Abstract
Brome mosaic bromovirus (BMV), a positive-stranded RNA virus, supports both homologous and nonhomologous RNA recombinations. Two BMV (temperature-sensitive) mutants with alterations in the 2a protein, the putative RNA polymerase component of the viral replicase, were tested for their ability to support both types of recombination. Here we report that one of these mutants with the Leu-486 substituted by Phe did not support nonhomologous recombination. Effect on homologous recombination was mainly on the location and precision of crossover events. The other 2a mutant with Asn-458 substituted by Asp did not negatively affect either type of recombination. Apparently, BMV RNA polymerase participates differently in the two types of recombination events.
Genetic RNA–RNA recombination has been described for animal (1, 2), plant (3–5), and bacterial (6) RNA viruses between virus strains, between different viruses, or between virus and host RNAs (2, 7). RNA recombination has an obvious role in RNA virus evolution (5), with nonhomologous crossovers leading to the generation of new viral species and homologous crossovers potentially eliminating errors of RNA replication (2, 5, 7). An unanswered question is: Why do certain RNA viruses show rapid recombination, and many others appear to recombine infrequently (1, 2)?
The RNA recombination processes are generally categorized as either homologous or nonhomologous. A general definition provided by Lai (2) describes homologous recombination as occurring between two related RNA molecules at corresponding sites, describes aberrant homologous recombination as occurring between related RNAs at noncorresponding sites, and describes nonhomologous recombination as occurring between unrelated RNA molecules.
Among experimental systems available to study viral RNA recombination (2, 5, 7), that of the brome mosaic bromovirus (BMV) is one of the most advanced (7). BMV is a tripartite, single-stranded RNA virus (8), whose RNA1 and RNA2 encode the 1a and 2a proteins, respectively, that form the viral replicase. RNA3 encodes the cell-to-cell movement (9) and the coat proteins (8). The 1a protein has N-terminal methyltransferase and C-terminal helicase domains (10), and the 2a has a central RNA-dependent RNA polymerase core domain (11) and an N-terminal region that is necessary for 1a/2a binding (12, 13).
All BMV RNA segments share a highly structured 200-nt region at their 3′ end (8). Inefficient recombination was observed when a viable BMV RNA3 3′ mutant was repaired by crossovers with wild-type (wt) RNA1 or RNA2 in vivo (3). The frequency of crossovers was greatly increased by duplication of the mutated 3′ sequence in RNA3 (14, 15) or by the use of special recombinationally active sequences (16, 17). Also, the use of Chenopodium quinoa plants has allowed us to characterize diverse populations of BMV recombinants (15, 16) accumulating independently in individual local lesions (18).
Both homologous and nonhomologous BMV RNA recombinants were identified (19). Nonhomologous crossovers were found to be directed to a double-stranded region formed between RNA1 and an engineered RNA3 (16, 20). The sites of crossovers were shifted after destabilizing this region, suggesting a template switching (copy choice) mechanism (16). In contrast, homologous crossovers depended on sequence homologies between the recombining BMV RNAs (17). A short RNA2 sequence efficiently directed homologous RNA2/RNA3 crossovers when introduced into RNA3 (17). A fraction of the homologous recombinants was imprecise, having sequence modifications located at the crossover sites (2, 17). This was probably the result of the involvement of the replicase in homologous recombination. All of these studies prompted us to define the two types of recombination events in BMV as follows: Homologous recombination is supported by sequence homology at crossover sites, and nonhomologous recombination can be supported by sequence complementarity at crossover sites (heteroduplex-driven recombination). We also have observed imprecise (aberrant) events for the two basic recombination types (16, 17, 21). We use these definitions, which are based on mechanistic observations, in this work.
In another well studied turnip crinkle carmovirus system, aberrant homologous recombination was observed between satellite RNAs (4, 22), while nonhomologous recombination occurred between satellite and genomic RNAs (23). Here, downstream crossovers clustered at the base of highly structured regions with sequences resembling the 5′ end and subgenomic promoter consensus sequences. This led to the suggestion that turnip crinkle carmovirus recombination occurs during synthesis of plus RNA strands (22).
