Abstract
GDP-l-fucose is the activated nucleotide sugar form of l-fucose, which is a constituent of many structural polysaccharides and glycoproteins in various organisms. The de novo synthesis of GDP-l-fucose from GDP-d-mannose encompasses three catalytic steps, a 4,6-dehydration, a 3,5-epimerization, and a 4-reduction. The mur1 mutant of Arabidopsis is deficient in l-fucose in the shoot and is rescued by growth in the presence of exogenously supplied l-fucose. Biochemical assays of the de novo pathway for the synthesis of GDP-l-fucose indicated that mur1 was blocked in the first nucleotide sugar interconversion step, a GDP-d-mannose-4,6-dehydratase. An expressed sequence tag was identified that showed significant sequence similarity to proposed bacterial GDP-d-mannose-4,6-dehydratases and was tightly linked to the mur1 locus. A full-length clone was isolated from a cDNA library, and its coding region was expressed in Escherichia coli. The recombinant protein exhibited GDP-d-mannose-4,6-dehydratase activity in vitro and was able to complement mur1 extracts in vitro to complete the pathway for the synthesis of GDP-l-fucose. All seven mur1 alleles investigated showed single point mutations in the coding region for the 4,6-dehydratase, confirming that it represents the MUR1 gene.
l-Fucose (6-deoxy-l-galactose) is a monosaccharide found in a diverse array of organisms. The sugar is a known component of bacterial lipopolysaccharides, mammalian and plant glycoproteins, and polysaccharides of plant cell walls such as xyloglucan and rhamnogalacturonans I and II. The precise function of l-fucose within these polysaccharides is not clear, but it may stabilize conformations of xyloglucan, which can efficiently bind to cellulose microfibrils (1), possibly aiding in cell wall integrity. Furthermore, xyloglucan fucosylation is essential for the biological activity of some xyloglucan-derived oligosaccharides (2). The pathway for the synthesis of l-fucose has been studied biochemically, but genes for the corresponding enzymes have not been cloned from any eukaryote.
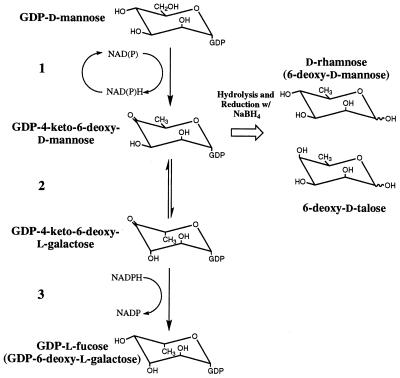
GDP-l-fucose (guanosine-diphospho-l-fucose) is the activated form of this sugar, synthesized de novo from GDP-d-mannose via a three-step mechanism or through a salvage pathway involving phosphorylation of free l-fucose and subsequent nucleoside-diphosphate attachment (3–5). The de novo pathway for GDP-l-fucose production is shown in Fig. 1. The first step is catalyzed by GDP-d-mannose-4,6-dehydratase and involves the formation of the intermediate GDP-4-keto-6-deoxy-d-mannose, which is then used in the second and third steps of the pathway by 3,5-epimerase and 4-reductase activities to yield GDP-l-fucose. The pathway was initially elucidated in bacteria but has since been characterized in mammalian and plant systems (6–11).
Figure 1.
Reaction scheme for the de novo synthesis of GDP-l-fucose from GDP-d-mannose, involving 4,6-dehydratase (step 1), which produces a 4-keto-6-deoxy intermediate, 3,5-epimerization (step 2), and the final NADPH-dependent reduction of the 4-keto group (step 3). Expected products from the chemical reduction and hydrolysis of the 4-keto-6-deoxy intermediate, are also included.
Recently an l-fucose-deficient cell wall mutant, mur1, was isolated from Arabidopsis thaliana and characterized phenotypically (12). Eight recessive mur1 alleles were obtained from this screen, most of which exhibit 50- to 200-fold reductions in l-fucose in the cell walls of shoot organs, while the l-fucose content in roots is only reduced by 40%. The mur1 mutation causes reduced tensile strength of elongating stem segments and slight dwarfism. When mur1 plants are grown in the presence of exogenous l-fucose, the l-fucose content is restored to wild type and all other mutant phenotypes are rescued. This suggests that the salvage pathway is intact and that the lack of l-fucose observed in the mur1 mutant may be due to a defect in the de novo synthesis of GDP-l-fucose.
