and Cys349-Cys364) in GCL. Mutation of either Cys-349 or Cys-364 to a Ser reduced reaction rate by twofold, but substitution of a Ser for either Cys-186 or Cys-406 decreased activity by 20-fold and abrogated the response to changes in redox environment. Redox titrations show that the regulatory disulfide bond has a midpoint potential comparable with other known redox-responsive plant proteins. Mutation of Cys-102, Cys-251, Cys-349, or Cys-364 did not alter the response to redox environment, indicating that modulation of activity depends on the Cys186-Cys406 disulfide bond. In vivo analysis of GCL in Arabidopsis root extracts revealed that multiple oxidative stresses altered the distribution of oxidized (active) and reduced (inactive) enzyme and that this change correlated with increased GCL activity. The thiol-based regulation of GCL provides a posttranslational mechanism for modulating enzyme activity in response to in vivo redox environment and suggests a role for oxidative signaling in the maintenance of glutathione homeostasis in plants.
INTRODUCTION
Molecular mechanisms that control the damage caused by reactive oxygen species are essential for normal cellular function in all organisms. Plants respond to environmental stresses, including temperature extremes, heavy metal–contaminated soils, drought, pathogens, and air pollutants by regulating pathways that counteract oxidative damage and maintain intracellular redox environment (Inzé and van Montagu, 1995; Noctor and Foyer, 1998; Foyer and Noctor, 2005; Meyer and Hell, 2005; Parisy et al., 2007). Multiple stress response pathways, including the detoxification of reactive oxygen species, xenobiotics, and heavy metals, require glutathione, which acts as an antioxidant and is involved in the ascorbate-glutathione cycle that eliminates peroxides (May and Leaver, 1993; Xiang et al., 2001; Meyer and Fricker, 2002; Mullineaux and Rausch, 2005).
The synthesis of glutathione occurs in two ATP-dependent steps (Meister, 1995). First, glutamate-cysteine ligase (GCL) catalyzes formation of γ-glutamylcysteine from Cys and Glu. This step is thought to be the rate-limiting reaction of the pathway. In the second step, glutathione synthetase (GS) adds Gly to γ-glutamylcysteine to yield glutathione. As synthesized, the reduced form of glutathione provides a substrate for multiple cellular reactions that yield oxidized glutathione (i.e., two glutathione molecules linked by a disulfide bond). The balance between the reduced and oxidized forms of glutathione is a central component in maintaining cellular redox state (Meister, 1995; Inzé and van Montagu, 1995; Noctor and Foyer, 1998; Foyer and Noctor, 2005).
Various stresses exert different effects on glutathione biosynthesis in plants, and these differences suggest regulation of the pathway at multiple layers, including (1) translational regulation, (2) substrate availability, (3) feedback inhibition by glutathione on GCL and GS, and (4) possible posttranscriptional modifications. Efforts to understand the regulation of glutathione biosynthesis in plants have primarily focused on the first three of these control mechanisms (Hell and Bergmann, 1990; May and Leaver, 1993; Foyer et al., 1997; May et al., 1998; Schafer et al., 1998; Xiang and Oliver, 1998; Xiang et al., 2001; Meyer and Fricker, 2002; Jez et al., 2004; Jez and Cahoon, 2004; Wachter et al., 2005; Herrera et al., 2007; Parisy et al., 2007). Interestingly, in Arabidopsis thaliana (thale cress) suspension cells, upregulation of GCL gene expression in response to various oxidative stresses was not observed, even though both GCL activity and cellular glutathione levels increased (May et al., 1998). These results implied the operation of a posttranscriptional mechanism for regulating GCL activity in plants.
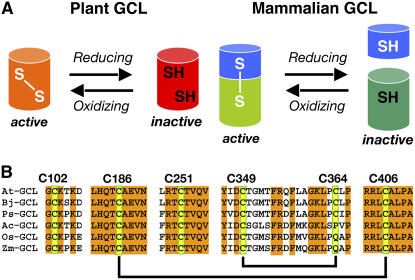
The plant GCLs differ in both amino acid sequence and overall structure from those in nonplant eukaryotes, such as mammals and Drosophila (Figure 1A). The mammalian GCLs consist of catalytic and regulatory proteins. Association of the two proteins increases affinity for Glu and decreases the inhibitory effect of glutathione (Seelig et al., 1984; Huang et al., 1993). Multiple intermolecular disulfide bonds mediate the reversible association of the catalytic and regulatory subunits to modulate GCL activity in response to changes in redox environment (Seelig et al., 1984; Tu and Anders, 1998; Fraser et al., 2002, 2003). By contrast, regulation of the plant GCLs, which share <25% amino acid sequence identity with the mammalian, yeast, and bacterial versions of the enzyme (May and Leaver, 1994), occurs through at least one intramolecular disulfide bond (Jez et al., 2004; Hothorn et al., 2006). Biochemical examination of Arabidopsis GCL (At GCL) demonstrated that reversible disulfide bond formation changes catalytic activity as a response to in vitro redox environment with the oxidized protein more active than reduced protein (Jez et al., 2004). Subsequent crystallographic studies of the GCL from Brassica juncea (Indian mustard) revealed the presence of two intramolecular disulfide bonds in the enzyme (Figure 1B) (Hothorn et al., 2006), but it is unclear which bond acts as a redox-responsive switch of enzyme activity or if the same bonds are found in the GCL from other plant species.
Figure 1.
Thiol-Based Changes in GCL.
(A) Comparison of plant and mammalian GCL responses to redox environment. The plant GCLs are regulated by intramolecular disulfide bonds. The mammalian GCL are regulated by intermolecular disulfided bonds formed between catalytic (green) and regulatory (blue) subunits.
(B) Cys residues in the plant GCL. Shown amino acid sequences correspond to regions around the Cys residues in the GCL from Arabidopsis (At GCL; NP194041), B. juncea (Bj GCL; EMB:CAD91713.1), Pisum sativum (Ps GCL; GB:AAF22127.1), Allium cepa (Ac GCL; GB:AAL61610.1), Oryza sativa (Os GCL; EMB:CAD48599.3), and Zea mays (Zm GCL; EMB:CAC83005.1). Yellow boxes highlight Cys residues in At GCL. Amino acid numbering corresponds to At GCL. Disulfide bonds found in At GCL and Bj GCL are indicated by black lines.