Kirkegaard and Baltimore (24) have provided genetic evidence for the replicase-mediated (copy choice) mechanism of RNA recombination in poliovirus. We have confirmed that mutations within the helicase domain of 1a altered the distribution of crossover sites during heteroduplex-primed nonhomologous recombination in BMV (25). In this work, we analyze the effects that alterations in the 2a protein have on both homologous and nonhomologous processes. We demonstrate that a single amino acid substitution in 2a inhibits heteroduplex-mediated nonhomologous recombination and increases homologous recombination whereas another 2a alteration does not affect either type of recombination. This confirms the participation of 2a in BMV RNA recombination and verifies that replication and heteroduplex-mediated nonhomologous and homologous recombinations can be functionally separated.
MATERIALS AND METHODS
Materials.
Plasmids pB1TP3, pB2TP5, and pB3TP7 (26) were used to synthesize infectious transcripts in vitro of wt BMV RNA components 1, 2, and 3. Plasmids pB2DR7 and pB2DR13 (a generous gift of P. Ahlquist, University of Wisconsin, Madison; ref. 27) were used to synthesize BMV RNA2 transcripts bearing the DR7 and DR13 mutations, respectively. Plasmid pPN8(−) (16) was used to obtain transcripts of the PN8(−) derivative of BMV RNA3. Plasmids pPN-H39 and pPN-H66 were used to synthesize transcripts of the PN-H39 and PN-H66 derivatives of BMV RNA3 (17). T7 RNA polymerase, Moloney murine leukemia virus reverse transcriptase, and restriction enzymes were from GIBCO/BRL, and the Sequenase kit was from United States Biochemical.
In Vitro Transcription and Whole Plant Infections.
Full length, capped RNA transcripts were made from EcoRI-linearized plasmids according to previously published procedures (26). C. quinoa leaves were inoculated with a mixture of the transcribed BMV RNA components (as described in ref. 15). In brief, a mixture of 1 μg of each transcript in 15 μl of inoculation buffer (10 mM Tris, pH 8.0/1 mM EDTA/0.1% Celite/0.1% bentonite) was inoculated on one fully expanded leaf. In each experiment, from six to nine separate leaves were inoculated. Each inoculation experiment was repeated two or three times. The inoculated C. quinoa plants were maintained in a growth chamber at 24°C. Local lesions were counted 14 days postinoculation.
RNA Extraction, Amplification of Recombinant RNAs by Reverse Transcriptase PCR (RT-PCR), Cloning, and Sequencing.
For each RNA2 mutant/RNA3 combination, total RNA was isolated from a number of lesions as described (15), and the 3′ regions in RNA3 recombinants were amplified with an RT-PCR procedure (as described in ref. 15). The first strand synthesis primer (primer 1: 5′-CAGTGAATTCTGGTCTCTTTTAGAGATTTACAG-3′; EcoRI site underlined) was complementary to the 3′-terminal 23-nt of all of the BMV virion RNAs; see ref. 14), and the second strand primer (primer 2: 5′-CTGAAGCAGTGCCTGCTAAGGCGGTC-3′) corresponded to nucleotides 392–367 from the 3′ end of wt BMV RNA3. By comparing the size of the obtained RT-PCR products with those synthesized using the RNA inoculation mixture, it was determined whether the accumulating RNA3 represents a recombinant or parental molecule. The resulting cDNA products were digested with EcoRI–XbaI restriction enzymes and ligated into the pGEM3zf(−) cloning vector (Promega). The sites of crossovers were localized by sequencing with the Sequenase kit (United States Biochemical). A similar methodology was used to check if the DR7 and DR13 mutants maintained their modified amino acids during infection.
RESULTS
Infectivity and Stability of 2a Mutants During Infection.
To study the role of BMV RNA polymerase (2a protein) in recombination, two RNA2 mutants, described by Kroner et al. (27) and designated DR7 and DR13, were used (Fig. 1). Both mutations were within the 2a core domain and both supported replication to wt levels in barley protoplasts at 24°C but were temperature-sensitive at 31°C (27). DR7 has a C-to-U substitution at position 1559, which corresponds to Leu-486 substituted by Phe. DR13 has an A-to-G substitution at position 1475, which corresponds to Asn-458 substituted by Asp.