Our knowledge of plant genes encoding enzymes involved in nucleotide sugar interconversions is limited with only two genes isolated so far, a UDP-d-glucose-4-epimerase from Arabidopsis (13) and a UDP-d-glucose dehydrogenase from soybean and Arabidopsis (14). Bacterial pathways have been studied in more detail and genes for putative GDP-d-mannose-4,6-dehydratases have been identified and are believed to play a role in the synthesis of GDP-l-fucose and/or GDP-colitose (GDP-3,6-dideoxy-l-galactose; refs. 15 and 16). However, no biochemical evidence for the postulated function of these enzymes has been described. Here, we report on the cloning of the MUR1 gene of A. thaliana and demonstrate that it encodes an isoform of GDP-d-mannose-4,6-dehydratase. This represents the first example of a gene in plant nucleotide sugar interconversions for which a mutant has been obtained.
MATERIALS AND METHODS
Plant Material.
All plants were grown in environmental chambers at 23°C and 60–70% humidity under continuous fluorescent light (60–70 μmol·m−2·s−1). Wild-type plants of the Columbia ecotype and mutant plants carrying the mur1-2 allele (12) were used for all biochemical assays.
Enzyme Extraction and Assay.
Crude extracts containing GDP-d-mannose-4,6-dehydratase and GDP-4-keto-6-deoxy-d-mannose-3,5-epimerase-4-reductase activities were obtained from freshly harvested leaf tissue from 14-day-old seedlings homogenized with a mortar and pestle in two volumes of buffer 1 (100 mM Tris·HCl buffer, pH 7.6/2 mM DTT) containing 1% (wt/vol) polyvinylpolypyrrolidone. The homogenate was strained through Miracloth (Calbiochem) and centrifuged at 5000 rpm for 5 min in Sorvall rotor SS34, and pelleted cell debris was discarded. Protein was precipitated from the soluble extract by (NH4)2SO4 fractionation, and the protein fraction precipitated between 25% and 75% saturation was collected by centrifugation at 15,000 rpm for 30 min in a Sorvall SS34 rotor. This material was then resuspended in a minimum volume of buffer 2 (20 mM Tris·HCl buffer, pH 7.6/0.5 mM DTT) and dialyzed (3500 Da molecular mass cutoff) against the same buffer. All steps were completed at 0–4°C. Both enzyme activities were stable for several months at −20°C.
Crude extracts from 50-ml cultures of Escherichia coli were prepared by sonication in 4 volumes of buffer 1 per g of fresh weight. The bacterial extracts were centrifuged at 10,000 rpm for 20 min in Sorvall rotor SS34, and the supernatant was dialyzed against buffer 2. All steps were completed at 0–4°C.
The combined activities of both dehydratase and epimerase-reductase converting GDP-d-mannose to GDP-l-fucose (steps 1–3 in Fig. 1) were measured in a reaction mixture comprising 20 μl of enzyme extract and 10 μl of Tris·HCl buffer (pH 7.6) containing 925 Bq of GDP-d-[U-14C]mannose (specific activity 10.4 GBq·mmol−1; Amersham), 0.15 μmol of NADPH, and disodium EDTA at a final assay concentration of 10 mM. Incubation was for 1 h at 37°C. The reaction was stopped by heat denaturation at 100°C for 3 min. Assay of the 4,6-dehydratase activity alone (step 1 in Fig. 1) was completed in the above reaction mix minus NADPH and the GDP-4-keto-6-deoxy-d-mannose intermediate was chemically reduced by the addition of 1 μmol of NaBH4 to the stopped reaction and a further incubation for 90 min at 37°C. GDP-4-keto-6-deoxy-d-mannose for the assay of the 3,5-epimerase-4-reductase (steps 2 and 3 in Fig. 1) was produced using the same conditions without subsequent chemical reduction, and the assay was completed by the addition of 0.15 μmol of NADPH and 20 μl of protein extract and a further 1-h incubation at 37°C.
Reaction products were analyzed by TLC on Baker-Flex 250-μm cellulose plates (J. T. Baker). GDP-d-mannose and GDP-l-fucose were analyzed directly in ethanol/1 M ammonium acetate, pH 7 (7:3 by volume). Nucleoside-diphospho sugars were hydrolyzed, where indicated, by addition of 100 μl of 2 M trifluoroacetic acid and incubation at 95°C for 20 min to produce free monosaccharides. Trifluoroacetic acid was removed in vacuo and monosaccharides analyzed in 1-butanol/acetic acid/water (12:3:5 by volume) followed by ethyl acetate/pyridine/water (8:2:1 by volume) in the same direction. Nucleoside-diphospho sugars were identified by radiolabeled standards, and monosaccharides by authentic sugars stained with aniline-hydrogen phthalate (17). Developed chromatograms were analyzed using the Bio-Rad Molecular Analyst phophorimaging system and software.
Isolation and Analysis of Arabidopsis and Plasmid DNA.