Given the central role of glutathione in maintaining intracellular redox balance, redox regulation of GCL activity offers a molecular control mechanism for glutathione biosynthesis. Here, we explore the role of Cys residues in At GCL as potential thiol-based regulatory switches and examine the response of GCL to redox environment in vitro and in vivo. The thiol-based regulation of GCL provides a posttranslational mechanism for modulating enzyme activity in response to the cellular redox environment and suggests a role for oxidative signaling in the maintenance of glutathione homeostasis in plants.
RESULTS
Analysis of Disulfide Bonds in At GCL by Mass Spectrometry
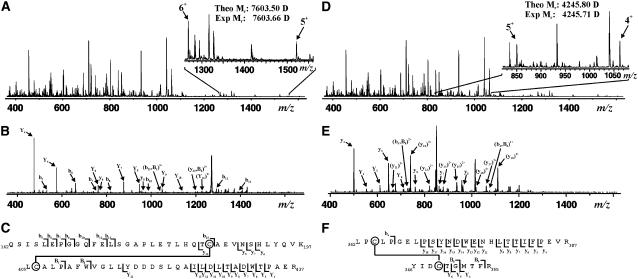
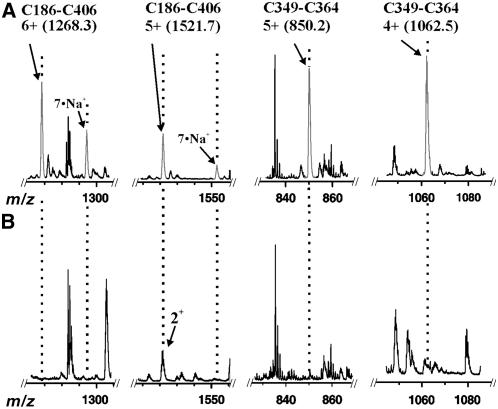
To map potential disulfide bonds in GCL, the enzyme was cleaved with trypsin under conditions that minimized disulfide bond scrambling and was analyzed by electrospray ionization–quadrupole time of flight (ESI-Q-TOF) mass spectrometry. Analysis of an unfractionated trypsin digestion of GCL yielded a species of 7603.66 D, correlating with the mass of the disulfide-bound tryptic dipeptide (Cys186-Cys406) containing Gln162-Lys197 and Leu405-Arg437 (Figure 2A). Direct fragmentation of this species using collisionally activated dissociation produced a series of fragment ions consistent with this dipeptide sequence (Figure 2B). As shown in Figure 2C, multiple fragment ions were assigned to the dipeptide containing the Cys186-Cys406 disulfide bond. Additionally, a species at 4245.71 D was detected that corresponds with the mass of a second disulfide-bound tryptic dipeptide (Cys349-Cys364) containing Tyr346-Arg355 and Leu362-Arg387 (Figure 2D). Fragmentation of this ion also confirmed the identity of this dipeptide (Figures 2E and 2F). To verify that each dipeptide contains a disulfide bond, a sample of At GCL was reduced in the presence of DTT before tryptic digestion and then was analyzed by ESI-Q-TOF mass spectrometry. As shown in Figure 3, the ion signals corresponding to the Cys186-Cys406 and Cys349-Cys364 dipeptides were not present, which is consistent with the disappearance of a disulfide bond in each dipeptide. After addition of DTT, all four of the reduced peptides (Gln162-Lys197, Leu405-Arg437, Tyr346-Arg355, and Leu362-Arg387) were detected with multiple charge states for each peptide (data not shown).
Figure 2.
Mapping of Disulfide Bonds in At GCL.
(A) ESI-Q-TOF mass spectrum (mass-to-charge ratio [m/z] of 400 to 1600) of an unfractionated tryptic digestion; the inset (m/z of 1260 to 1560) highlights the 6+ and 5+ charge states corresponding to the disulfide-containing peptide of interest (theoretical Mr: 7603.50 D).
(B) Tandem mass spectrometry (MS/MS) fragmentation of the 6+ charge state (m/z of 1268).
(C) Graphical fragment map correlating the fragmentation ions to the sequence of the disulfide-containing peptide. The disulfide-linked Cys residues are circled.
(D) ESI-Q-TOF mass spectrum (m/z of 400 to 1600) of an unfractionated tryptic digestion; the inset (m/z of 820 to 1075) highlights the 5+ and 4+ charge states corresponding to the disulfide-containing peptide of interest (theoretical Mr: 4245.80 D).
(E) MS/MS fragmentation of the 5+ charge state (m/z of 850).
(F) Graphical fragment map correlating the fragmentation ions to the sequence of the disulfide-containing peptide. The disulfide-linked Cys residues are circled.
Figure 3.
Comparison of Unreduced versus Reduced At GCL.
(A) ESI-Q-TOF mass spectrum of an unfractionated tryptic digestion highlighting various regions of the m/z range displaying charge states of the disulfide-linked dipeptides.
(B) ESI-Q-TOF mass spectrum of a reduced, unfractionated tryptic digestion highlighting the disappearance of the dipeptide species (gray) visible in (A).
Mutagenesis of Cys Residues in GCL
At GCL (lacking the chloroplast transit peptide sequence) contains six Cys residues (Cys-102, Cys-186, Cys-251, Cys-349, Cys-364, and Cys-406), of which four form two disulfide bonds (Cys186-Cys406 and Cys349-Cys364). The residues corresponding to Cys-102 and Cys-251 in the B. juncea GCL structure are located on a surface-exposed loop and in the active site (Hothorn et al., 2006). All of the Cys residues, except Cys-364, are conserved in the GCL from various plant species (Figure 1B). Moreover, there is no positional correlation between the Cys residues in the plant GCL and in the mammalian catalytic subunit (May and Leaver, 1994; Jez et al., 2004).
To probe the role of these residues in regulating At GCL, each Cys residue was mutated to a Ser residue. Mutant GCL were expressed, purified, and assayed using published methods (Jez et al., 2004). The C102S and C251S mutants were kinetically similar to wild-type enzyme (Table 1). The C349S and C364S mutants displayed up to twofold reductions in turnover rate. Likewise, under nonreducing conditions, the specific activities of these mutants paralleled their kcat values. Wild-type, C102S, and C251S GCL had specific activities of 150 to 200 nmol min−1 mg protein−1. The specific activities of the C349S and C364S mutants were 70 to 80 nmol min−1 mg protein−1. By contrast, the C186S and C406S mutants had specific activities (5 to 10 nmol min−1 mg protein−1) similar to those of reduced wild-type enzyme (Jez et al., 2004). The low specific activity of these two mutants precluded the accurate determination of steady state kinetic parameters. Following treatment with either DTT or β-mercaptoethanol (βME), the wild type and all of the mutants displayed similar specific activities (5 to 10 nmol min−1 mg protein−1).