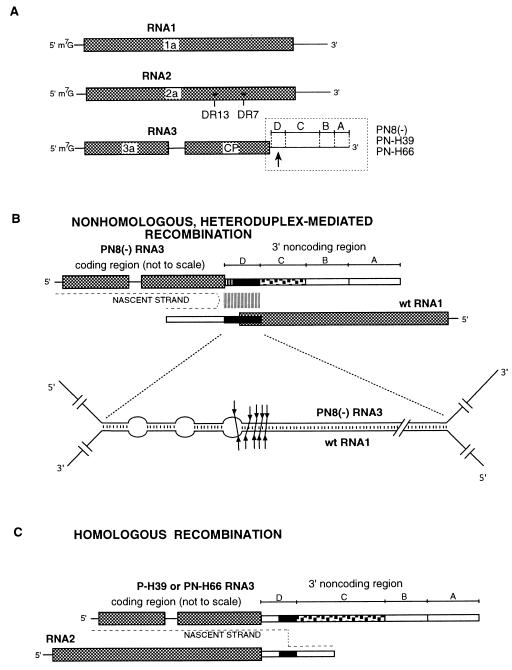
Figure 1.
The BMV recombination systems used in this study. (A) The BMV genome. The ORF are boxed and labeled, and the 3′- and 5′-terminal sequences are represented by lines. The 3′-noncoding region in the RNA3 components used in this work is comprised of four regions marked by letters A-D (see below for details). The crossovers directed into region D are denoted by a bold arrow below the RNA3 molecule. The position of DR7 and DR13 mutations in RNA2 are marked by asterisks. DR7 has an L-to-F amino acid substitution at position 486, and DR13 has an N-to-D substitution at position 458. (B) Heteroduplex-mediated, nonhomologous recombination system. The wt BMV RNA1–3′-terminal sequence (lower line) is hybridized through a complementary insert (region D, represented by black boxes) to the engineered 3′-noncoding region of the PN8(−) RNA3 construct (upper line). In PN8(−) RNA3, regions A-D are as follows: region A is 216 nt long and consists of the conserved BMV 3′ end (positions 1–236 from the 3′ end); region B contains a partial duplication of region A between positions 7 and 200. Additionally, the A and B regions have a 20-nt deletion between positions 81 and 100. The 197-nt region C (shaded) is derived from the 3′-terminal sequence of CCMV RNA3 except for the last 23 nt, and region D (black box) is the 141-nt sequence complementary to the RNA1 3′-noncoding fragment between positions 2845 and 2985. The upstream portion of this 141-nt RNA1 sequence has three 6-nt mismatch inserts to generate the heteroduplex (magnified below) with three bubble structures. The clustered arrows in the middle of the heteroduplex indicate the location of the crossover hot spots. The nascent strand of the recombinant RNA molecule is marked by a dotted line. The construction of PN8(−) RNA3 has been described by Nagy and Bujarski (16). (C) Homologous recombination system. The parallel-arranged wt RNA2 and the engineered RNA3 molecules (PN-H39 or PN-H66) are shown as, respectively, lower and upper lines. The organization of the 3′-noncoding region in PN-H39 is similar to that of PN8(−) except that region C contains a 765-nt CCMV 3′ sequence (positions 24–788, counting from the 3′ end), and region D consists of both the wt RNA2 sequence (in direct orientation) located between positions 160 and 219 (black box) and the upstream wt RNA3 sequence between positions 220 and 297. PN-H66 contains a 3′ sequence identical with that of PN-H39, except that the RNA2-derived fragment of region D is shorter (between position 197 and 219). The nascent strand of the recombinant RNA molecule is marked with a dotted line. The construction of PN-H39 and PN-H65 RNA3 has been described (17).
To determine the infectivity of the RNA2 mutants, the leaves of C. quinoa plants were mechanically inoculated with a mixture of wt RNA1, wt or mutated RNA2, and an RNA3 construct (PN8−, PN-H39, or PN-H66; see Figs. 1, 2, 3). The infections with DR7 as the RNA2 component produced a similar number of local lesions as that produced with wt RNA2, and DR13 produced ≈50% fewer lesions (data not shown). The organization of the 3′ noncoding regions of mutants PN8(−), PN-H39, and PN-H66 is described in the next section of Results.
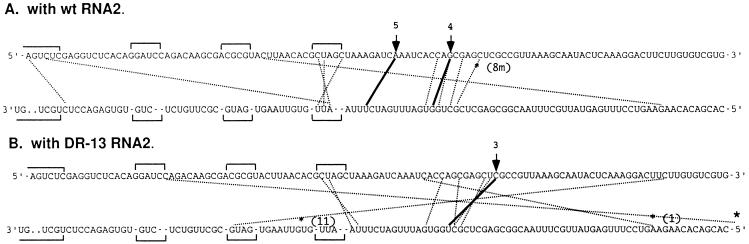
Figure 2.