Arabidopsis DNA was isolated from 3-week-old plants (18) and further purified using anion exchange columns according to the manufacturer’s protocol (Qiagen, Chatsworth, CA). Restriction enzyme digests were performed using the manufacturer’s specifications (New England Biolabs). For Southern blots, ≈1 μg of digested genomic DNA was loaded per lane and separated by electrophoresis through 0.8% agarose gels. Radiolabeled probes were created by excision of the cDNA insert from pZL1 clones using EcoRI and BamHI, purification via the Qiaquick gel extraction kit (Qiagen), and random primer labeling with [α-32P]dCTP (specific activity 111 TBq·mmol−1; Amersham). Nucleic acid hybridizations were done as described (19). Overnight exposure at −80°C using BioMax film with a BioMax intensifying screen was performed according to the manufacturer’s specifications (Kodak).
Plasmid DNA from E. coli cells was extracted and purified using the Qiaprep spin miniprep or midiprep kit following the protocol suggested by the manufacturer (Qiagen).
Isolation of cDNA Clones.
A digoxigenin-labeled probe corresponding to expressed sequence tag (EST) clone 90A12T7 was created via PCR using the T7 and SP6 primers flanking the multiple cloning site of pZL1 (GIBCO/BRL). Labeling conditions were as described in the manufacturer’s protocol (Boehringer Mannheim), except that the ratio between dTTP and digoxigenin-dUTP was adjusted to 4:1. The digoxigenin-labeled probe was used to screen a lambda PRL-2 library (20) obtained from the Arabidopsis Biological Resource Center (Ohio State University) via a colorimetric detection procedure, as specified by Boehringer Mannheim.
Genetic Mapping.
The mur1-2 mutant (background Columbia) of A. thaliana was crossed to wild-type Landsberg erecta (Ler) plants, F1 plants were obtained, and these were selfed to obtain an F2 population. Identification of mur1 plants via gas–liquid chromatography of alditol acetates was done as described previously (12).
Restriction fragment length polymorphisms were identified between Columbia and Ler DNA by probing Southern blots with 32P-labeled cDNA clones GMD1 and GMD2. Total genomic DNA from mur1 plants obtained from the F2 population was digested with HpaI and ScrFI, respectively, and probed with their corresponding 32P-labeled cDNA (GMD1 and GMD2).
RNA Isolation and Northern Blots.
RNA was isolated from leaves of 3-week-old Arabidopsis plants by grinding in liquid nitrogen, phenol extraction, and precipitation with LiCl (21). Approximately 10 μg of RNA was prepared for agarose gel electrophoresis by glyoxylation, run at 3–4 V/cm on an Owl Scientific (Woburn, MA) Buffer Puffer recirculating electrophoresis system, blotted to a positively charged nylon membrane (Hybond N+; Amersham), and hybridized with 32P-labeled GMD2 via standard procedures (21). To determine the approximate size of the GMD2 mRNA, bacteriophage lambda DNA digested with HindIII was heat-denatured, glyoxylated, and probed separately with 32P-labeled lambda DNA after blotting.
PCR Conditions and Cloning.
The sequences of the oligonucleotide primers used for subcloning coding regions of the MUR1 gene into pBluescript (KS+) and the pET11d expression vector (Stratagene) were as follows: MUR1/pBlue/pET11d, upper, 5′-ACACTGCAGCCATGGCGTCAGAGAACAACG-3′, and MUR1/pBlue/pET11d, lower, 5′-ACAGATATCAAGGTTGCTGCTTAGCATCC-3′, with NcoI and PstI sites engineered into the upper primer and an EcoRV site engineered into the lower primer. PCR was performed using a model 2400 Gene Amp PCR System and PCR Core Reagents (Perkin–Elmer) with the following conditions: denaturation at 94°C for 10 min, cooling to 20°C, and addition of reaction mix, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 59°C for 1 min, extension at 72°C for 1.5 min, and final extension at 72°C for 10 min. The final concentration of MgCl2 was 2 mM.
Cloning of PCR products into pBluescript KS+ was done as follows: PCR products gel-purified using the Qiaquick gel extraction kit from Qiagen were digested with PstI and EcoRV. Following ligation to pBluescript KS+ cleaved with PstI and EcoRV, the DNA was used for transformation into E. coli XL1 Blue MRF′ supercompetent cells according to the manufacturer’s protocol (Stratagene). Seven mur1 alleles and the wild-type allele were cloned by this procedure.
To clone the MUR1 coding region into the pET11d expression vector, 3′ recessed ends of BamHI-digested vector were filled in with Klenow enzyme (GIBCO/BRL) using standard methods (21), followed by digestion with NcoI. The pBluescript clone containing the wild-type MUR1 gene was digested with NcoI and EcoRV. The insert DNA was gel-purified and ligated to the pET11d vector as described above. Transformation into E. coli BL21 (DE3) competent cells was performed according to the manufacturer’s protocol (Stratagene).
Nucleic Acid Sequence Determination and Analysis.