Table 1.
Steady State Kinetic Parameters
| Enzyme | Substrate | kcat (min−1) | Km (mM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| At GCL | Glu | 6.1 | 9.1 | 10.1 |
| Cys | 4.5 | 1.6 | 46.9 | |
| ATP | 6.8 | 2.7 | 42.0 | |
| C102S | Glu | 4.6 | 9.0 | 8.5 |
| Cys | 5.5 | 1.6 | 57.3 | |
| ATP | 5.8 | 2.4 | 40.3 | |
| C251S | Glu | 7.1 | 8.5 | 13.9 |
| Cys | 4.0 | 1.7 | 39.2 | |
| ATP | 6.3 | 2.8 | 37.5 | |
| C349S | Glu | 4.7 | 7.0 | 11.2 |
| Cys | 3.6 | 1.3 | 46.1 | |
| ATP | 5.5 | 2.9 | 31.6 | |
| C364S | Glu | 4.1 | 9.7 | 7.0 |
| Cys | 4.1 | 1.5 | 45.5 | |
| ATP | 4.4 | 2.9 | 25.3 |
All reactions were performed as described in Methods. Values shown are means (n = 3) with se <15%.
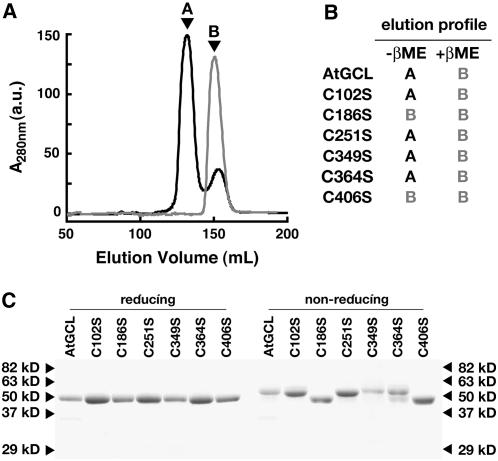
As previously reported (Jez et al., 2004), analysis of wild-type and mutant At GCL by size exclusion chromatography showed changes in the elution profile and enzymatic activity depending on the absence or presence of reducing agents (Figures 4A and 4B). The C102S, C251S, C349S, and C364S mutants displayed identical elution profiles as the wild-type enzyme under either reducing or nonreducing conditions (Figure 4B). The C186S and C406S mutants eluted at a volume under either reducing or nonreducing conditions that corresponds to reduced GCL (Figures 4A and 4B).
Figure 4.
In Vitro Redox Sensitivity of At GCL.
(A) Size exclusion chromatography of wild-type GCL and the C186S mutant in the absence of reducing agents. Purified GCL (black) or C186S mutant (gray) protein (1 mg mL−1) was chromatographed in 50 mM HEPES, pH 7.5, 300 mM NaCl, and 5 mM MgCl2. Peak A corresponds to 60 kD and peak B to 30 kD using gel filtration molecular mass standards. a.u., arbitrary units.
(B) Summary of elution profiles of wild-type and mutant GCL in the absence or presence of 10 mM βME. The corresponding peak (either A or B) observed during size exclusion chromatography, as performed in (A), is indicated.
(C) Comparison of wild-type and mutant GCL under reducing (+βME) or nonreducing (−βME) SDS-PAGE conditions. Protein (3 μg) was stained with Coomassie blue. Arrowheads correspond to molecular mass markers.
Comparison of wild-type and mutant GCL by reducing and nonreducing SDS-PAGE also revealed differences between the C186S and C406S mutant enzymes and the other GCL mutants (Figure 4C). GCL migrates at different molecular weights, which correspond to the reduced and oxidized forms of the protein under reducing or nonreducing SDS-PAGE conditions, respectively. The C102S, C251S, C349S, and C364S mutants displayed the same electrophoretic properties as wild-type GCL under either condition. By contrast, the C186S and C406S mutants behave the same as reduced GCL when analyzed under either reducing or nonreducing conditions (Figure 4C). These experiments indicate that mutation of either Cys-186 or Cys-406 locks the enzyme in the less active or reduced form and suggest that the disulfide bond formed between these residues functions as a redox-sensitive switch for GCL.
Redox Titrations of Wild-Type and Mutant GCL
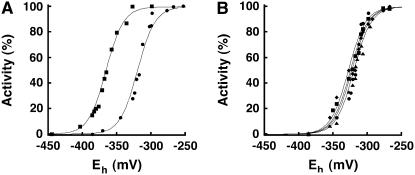
Although different labs report the effect of redox environment on the plant and mammalian GCL (Seelig et al., 1984; Hell and Bergmann, 1990; Fraser et al., 2002, 2003; Jez et al., 2004; Hothorn et al., 2006), no measurements of the redox midpoint potential (Em) of a GCL from any source have been described. To determine the equilibrium midpoint potential of the regulatory sulfhydryl groups of GCL, purified protein was incubated in solutions containing defined ratios of DTT (DTTred) and trans-4,5-dihydroxy-1,2-dithiane (DTTox) (10 mM total concentration) and then assayed for activity. Multiple titrations of GCL at pH 7.0 and pH 7.9 yielded average Em values of −318 ± 11 mV and −365 ± 3 mV, respectively, when fit to the Nernst equation for a single, two-electron (n = 2) redox reaction, as expected for the reversible cleavage and formation of a single regulatory disulfide bond (Figure 5A).
Figure 5.
Redox Titration of Wild-Type and Mutant At GCL.
(A) Purified wild-type GCL was incubated at either pH 7.0 (circles) or pH 7.9 (squares) for 90 min in the presence of 10 mM DTT (total) at defined thiol/disulfide ratios. GCL activity was measured as a function of ambient redox environment (Eh). Solid lines are fits of the data to the Nernst equation (n = 2). Data shown are the average of three replicates.
(B) Purified C102S (circles), C251S (squares), C349S (triangles), and C364S (diamonds) At GCL mutants were incubated at pH 7.0 for 90 min in the presence of 10 mM DTT (total) at defined thiol/disulfide ratios. GCL activity was measured as a function of ambient redox environment (Eh). Solid lines are fits of the data to the Nernst equation (n = 2). Data points shown are the average of three replicates.