Distribution of crossover sites in nonhomologous recombinants. Each recombinant was isolated from a separate local lesion. Upper sequence lines represent antisense RNA1 sequences (of segment D) of the positive strand of PN8(−) RNA3, and the lower lines represent the corresponding (complementary) sequences in wt RNA1. The lines inside each heteroduplex connect the last nucleotide of PN8(−) RNA3 and the first nucleotide of wt RNA1. The thickness of each line is proportional to the number of independent lesions on C. quinoa containing that particular recombinant; broken lines symbolize unique crossovers whereas solid lines symbolize those identified in a larger number of local lesions (as indicated by the numbers above arrows). Three recombinants that were tested for accumulation activity (see Fig. 4) are indicated by asterisks and numbers: *(1), *(11), and *(8m). A large asterisk on the right hand side, bottom line, indicates that the RNA3 cross site extends beyond the figure toward the 5′ end. Horizontal brackets depict unpaired regions (25).
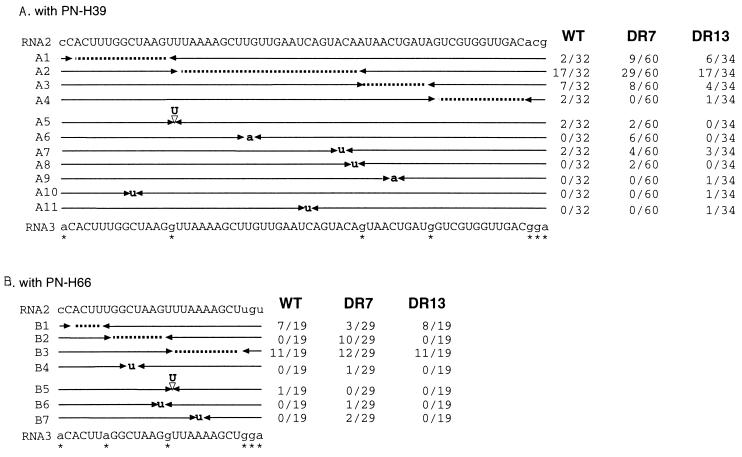
Figure 3.
Distribution of crossover sites in homologous recombinants. Each recombinant has been isolated from a separate local lesion. (A) The locations of crossover sites and of the marker nucleotide substitutions within the common sequence in the homologous RNA2/RNA3 recombinants isolated from infections with: wt RNA1; wt, DR7, or DR13 RNA2; and PN-H39 RNA3. (B) The same locations using PN-H66 RNA3. Capital letters depict the homologous nucleotides. Marker mutations are indicated by asterisks. Each recombinant contains 3′-terminal sequences derived from RNA2 on the right side and 5′ sequences from RNA3 on the left side. Arrowheads indicate the last RNA2 and the first RNA3 nucleotides. Ambiguous regions are marked by dotted lines, and sequence insertions are represented by triangles. Capital letters mark nucleotide insertions, and small letters mark nucleotide substitutions. The incidence of each RNA3 recombinant per the total number of homologous recombinants sequenced for wt, DR7, and DR13 variants is shown in the right three columns.
To determine if DR7 and DR13 mutations were maintained stably in local lesions, total RNA was extracted from separate lesions (10 per infection) 14 days postinoculation, the mutated region in RNA2 was amplified with RT-PCR using flanking oligonucleotide primers, and the cDNA products were cloned and sequenced. Both RNA2 mutations were maintained during infection after coinoculation with wt RNA1 and either wt or modified [PN8(−), PN-H39, or PN-H66] RNA3 (data not shown).
Effect of Mutations in the 2a ORF on Heteroduplex-Mediated, Nonhomologous Recombination.
The three RNA3 constructs used in this work carry a long 3′ noncoding region comprised of a duplicated promoter region (regions A and B, Fig. 1A), which are the same in the three constructs. The A/B duplication and the presence of a small deletion in both A and B sequences were found to debilitate BMV RNA replication, which allows the progeny recombinants to take over during infection (14, 15). More upstream, the three constructs have a heterologous 3′ sequence from cowpea chlorotic mottle virus (CCMV) RNA3, which is 197 nt long in PN8(−) (16) and 765 nt long in PN-H39 or PN-H66 (17). The CCMV inserts function as spacers between the initiation site of RNA replication and the site of crossovers (region D) plus provide an inactive heterologous replication promoter that eliminates unwanted recombinants. This directs the cross sites effectively toward the upstream region D.
Among several RNA3 constructs that were found to support heteroduplex-mediated nonhomologous recombination (16), in this work we used the PN8(−) RNA3. In PN8(−) region D represents a 141-nt antisense 3′ sequence from BMV RNA1 that has three 6-nt inserts within its upstream area (16, 25). Thus, region D forms a heteroduplex with the plus strand of BMV RNA3, whose upstream portion has three unhybridized “bubbles” (Fig. 1B; ref. 28). Such an arrangement was found to shift the crossovers toward inner parts of heteroduplex and to support moderate (78%) recombination frequency, and it was chosen in this work to allow better the observation of changes in crossover frequency and in location of crossover sites.
To determine the profiles of recombinants accumulating in local lesions, the progeny RNA3 sequences were amplified by RT-PCR (25), and cloned cDNA was sequenced. At least five clones per lesion were sequenced in preliminary experiments. This demonstrated that the progeny virus in the majority of local lesions tested accumulates only one type of recombinant RNA3 components (17, 25); lesions rarely accumulate two types of recombinants (25). Also, comparisons of RT-PCR and Northern blot analysis results have revealed that, in addition to progeny recombinants, the lesions accumulated sometimes small amounts of unrecombined parental RNA3 constructs and that the cDNA amplification by RT-PCR was proportional to the relative concentration of parental and recombinant RNA3 variants (data not shown).
The heteroduplex-mediated, nonhomologous recombination frequency between RNA1 and RNA3 was 78%, when using wt RNA2 (Table 1). It was increased to 100% when wt RNA2 was replaced by DR13 RNA2 (per 30 lesions analyzed) and dropped to 0% for DR7 RNA2 (per 70 lesions analyzed) (Table 1). Apparently, 2a protein made by the DR7 RNA2 cannot support heteroduplex-primed recombination to a detectable level.
Table 1.
The effect of RNA2 mutations on BMV infectivity and on the ability to support heteroduplex-mediated, nonhomologous and homologous recombination
| RNA 3 derivative | RNA2 | Recombination activity
|
||
|---|---|---|---|---|
| Local lesions analyzed, n | Recombinants, n | Recombination frequency, %* | ||
| Nonhomologous recombination | ||||
| PN8(−) | ||||
| wt | 50 | 38 | 78 | |
| DR7 | 70 | 0 | 0 | |
| DR13 | 30 | 30 | 100 | |
| Homologous recombination | ||||
| PN-H39 | ||||
| wt | 36 | 36 | 100 | |
| DR7 | 62 | 62 | 100 | |
| DR13 | 36 | 36 | 100 | |
| Homologous recombination | ||||
| PN-H66 | ||||
| wt | 36 | 20 | 56 | |
| DR7 | 40 | 31 | 77 | |
| DR13 | 32 | 21 | 66 | |
Recombination frequency is defined as the fraction of local lesions that accumulates recombinant RNA3 progeny, either homologous RNA2/RNA3 recombinants or nonhomologous (background) RNA1/RNA3 recombinants (see refs. 17 and 21), as detected by Northern blot analysis and confirmed by RT-PCR (see Materials and Methods).
Previously, we observed that mutations in the helicase domain of 1a can shift the mean position of nonhomologous crossovers within the heteroduplex (25). To determine if DR13 also altered the distribution of crossover sites, 15 recombinants were sequenced. The positions of crossovers within the heteroduplex formed by the recombining RNA1 and RNA3 substrates are shown in Fig. 2. For both wt and DR13 RNA2, the crossovers occurred at similar locations, demonstrating no significant differences in the distribution of nonhomologous crossover sites.
Effect of Mutations in the 2a ORF on Homologous Recombination.
Unlike in PN8(−), region D in PN-H39 or PN-H66 RNA3 constructs is comprised of two different sense RNA2-derived sequences that are, respectively, 60 and 23 nt long (ref. 17; Fig. 3C). In addition, region D has an upstream 80-nt sequence, present also in wt RNA3, that is heterologous with RNA2. Previously, we demonstrated that PN-H39 and PN-H66 BMV RNA3 constructs can support efficient homologous crossovers with RNA2 (17). Two types of homologous recombinants were observed: precise and imprecise (17, 21). Although in precise recombinants the entire homologous portion of region D was preserved, the imprecise recombinants contained nucleotide insertions or substitutions within this region.