To examine the effect of the Cys mutations on the Em value of GCL, additional titrations were performed (Figure 5B). Redox titration experiments with four of the Cys mutants yielded Em values similar to the wild-type enzyme as follows: C102S (Em = −322 ± 10 mV), C251S (Em = −318 ± 11 mV), C349S (Em = −325 ± 13 mV), and C364S (Em = −323 ± 9 mV). Mutation of either Cys residue in the Cys349-Cys364 disulfide bond did not significantly change the midpoint potential of GCL, indicating that this is not a redox regulatory disulfide bond. The low specific activities of the C186S and C406S mutant proteins precluded accurate redox titration experiments; however, incubation of each mutant with either 10 mM DTTred or 10 mM DTTox did not change the observed specific activities of these mutant proteins, which is also consistent with the Cys186-Cys406 linkage as the redox-responsive switch for GCL.
In Vivo Response of GCL to Oxidative Stresses
In vitro experiments demonstrate that the plant GCLs respond to changes in redox environment (Jez et al., 2004; Hothorn et al., 2006). As shown above, GCL migrates differently under reducing and nonreducing SDS-PAGE conditions, with the two observed bands corresponding to the reduction or oxidized forms of the protein (Figure 4C). This difference also provides a means to evaluate the in vivo state of GCL by protein gel blot analysis.
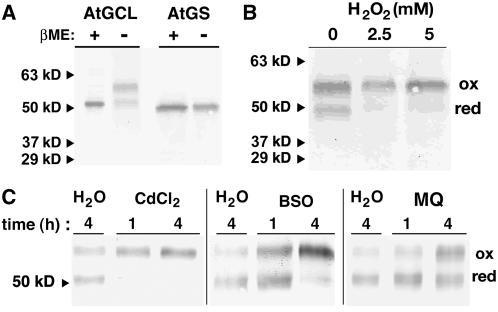
Antibodies were raised in rabbits against purified recombinant At GCL and At GS. Testing of the GCL and GS antisera using purified recombinant proteins showed no cross-reactivity between the antisera, and no bands were detected when either preimmune serum was used as the primary antibody (data not shown). Immunodetection of GCL and GS in root extracts from Arabidopsis seedling extracts confirm the effectiveness of each antiserum (Figure 6A). Under reducing conditions, GCL and GS each migrated as a single immunoreactive band of appropriate molecular weight; however, under nonreducing conditions, GCL was observed as two immunoreactive species, which represent the oxidized and reduced forms of the enzyme. Although similar results were obtained using extracts from leaf tissue (data not shown), cell-free extracts from root tissue were used for subsequent oxidative stress experiments to minimize possible changes in redox state resulting from photosynthesis (Mullineaux and Rausch, 2005).
Figure 6.
In Vivo Redox Sensitivity of At GCL.
(A) Detection of oxidized and reduced GCL. Total protein (10 μg) from cell-free protein extracts of roots from Arabidopsis seedlings was separated by SDS-PAGE under reducing (+βME) or nonreducing (−βME) conditions and electroblotted to nitrocellulose. At GCL and At GS antisera were used for protein gel blot analysis. Arrowheads correspond to molecular mass markers.
(B) In vivo oxidation of At GCL following H2O2 treatment. Arabidopsis seedlings were grown in liquid culture and treated with H2O2 (0 to 5 mM) for 1 h. Total protein (10 μg) from cell-free protein extracts of roots from seedlings was separated by SDS-PAGE under nonreducing (−βME) conditions and electroblotted to nitrocellulose. At GCL antisera were used for protein gel blot analysis. Positions of oxidized (ox) and reduced (red) GCL are indicated.
(C) Effect of oxidative stresses on the in vivo redox state of At GCL. Arabidopsis seedlings were grown in liquid culture and treated with either water (H2O), 50 μM CdCl2, 50 μM BSO, or 200 μM menadione (MQ) as indicated. Total protein (10 μg) from cell-free protein extracts of roots from seedlings was separated by SDS-PAGE under nonreducing (−βME) conditions and electroblotted to nitrocellulose. At GCL antisera were used for protein gel blot analysis. Positions of oxidized (ox) and reduced (red) GCL are indicated.
To examine whether GCL changes its disulfide-bonded state in vivo as a response to oxidation state, Arabidopsis seedlings were grown in liquid culture medium and then treated with either water (control) or hydrogen peroxide. Root extracts were prepared and examined by protein gel blot analysis (Figure 6B). Treatment of seedlings with hydrogen peroxide shifted the distribution of GCL to the oxidized form compared with control treatments. No changes in the form of GS were observed (data not shown). Assays of GCL and GS activity in cell-free root extracts were also performed. Following hydrogen peroxide treatment, the specific activity of GS in both the control and treated tissues (9.0 ± 1 nmol min−1 mg protein−1) was unchanged; however, a twofold increase in GCL activity (control, 3.1 ± 0.5 nmol min−1 mg protein−1; 5 mM H2O2, 6.0 ± 1.3 nmol min−1 mg total protein−1) was observed. The twofold change in GCL activity in the root tissue corresponds with the change from ∼50% oxidized GCL to nearly 100% oxidized enzyme, as observed by protein gel blot analysis.
The effect of other oxidative stresses on the distribution of oxidized and reduced GCL in plants was also tested. Arabidopsis seedlings were treated with water (control), heavy metal (CdCl2), buthionine sulfoximine (BSO), or menadione, and then the protein extracts were analyzed by protein gel blots (Figure 6C). Heavy metals, like cadmium, induce changes in cellular redox state (Schutzendubel and Polle, 2002). BSO specifically and irreversibly inactivates GCL and induces oxidative stress by decreasing glutathione levels (Griffith and Meister, 1979; Jez et al., 2004). Menadione, a polycyclic aromatic quinone, causes oxidative stress by producing reactive oxygen species through redox cycling and by alkylating glutathione (Jamieson, 1998; Zadzinski et al., 1998; Gutierrez, 2000). As observed with hydrogen peroxide treatment, exposure of Arabidopsis to cadmium, BSO, or menadione shifts the distribution of GCL in favor of the oxidized form (Figure 6C).
DISCUSSION
Although glutathione biosynthesis is a central component in the network of plant stress responses, the molecular basis for the biochemical regulation of GCL is an open question. Using a combination of mass spectrometry, site-directed mutagenesis, and biochemical assays, we examined the response of At GCL to redox environment. Our results indicate that modulation of GCL activity primarily depends on disulfide bond formation between Cys-186 and Cys-406. Importantly, in vivo experiments support a mechanism for the physiologic redox regulation of glutathione biosynthesis. Early studies showed that increased glutathione levels are triggered by oxidative stress in plants but that steady state mRNA levels for GCL were unchanged after treatment with either hydrogen peroxide, aminotriazole (a catalase inhibitor), or menadione (a superoxide-generating quinone), even though GCL activity was enhanced (May and Leaver, 1993; May et al., 1998; Xiang and Oliver, 1998; Meyer and Fricker, 2002). Thiol-based regulation of GCL provides a posttranscriptional mechanism that explains these earlier observations.