The frequency and distribution of crossovers in homologous recombinants generated during infection with combinations of wt RNA1, DR7, DR13, or wt RNA2 and PN-H39 (Fig. 3A) or PN-H66 (Fig. 3B) RNA3 were determined. As shown in Table 1, for PN-H39, the observed recombination frequency was 100% using wt, DR7, or DR13 RNA2 variants. Eleven types of recombinants were found, which are listed as A1–A11 in Fig. 3A. In precise recombinants A1–A4, the crossovers occurred between marker mutations at an upstream region (A1), central region (A2), or downstream regions (A3 and A4). Recombinants A5–A11 were imprecise, carrying either an extra nucleotide (A5) or nucleotide substitutions (A6–A11) within the region of crossovers. Of the 36 lesions induced with wt RNA2, 62 with DR7 RNA2, and 36 with DR13 RNA2, 32, 60, and 34 accumulated homologous RNA2/RNA3 recombinants and 4, 2, and 2 lesions, respectively, accumulated the nonhomologous (background) RNA1/RNA3 recombinants. Of the homologous RNA2/RNA3 recombinants, 17, 29, and 17 lesions, respectively, accumulated recombinant type A2 (Fig. 3A). Recombinant type A1 accumulated in 2, 9, and 6 of the lesions, respectively, and types A3 and A4 accumulated in 9, 8, and 5 of the lesions, respectively. The fraction of imprecise homologous recombinants was similar for the wt, DR7, and DR13 RNA2 variants. However, DR7 was the only mutant that generated imprecise recombinant A6, which had a novel U-to-A substitution hotspot.
In the case of PN-H66 RNA3, the recombination frequency was 56% for wt, 77% for DR7, and 66% for DR13 RNA2 (Table 1). Here, respectively, 19, 29, and 19 local lesions accumulated RNA2/RNA3 homologous recombinants whereas, respectively, 1, 2, and 2 local lesions accumulated background RNA1/RNA3 recombinants. The distribution of the precise homologous crossovers was similar for wt RNA2 and for DR13 RNA2: of 19 local lesions analyzed, 18 and 14, respectively, contained either recombinant B1 or B3 (Fig. 3B). For DR7, however, 15 of the 29 lesions analyzed accumulated precise recombinants B1 or B3, and 10 lesions accumulated a novel precise recombinant B2. Also, for DR7, 4 lesions contained the imprecise recombinants B4, B6, and B7 whereas for wt RNA2 only one lesion had an imprecise recombinant, B5. For DR13m no imprecise recombinants were detected. Apparently, the DR7 mutant generated the highest number of imprecise recombinants and shifted the location of crossovers to the central portion of the homologous region.
Growth Characteristics of Reconstructed, Nonhomologous Recombinants.
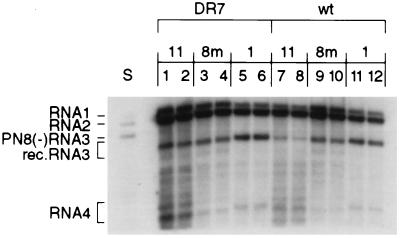
To check for possible differences in selection pressure for or against different recombinants, the growth properties of viruses containing selected, nonhomologous, recombinant RNA3 components were determined. Specifically, three different recombinants, which were isolated from infections with PN8(−) RNA3, were tested: 1, 8m, and 11 (indicated in Fig. 2). Recombinant 1 was derived by an asymmetric crossover, with the RNA3 sequence located toward the 5′ end and the RNA2 sequence toward the 3′ end within the heteroduplex structures shown in Fig. 2. Recombinant 11 also was derived by an asymmetric crossover, but the RNA3 sequences were toward the 3′ end and the RNA2 sequences were toward the 5′ end within the heteroduplex. In contrast, for recombinant 8m, the crossover sites on RNAs 2 and 3 were separated by only 3 nt and occurred at a hot spot area.
The leaves of C. quinoa were inoculated with wt RNA1, either DR7 RNA2 (Fig. 4B, lanes 1–6) or wt RNA2 (lanes 7–12), and one of the above recombinant RNA3 derivatives. The RNA was extracted from 10 local lesions for each combination 10 days postinoculation and analyzed by Northern blot analysis. Fig. 4 shows a typical Northern hybridization with two lesions for each. This confirms that wt and DR7 RNA2 can support accumulation of the RNA3 recombinants to comparable levels. Thus, there were no major constraints against accumulation of recombinants by DR7 if these recombinants had been formed.