Multiple reports describe the sensitivity of GCL from plants and other eukaryotes to in vitro redox changes (Seelig et al., 1984; Hell and Bergmann, 1990; Fraser et al., 2002, 2003; Jez et al., 2004; Hothorn et al., 2006). Although the plant and mammalian GCL catalyze a common reaction, the enzymes from Arabidopsis and B. juncea are regulated by redox environment via intramolecular disulfide bonds and not intermolecular disulfide bonds, as occur in the mammalian enzymes (Figure 1A). Mass spectrometry (Figures 2 and 3) and protein crystallography (Hothorn et al., 2006) experiments clearly demonstrate that At GCL and Bj GCL contain two analogous disulfide bonds (Cys186-Cys406 and Cys349-Cys364) and that the redox state of these Cys residues regulates enzyme activity; however, the assessment of the contribution of each disulfide bond to redox regulation and the possible oligomerization state of the enzyme differ based on experimental methodology.
The Cys186-Cys406 disulfide bond plays a dominant regulatory role. Mutation of either Cys residue decreases activity, abrogates redox responsiveness, and changes how the oxidized and reduced forms migrate in either size exclusion chromatography or SDS-PAGE (Figure 4); these effects do not occur with disruption of the Cys349-Cys364 disulfide bond. Hothorn et al. (2006) reported that mutation of either residue in the corresponding Cys186-Cys406 bond led to protein aggregation, but modification of solution conditions can yield folded, soluble protein (Figures 4A and 4B). Moreover, evolutionary conservation of the Cys186-Cys406 disulfide bond in the GCL from multiple plant species strongly implies a universal role for this linkage (Figure 1B). By contrast, residues corresponding to Cys-349 and Cys-364 are not conserved. The three-dimensional structure of the Brassica enzyme showed that the corresponding Cys186-Cys406 disulfide bond links two α-helices together, which suggests that reduction/oxidation of the bond may result in reversible structural changes that modulate GCL activity (Hothorn et al., 2006).
The crystal structure of Bj GCL reveals that the corresponding Cys349-Cys364 disulfide bond is in a β-hairpin structure located 20 Å away from the active site (Hothorn et al., 2006), suggesting that reduction of this bond may reorient the β-hairpin to disrupt substrate binding and/or entry to the active site; however, analysis of At GCL suggests that the proposed role of the Cys349-Cys364 linkage needs reevaluation. Mutation of Cys-349 or Cys-364 reduces turnover rate (kcat) to 50 to 60% of wild-type At GCL (Table 1) and not to the 10 to 20% activity reported for the corresponding Bj GCL mutants (Hothorn et al., 2006). This difference likely reflects assay conditions, inherent variation between proteins, and/or data fitting protocols. Variability in specific activities and kinetic constants is commonly observed between isoforms from a given species or homologs from assorted species. Moreover, the C349S and C364S At GCL mutants display redox titration curves nearly identical to wild-type enzyme (Figure 5B), indicating that the Cys349-Cys364 linkage is not responsible for modulating activity over the range of physiologically relevant redox potentials examined.
A functional role for the Cys349-Cys364 disulfide bond was also suggested because of the fast reduction of this linkage compared with the slow reduction of the Cys186-Cys406 disulfide bond by tris(2-carboxyethyl)phosphine (TCEP) (Hothorn et al., 2006); however, the chemical nature of TCEP does not allow for direct comparison of disulfide bond reduction rates in the same protein (Burns et al., 1991; Cline et al., 2004). Reduction of sulfhydryl groups by TCEP requires space around the target disulfide. Consequently, the larger size of TCEP compared with DTT makes the phosphine selective for solvent-accessible protein disulfides (Cline et al., 2004), such as the Cys349-Cys364 linkage. Thus, the fast reduction of the solvent-exposed Cys349-Cys364 disulfide versus the slow reduction of the less accessible Cys186-Cys406 linkage (Hothorn et al., 2006) is likely an artifact of using TCEP as a reductant. The Cys349-Cys364 disulfide bond is needed for maximal activity, but the work described here and previously demonstrates that removal of this linkage yields an enzyme that remains redox regulated under physiological conditions through the Cys186-Cys406 disulfide bond.
Two structural models for the redox regulation of GCL have been proposed, but the oligomerization state of the enzyme remains equivocal (Jez et al., 2004; Hothorn et al., 2006). In the first model, the protein functions as a monomer that undergoes a conformational change upon reduction of the Cys186-Cys406 disulfide bond. Alternatively, the plant GCL homodimers disassociate into monomers under reducing conditions. Because each model is supported by experimental evidence, further examination by biophysical and structural approaches is required to establish the nature of the transition between the oxidized and reduced forms of the enzyme. Nonetheless, the plant GCLs are modulated by physiologically relevant changes in redox state.
The midpoint redox potential of the regulatory disulfide in At GCL (Em = −318 mV) falls within the range of values (Em = −290 to −330 mV) reported for other redox-active proteins, including thioredoxins, ferredoxin-thioredoxin reductase, and thioredoxin-regulated enzymes (Hutchinson and Ort, 1995; Hirasawa et al., 1999; Hutchinson et al., 2000). The difference in the Em values of At GCL over the measured pH range is within error for the expected −60 mV per pH unit for the two-electron reduction of a single disulfide to two thiols (Hutchinson and Ort, 1995; Hirasawa et al., 1999; Hutchinson et al., 2000). These results indicate that GCL is as sensitive to changes in intracellular environment as other plant proteins with established roles in redox-balancing systems.
Multiple oxidative stresses (hydrogen peroxide treatment, exposure to cadmium, inhibition of glutathione biosynthesis by BSO, and quinone-generated reactive oxygen species) shift the distribution of GCL toward the active (oxidized) form of the enzyme (Figure 6), which corresponds with increased GCL activity in root extracts. These results demonstrate that the in vivo pool of GCL is sensitive to changes in cellular redox state, which is also consistent with the behavior of the enzyme in redox titrations in vitro. Experiments on Arabidopsis transformed with an engineered redox-sensing green fluorescent protein with expression targeted to either the cytoplasm or mitochondria allowed real-time monitoring of in vivo redox potential (Jiang et al., 2006). In that study (Jiang et al., 2006), the ambient redox potentials of the root cytoplasm and mitochondria were estimated as −318 mV and −362 mV, respectively. After hydrogen peroxide stress, the cytoplasmic and mitochondrial potentials were −252 and −327 mV, respectively. Although measurements in the plastid were not reported, similar changes in redox potential following oxidative stress would also activate GCL. For example, if a stress condition changed the redox potential of an organelle, such as the plastid from −360 to −320 mV, then the ratio of oxidized to reduced GCL would shift from ∼50 to 100% active enzyme at pH 7.9 or from <5 to 50% oxidized form at pH 7.0 (Figure 5).
Because of the redox sensitivity of GCL, the cellular environment in which the enzyme resides will be an important factor in modulating activity. Biochemical studies in maize and B. juncea suggest that GCL activity is primarily localized in the plastids (Rüegsegger and Brunold, 1993; Wachter et al., 2005). No information on either the subcellular or suborganelle localization of GCL in Arabidopsis is available. Likewise, the possible effects of oxidative stress on localization remain unexamined. It is also unclear if the transition between oxidized and reduced forms of GCL occurs directly or is mediated by other redox-sensing proteins like thioredoxin or glutaredoxin. Comparison of the in vitro redox titrations of GCL (Figure 5) to the changes in cellular redox potential reported by Jiang et al. (2006) suggest that modulation of GCL does not require a redox-coupling system. Moreover, GCL has not been identified as a target of thioredoxin in plants (Buchanan and Balmer, 2005), but this does not rule out possible interaction with the glutaredoxin system.
During oxidative stress, glutathione is used for the elimination of reactive oxygen species. If production of reactive oxygen species exceeds the capacity to maintain the balance of reduced to oxidized glutathione, then accumulation of oxidized glutathione and/or reactive oxygen species would activate GCL by directly oxidizing the redox-active disulfide bond and increasing the capacity for glutathione biosynthesis. Such a situation occurs during hydrogen peroxide treatment or production of reactive oxygen species following treatment with menadione. Exposure to heavy metals, such as cadmium, also results in depletion of glutathione by initiating production of phytochelatins, which are heavy metal chelating peptides (Grill et al., 1985). Likewise, inactivation of GCL by BSO will decrease the rate of glutathione synthesis and cause changes in redox state (Griffith and Meister, 1979). As the redox environment becomes more reducing, GCL activity would be downregulated. Therefore, direct modulation of the rate-limiting enzyme in glutathione production provides a posttranscriptional switching mechanism for responding to intracellular oxidative signals.
By sensing changes in the cellular redox environment, disulfide bonds provide an efficient mechanism for modulating protein function. Oxidative signal-induced disulfide bond formation in yeast and bacteria alters the activity of transcription factors (OxyR, HSF-1, and YAP-1) and the molecular chaperone Hsp33 (Zheng et al., 1998; Graumann et al., 2001; Ahn and Thiele, 2003; Wood et al., 2004). In plants, changes in redox environment activate the transcription factor NPR1, an essential regulator of plant systemic acquired resistance that confers immunity to a range of pathogens, by reducing the disulfide-linked oligomeric protein to a monomer, which undergoes translocation to the nucleus (Mou et al., 2003). Likewise, certain enzymes in plants, including glutathione reductase, glucose-6-phosphate dehydrogenase, adenylylsulfate reductase, and superoxide dismutase, are also redox responsive (Hérouart et al., 1993; Hausladen and Alscher, 1994; Wenderoth et al., 1997; Bick et al., 2001). Our work suggests that GCL belongs to this class of proteins that sense in vivo redox conditions and modulate function by oxidative signaling.
METHODS
Protein Expression and Purification
Wild-type and mutant At GCL was overexpressed in Escherichia coli and purified using affinity and size exclusion chromatographies as described previously (Jez et al., 2004).
Mass Spectrometry
Protein digestion and ESI-Q-TOF mass spectrometric analysis of At GCL was performed as follows. For At GCL reduction (where applicable), 1 M DTT (1 μL) was added to At GCL (100 μL reactions) in the digestion buffer (50 mM NH4HCO3, pH 7.8) and allowed to incubate for 30 min before the addition of trypsin. Proteolysis was performed by the addition of TPCK-treated trypsin (Promega) to purified At GCL (100 μg) at a protease-to-substrate ratio of 1:5 (w/w) in 50 mM NH4HCO3, pH 7.8, and incubated at 30°C for 10 min. Reactions were quenched by the addition of 20% (v/v) formic acid (15 μL) and subjected to C18 Zip-Tip (Millipore) purification for desalting before analysis by mass spectrometry. Sample analysis proceeded with an ABI QSTAR XL hybrid Q-TOF MS/MS mass spectrometer (Applied Biosystems/MDS Sciex) equipped with a nanoelectrospray source (Protana XYZ manipulator). Positive mode nanoelectrospray was generated from borosilicate nanoelectrospray needles at 1.5 kV. TOF mass spectra were obtained using the Analyst QS software with an m/z range of 100 to 2000. The m/z response of the instrument was calibrated daily with standards from the manufacturer. Species of interest were selected using the quadrupole and subjected to collisionally activated dissociation (MS/MS) for confirmation of peptide identification. The fragmentation data were manually interpreted and correlated to the peptide primary amino acid sequence.
Mutagenesis
The At GCL C102S, C186S, C251S, C349S, C364S, and C406S mutants were generated using the QuikChange PCR method (Stratagene) with the pHIS8-At GCLΔ85 bacterial expression vector as the template (Jez et al., 2000, 2004). All mutant expression constructs were sequenced to verify that only the desired mutation was generated (Washington University DNA Sequencing Facility).
Enzyme Assays
Enzymatic activity of purified wild-type or mutant At GCL was determined spectrophotometrically using a coupled assay with pyruvate kinase and lactate dehydrogenase (Jez et al., 2004). Steady state kinetic parameters were determined by initial velocity experiments with measurements performed and data fitted as described previously (Jez et al., 2004). To determine GCL and GS activity in Arabidopsis thaliana roots, extracts were prepared by homogenizing roots in 100 mM MOPSO, pH 7, 500 mM NaCl, 2 mM MgCl2, and 1 mM EDTA. Homogenates were centrifuged and the supernatant collected. Protein concentration was determined by Bradford assay. GS activity in root tissue extracts was measured using a colorimetric assay (Jez and Cahoon, 2004). GCL activity in root tissue extracts was determined as described by Hell and Bergmann (1990) and Rüegsegger and Brunold (1992). Reaction products were analyzed by HPLC analysis of samples derivatized with monobromobimane and separated with a Vydax C18 silica column on an Agilent 1100 HPLC coupled to a scanning fluorescence detector (λex = 380 nm; λem = 480 nm).