Figure 4.
Northern blot analysis of the accumulation of reconstructed BMV RNA3 recombinants in local lesions on C. quinoa. The leaves were coinoculated mechanically with in vitro-transcribed wt RNA1, either DR7 RNA2 (lanes 1–6) or wt RNA2 (lanes 7–12), and three recombinant RNA3 (1, 8m, and 11 of Fig. 2). The full length cDNA clones of the RNA3 recombinants were obtained by substituting the 3′ noncoding region in PN8(−) plasmid (XbaI–EcoRI fragment) with the corresponding fragment of RT-PCR-amplified recombinant cDNA. The RNAs were separated electrophoretically in a 1% agarose gel, blotted to a nylon membrane (HybondN+, Amersham), and probed with a 32P-labeled RNA complementary to the 3′ 200 nt of the plus strand. A sample of the RNA inoculation mixture containing wt RNA1, RNA2, and PN8(−) RNA3 in vitro transcripts is shown in lane S.
DISCUSSION
In this work, we show that DR7 RNA2 does not support nonhomologous (heteroduplex-mediated) recombination. In contrast, DR13 RNA2 supports this type of recombination in 100% of the lesions compared with 78% obtained with the wt RNA2. Essentially, this reveals that it is possible to de-couple recombination and replication functions by mutations in a replicase protein. In control experiments, we confirmed that (i) the parental RNA2 mutants were stable during infection and that (ii) the reconstructed RNA3 recombinants can stably accumulate in local lesions. We conclude that the lack of recombinants observed for DR7 must result from a defect at the initial stage of recombination, probably during generation of recombinant RNAs.
There are several implications of our results. The sequences of DR7, DR13, and wt RNA2 differ only in one nucleotide, so it is likely that the observed effects are at the protein level rather than at the RNA level. Consequently, similar to the 1a protein (25), BMV 2a is likely involved in heteroduplex-mediated recombination. We speculate that, in DR7, the modified 2a forms a replicase complex that has limited RNA template-switching ability. Crystal structures reveal common architecture of core regions among various types of nucleic acid polymerases, which contain “palm,” “fingers,” and “thumb” domains (29–31). Based on sequence comparisons (data not shown), we speculate that the mutation in DR7 is in the fingers domain, which has been proposed to bind and discriminate between DNA and RNA and to position the template within the catalytic groove. Perhaps one of these properties is crucial for RNA template switching near heteroduplex regions. Actually, the replicase, the donor template, the nascent strand, and the acceptor template all may form an intermediate complex (for which we propose the name “recombinosome”; see Fig. 1B), which transfers the nascent RNA strand from one template to another (16, 28). The DR7 mutation might somehow change the efficiency and/or destabilize the “recombinosome.”
The mutation in DR13 is in the putative palm region that contains the conserved D and GDD amino acid motifs (not shown), the likely catalytic center of the enzyme involved in RNA chain polymerization. The mutation may possibly alter the rate of polymerization, thus changing the processivity of the replicase, which could lead to an increased frequency of recombination.
A noteworthy observation of this work is the possibility of de-coupling homologous and nonhomologous recombination. A high activity in homologous recombination for DR7 may reflect differences in the mechanism of homologous and heteroduplex-mediated nonhomologous recombination. Here, we propose that the former occurs as a two-stage process which thus does not require the formation of the recombinosome complex. The acceptor RNA does not need to be close to the donor “launching site” when the replication complex leaves the primary template (5, 19). Once detached, the nascent RNA strand, and the enzyme can interact with the acceptor RNA at the “docking site” to resume RNA synthesis (Fig. 1C). In fact, an increase in homologous recombination for DR7 is predicted if, indeed, the interactions at the “launching” site are reduced. That tendency was observed during infection with wt RNA1 and PN-H66 RNA3 (see Table 1). Also, a shift of the crossover sites toward a central portion of the homologous region observed for DR7 (Fig. 3) may arise from an increase in the affinity for the template RNA, or from faster migration of the replicase before the detachment (21). Finally, an increase in the number of imprecise crossovers observed for DR7 (Fig. 3) may result from increased pausing by the replicase and, consequently, increased nucleotide misincorporation (21). Further mutagenesis and biochemical analyses are required to map regions on 1a and 2a involved in recombination.