Redox Titrations
The dependence of At GCL activity on redox potential was examined using redox buffers containing defined ratios of DTT (DTTred) and trans-4,5-dihydroxy-1,2-dithiane (DTTox) (Sigma-Aldrich). Protein samples were equilibrated in solutions containing 100 mM MOPSO, pH 7, 10 mM MgCl2, 100 mM NaCl, and 10 mM total DTTred/DTTox for 90 min at ambient temperature. At the end of the equilibration period, aliquots were removed and assayed for activity. Values for Em were determined by fitting titration data to the Nernst equation, Eh = Em + (RT/nF)(ln([ox]/[red]), with RT/F = 25.693 mV and the value of n = 2, as expected for a two-electron transfer process, using Kaleidagraph (Synergy Software) (Hutchinson and Ort, 1995; Bick et al., 2001). Calculation of the Em value is based on the Em value of DTT at pH 7.0 (−327 mV) (Lees and Whitesides, 1993).
Plant Material
Seeds of Arabidopsis ecotype Columbia were germinated and grown in Murashige and Skoog liquid medium (Sigma-Aldrich) in six-well plates on a rotary shaker (90 rpm) at 26°C. After 14 d, seedlings were treated with water, hydrogen peroxide, CdCl2, BSO, or menadione for 0 to 4 h. Seedlings were collected. Roots were rinsed with deionized distilled water and immediately frozen in liquid nitrogen for storage at −80°C.
Protein Gel Blots
Polyclonal antibodies were raised in rabbits against purified recombinant At GCL and At GS (Washington University Antibody Facility). Roots from Arabidopsis seedlings grown and treated as above were ground in liquid nitrogen, mixed with extraction buffer, and centrifuged. Total protein extracts were fractionated by SDS-PAGE (±βME). Proteins were transferred to an Immobilon-P PVDF membrane (Millipore). The membrane was washed with Tris-buffered saline (80 mM Tris and 200 mM NaCl, pH 7.5) and then incubated in Tris-buffered saline containing 5% (w/v) nonfat milk. Following this, membranes were incubated in a 1:5000 dilution of either At GCL or At GS antisera. Reactive proteins were identified using a 1:5000 dilution of goat anti-rabbit IgG-alkaline phosphatase antibody (Sigma-Aldrich) with color detection using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (Sigma-Aldrich). Color development was quenched by addition of 100 mM EDTA.
Acknowledgments
This work was supported by funds from the Donald Danforth Plant Science Center and an American Chemical Society Petroleum Research Fund grant (PRF-43012-AC4) to J.M.J. R.S.R. was supported by a Pfizer-Solutia Students and Teachers as Research Scientists internship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Joseph M. Jez (jjez@danforthcenter.org).
References
- Ahn, S.G., and Thiele, D.J. (2003). Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick, J.A., Setterdahl, A.T., Knaff, D.B., Chen, Y., Pitcher, L.H., Ziliniskas, B.A., and Leustek, T. (2001). Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry 40 9040–9048. [DOI] [PubMed] [Google Scholar]
- Buchanan, B.B., and Balmer, Y. (2005). Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 56 187–220. [DOI] [PubMed] [Google Scholar]
- Burns, J.A., Butler, J.C., Moran, J., and Whitesides, G.M. (1991). Selective reduction of disulfides by tris(2-carboxyethyl)phosphine. J. Org. Chem. 56 2648–2650. [Google Scholar]
- Cline, D.J., Redding, S.E., Brohawn, S.G., Psathas, J.N., Schneider, J.P., and Thorpe, C. (2004). New water-soluble phosphines as reductants of peptide and protein disulfide bonds: Reactivity and membrane permeability. Biochemistry 43 15195–15203. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H., Lopez-Delgado, H., Dat, J.F., and Scott, I.M. (1997). Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signalling. Physiol. Plant 100 241–254. [Google Scholar]
- Foyer, C.H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, J.A., Kansagra, P., Kotecki, C., Saunders, R.D., and McLellan, L.I. (2003). The modifier subunit of Drosophila glutamate-cysteine ligase regulates catalytic activity by covalent and noncovalent interactions and influences glutathione homeostasis in vivo. J. Biol. Chem. 278 46369–46377. [DOI] [PubMed] [Google Scholar]
- Fraser, J.A., Saunders, R.D., and McLellan, L.I. (2002). Drosophila melanogaster glutamate-cysteine ligase activity is regulated by a modifier subunit with a mechanism of action similar to that of the mammalian form. J. Biol. Chem. 277 1158–1165. [DOI] [PubMed] [Google Scholar]
- Graumann, J., Lilie, H., Tang, X., Tucker, K.A., Hoffman, J.H., Vijayalakshmi, J., Saper, M., Bardwell, J.C., and Jakob, U. (2001). Activation of the redox-regulated molecular chaperone Hsp33: A two-step mechanism. Structure 9 377–387. [DOI] [PubMed] [Google Scholar]
- Griffith, O.W., and Meister, A. (1979). Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine). J. Biol. Chem. 254 7558–7560. [PubMed] [Google Scholar]
- Grill, E., Winnacker, E.L., and Zenk, M.H. (1985). Phytochelatins: The principal heavy-metal complexing peptides of higher plants. Science 230 674–676. [DOI] [PubMed] [Google Scholar]
- Gutierrez, P.L. (2000). The metabolism of quinone-containing alkylating agents: Free radical production and measurement. Front. Biosci. 5 D629–D638. [DOI] [PubMed] [Google Scholar]
- Hausladen, A., and Alscher, R.G. (1994). Cold-hardiness-specific glutathione reductase isozymes in red spruce: Thermal dependence of kinetic parameters and possible regulatory mechanisms. Plant Physiol. 105 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell, R., and Bergmann, L. (1990). γ-Glutamylcysteine synthetase in higher plants: Catalytic properties and subcellular localization. Planta 180 603–612. [DOI] [PubMed] [Google Scholar]
- Hérouart, D., van Montagu, M., and Inzé, D. (1993). Redox-activated expression of the cytosolic copper/zinc superoxide dismutase gene in Nicotiana. Proc. Natl. Acad. Sci. USA 90 3108–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, K., Cahoon, R.E., Kumaran, S., and Jez, J.M. (2007). Reaction mechanism of glutathione synthetase from Arabidopsis thaliana: Site-directed mutagenesis of active site residues. J. Biol. Chem. 282 17157–17165. [DOI] [PubMed] [Google Scholar]
- Hirasawa, M., Schürmann, P., Jacquot, J.P., Manieri, W., Jacquot, P., Keryer, E., Hartman, F.C., and Knaff, D.B. (1999). Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38 5200–5205. [DOI] [PubMed] [Google Scholar]