The BMV recombination system is a useful model for the alphavirus-like supergroup, a group that includes important pathogens of humans, plants, and animals. Whether the replicase proteins are involved in genetic recombination of other viruses within this and different virus groups remains to be demonstrated.
Acknowledgments
We thank M. V. Graves, K. Kirkegaard, and K. Richards for comments and discussions. This work was supported by grants from the National Institute for Allergy and Infectious Diseases (3RO1 AI26769) and the National Science Foundation (MCB-9630794) and by the Plant Molecular Biology Center at Northern Illinois University.
ABBREVIATIONS
- BMV
brome mosaic bromovirus
- wt
wild-type
- RT-PCR
reverse transcriptase PCR
- CCMV
cowpea chlorotic mottle virus
References
- 1.King A M Q. In: RNA Genetics. Domingo E, Holland J J, Ahlquist P, editors. Vol. 2. Boca Raton, FL: CRC; 1988. pp. 149–185. [Google Scholar]
- 2.Lai M C M. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujarski J J, Kaesberg P. Nature (London) 1986;321:528–531. doi: 10.1038/321528a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascone P J, Carpenter C D, Li X H, Simon A E. EMBO J. 1990;9:1709–1715. doi: 10.1002/j.1460-2075.1990.tb08294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon A, Bujarski J J. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 6.Munishkin A V, Voronin L A, Chetverin A B. Nature (London) 1988;333:473–475. doi: 10.1038/333473a0. [DOI] [PubMed] [Google Scholar]
- 7.Bujarski J J, Nagy P D. In: Homologous Recombination in Plants. Paszkowski J, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 1–20. [Google Scholar]
- 8.Ahlquist P. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 9.Mise K, Allison R F, Janda M, Ahlquist P. J Virol. 1993;67:2815–2820. doi: 10.1128/jvi.67.5.2815-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroner P A, Young B M, Ahlquist P. J Virol. 1990;64:6110–6115. doi: 10.1128/jvi.64.12.6110-6120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traynor P, Young B M, Ahlquist P. J Virol. 1991;65:2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao C C, Quadt R, Hershberger R P, Ahlquist P. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao C C, Ahlquist P. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bujarski J J, Dzianott A M. J Virol. 1991;65:4153–4159. doi: 10.1128/jvi.65.8.4153-4159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy P D, Bujarski J J. J Virol. 1992;66:6824–6828. doi: 10.1128/jvi.66.11.6824-6828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy P D, Bujarski J J. Proc Natl Acad Sci USA. 1993;90:6390–6394. doi: 10.1073/pnas.90.14.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy P D, Bujarski J J. J Virol. 1995;69:131–140. doi: 10.1128/jvi.69.1.131-140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao A L N, Sullivan B P, Hall T C. J Gen Virol. 1990;71:1403–1407. doi: 10.1099/0022-1317-71-6-1403. [DOI] [PubMed] [Google Scholar]
- 19.Bujarski J J, Nagy P D, Flasinski S. Adv Virus Res. 1994;43:275–302. doi: 10.1016/S0065-3527(08)60051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzianott A, Flasinski S, Pratt S, Bujarski J. Virology. 1995;208:370–375. doi: 10.1006/viro.1995.1163. [DOI] [PubMed] [Google Scholar]
- 21.Nagy P D, Bujarski J J. J Virol. 1996;70:415–426. doi: 10.1128/jvi.70.1.415-426.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cascone P J, Haydar T F, Simon A E. Science. 1993;260:801–805. doi: 10.1126/science.8484119. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Cascone P J, Simon A E. Virology. 1991;184:791–794. doi: 10.1016/0042-6822(91)90454-j. [DOI] [PubMed] [Google Scholar]
- 24.Kirkegaard K, Baltimore D. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy P D, Dzianott A, Ahlquist P, Bujarski J J. J Virol. 1995;69:2547–2556. doi: 10.1128/jvi.69.4.2547-2556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janda M, French R, Ahlquist P. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 27.Kroner P A, Richards D, Traynor P, Ahlquist P. J Virol. 1989;63:5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bujarski, J. J. & Nagy, P. D. (1994) Arch. Virol. 9, Suppl., 231–238. [DOI] [PubMed]
- 29.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 30.Molina A, Ding J, Nanni R, Clark A, Lu X, Tantillo C, Williams R, Kamer G, Ferris A, Clark P, Hizi A, Hughes S, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poch O, Sanvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]