- Hothorn, M., Wachter, A., Gromes, R., Stuwe, T., Rausch, T., and Scheffzek, K. (2006). Structural basis for the redox control of plant glutamate cysteine ligase. J. Biol. Chem. 281 27557–27565. [DOI] [PubMed] [Google Scholar]
- Huang, C.S., Chang, L.S., Anderson, M.E., and Meister, A. (1993). Catalytic and regulatory properties of the heavy subunit of rat kidney γ-glutamylcysteine synthetase. J. Biol. Chem. 268 19675–19680. [PubMed] [Google Scholar]
- Hutchinson, R.S., Groom, Q., and Ort, D.R. (2000). Differential effects of chilling-induced photooxidation on the redox regulation of photosynthetic enzymes. Biochemistry 39 6679–6688. [DOI] [PubMed] [Google Scholar]
- Hutchinson, R.S., and Ort, D.R. (1995). Measurement of equilibrium midpoint potentials of thiol/disulfide regulatory groups on thioredoxin-activated chloroplast enzymes. Methods Enzymol. 252 220–228. [DOI] [PubMed] [Google Scholar]
- Inzé, D., and van Montagu, M. (1995). Oxidative stress in plants. Curr. Opin. Biotechnol. 6 153–158. [Google Scholar]
- Jamieson, D.J. (1998). Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14 1511–1527. [DOI] [PubMed] [Google Scholar]
- Jez, J.M., and Cahoon, R.E. (2004). Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana. J. Biol. Chem. 279 42726–42731. [DOI] [PubMed] [Google Scholar]
- Jez, J.M., Cahoon, R.E., and Chen, S. (2004). Arabidopsis thaliana glutamate-cysteine ligase: Functional properties, kinetic mechanism, and regulation of activity. J. Biol. Chem. 279 33463–33470. [DOI] [PubMed] [Google Scholar]
- Jez, J.M., Ferrer, J.L., Bowman, M.E., Dixon, R.A., and Noel, J.P. (2000). Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39 890–902. [DOI] [PubMed] [Google Scholar]
- Jiang, K., Schwarzer, C., Lally, E., Zhang, S., Ruzin, S., Machen, T., Remington, S.J., and Feldman, L. (2006). Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 141 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees, W.J., and Whitesides, G.M. (1993). Equilibrium-constants for thiol disulfide interchange reactions: A coherent, corrected set. J. Org. Chem. 58 642–647. [Google Scholar]
- May, M.J., and Leaver, C.J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, M.J., and Leaver, C.J. (1994). Arabidopsis thaliana γ-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc. Natl. Acad. Sci. USA 91 10059–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, M.J., Vernoux, T., Sanchez-Fernandez, R., van Montagu, M., and Inzé, D. (1998). Evidence for posttranscriptional activation of γ-glutamylcysteine synthetase during plant stress responses. Proc. Natl. Acad. Sci. USA 95 12049–12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, A. (1995). Glutathione metabolism. Methods Enzymol. 252 26–30. [DOI] [PubMed] [Google Scholar]
- Meyer, A.J., and Fricker, M.D. (2002). Control of demand-driven biosynthesis of glutathione in green Arabidopsis suspension culture cells. Plant Physiol. 130 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, A.J., and Hell, R. (2005). Glutathione homeostasis and redox-regulation by sulfhydryl groups. Photosynth. Res. 86 435–457. [DOI] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944. [DOI] [PubMed] [Google Scholar]
- Mullineaux, P.M., and Rausch, T. (2005). Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth. Res. 86 459–474. [DOI] [PubMed] [Google Scholar]
- Noctor, G., and Foyer, C.H. (1998). Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 249–279. [DOI] [PubMed] [Google Scholar]
- Parisy, V., Poinssot, B., Owsianowski, L., Buchala, A., Glazebrook, J., and Mauch, F. (2007). Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49 159–172. [DOI] [PubMed] [Google Scholar]
- Rüegsegger, A., and Brunold, C. (1992). Effect of cadmium on γ-glutamylcysteine synthesis in maize seedlings. Plant Physiol. 99 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger, A., and Brunold, C. (1993). Localization of γ-glutamylcysteine synthetase and glutathione synthetase activity in maize seedlings. Plant Physiol. 101 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, H.J., Haag-Kerwer, A., and Rausch, T. (1998). cDNA cloning and expression analysis of genes encoding GSH synthesis in roots of the heavy-metal accumulator Brassica juncea L.: Evidence for Cd-induction of a putative mitochondrial gamma-glutamylcysteine synthetase isoform. Plant Mol. Biol. 37 87–97. [DOI] [PubMed] [Google Scholar]
- Schutzendubel, A., and Polle, A. (2002). Plant responses to abiotic stresses: Heavy-metal induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 53 1351–1365. [PubMed] [Google Scholar]
- Seelig, G.F., Simondsen, R.P., and Meister, A. (1984). Reversible dissociation of γ-glutamylcysteine synthetase into two subunits. J. Biol. Chem. 259 9345–9347. [PubMed] [Google Scholar]
- Tu, Z., and Anders, M.W. (1998). Identification of an important cysteine residue in human glutamate-cysteine ligase catalytic subunit by site-directed mutagenesis. Biochem. J. 336 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter, A., Wolf, S., Steininger, H., Bogs, J., and Rausch, T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: Implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41 15–30. [DOI] [PubMed] [Google Scholar]
- Wenderoth, I., Scheibe, R., and von Schaewen, A. (1997). Identification of the cysteine residues involved in redox modification of plant plastidic glucose-6-phosphate dehydrogenase. J. Biol. Chem. 272 26985–26990. [DOI] [PubMed] [Google Scholar]
- Wood, M.J., Storz, G., and Tjandra, N. (2004). Structural basis for redox regulation of Yap1 transcription factor localization. Nature 430 917–921. [DOI] [PubMed] [Google Scholar]
- Xiang, C., and Oliver, D.J. (1998). Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10 1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Werner, B.L., Christensen, E.M., and Oliver, D.J. (2001). The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 126 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadzinski, R., Fortuniak, A., Bilinski, T., Grey, M., and Bartosz, G. (1998). Menadione toxicity in Saccharomyces cerevisiae cells: Activation by conjugation with glutathione. Biochem. Mol. Biol. Int. 44 747–759. [DOI] [PubMed] [Google Scholar]
- Zheng, M., Aslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279 1718–1721. [DOI] [PubMed] [Google Scholar]