All sequencing reactions were done by the enzymatic method with ≈10 μg of template DNA labeled using the CY5 Auto Read Labeling kit (Pharmacia) and an automated laser fluorescent sequencer (Pharmacia). Data analysis of nucleic acid and amino acid sequences and hydropathy plots of the derived MUR1 amino acid sequence were carried out with macvector software (Oxford Molecular Group, Oxford, U.K.).
To determine the DNA sequences from PCR products, cultures from at least six independent clones were pooled before plasmid purification. All DNA sequences were determined from both strands.
SDS/PAGE.
Protein from control cultures containing pET11d vector with no insert and pET11d vector containing the MUR1 coding region were boiled in sample buffer and run on a 10% SDS/polyacrylamide gel (22) for 3 h at 135 V using the Penguin vertical gel system (Owl Scientific).
RESULTS
The mur1 Mutant Lacks GDP-d-Mannose-4,6-Dehydratase Activity but Exhibits GDP-4-Keto-6-Deoxy-d-Mannose-3,5-Epimerase-4-Reductase Activity in Vitro.
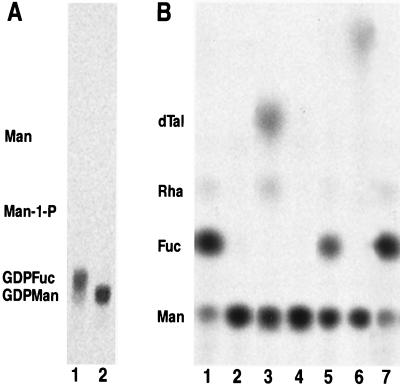
To determine the biochemical defect in the mur1 mutant, protein was extracted from leaves of wild-type and mur1 leaves and assayed for the ability to produce GDP-l-[14C]fucose from GDP-d-[14C]mannose in vitro. The expected products from this assay are shown in Fig. 1 and the results displayed in Fig. 2 A and B, lanes 1 and 2, indicate that protein extracts from wild-type leaves were able to convert GDP-d-[14C]mannose to GDP-l-[14C]fucose, while mur1 extracts lacked this activity.
Figure 2.
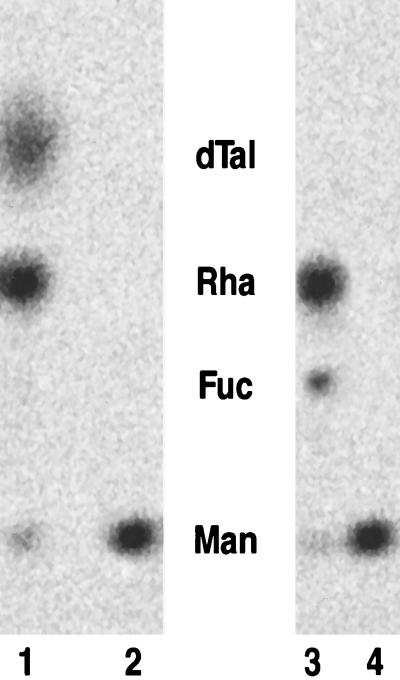
Assay of enzyme activities in the de novo synthesis of GDP-l-fucose. (A) Thin layer chromatogram of nucleotide diphospho-sugar products from the in vitro assay, following incubation of GDP-d-[14C]mannose with wild-type protein extract (lane 1) or mur1 protein extract (lane 2). (B) Thin layer chromatogram of the monosaccharides from the acid hydrolysis of in vitro assay products using GDP-d-[14C]mannose (lanes 1–4) or a mixture of GDP-d-[14C]mannose and GDP-4-keto-6-deoxy-d-[14C]mannose (lanes 5–7) as substrates. Lane 1, wild-type protein extract with exogenous NADPH; lane 2, mur1 protein extract with exogenous NADPH; lane 3, wild-type protein extract without exogenous NADPH; lane 4, mur1 protein extract without exogenous NADPH; lane 5, mur1 protein extract in the presence of NADPH; lane 6, same as lane 5 but using heat-inactivated mur1 protein; and lane 7, same as lane 5 but using wild-type extract. Samples in lanes 3 and 4 were reduced with NaBH4 before hydrolysis. Positions of authentic standards are shown. Man, mannose; Man-1-P, mannose-1-phosphate; Fuc, fucose; Rha, rhamnose; and dTal, 6-deoxy-talose.
Since the de novo synthesis of GDP-l-fucose comprises three steps, probably using two separate enzymes (a 4,6-dehydratase and a 3,5-epimerase-4-reductase) in eukaryotes, we wished to determine which step was blocked in the mur1 mutant. We found that, in agreement with previously published data (6), the final stereospecific reduction of the 4-keto group into the l-galacto configuration required exogenous reduced pyridine dinucleotide (NADPH). Omission of NADPH from the assay mixture resulted in the buildup of the GDP-4-keto-6-deoxy-d-mannose intermediate and allowed us to assay the 4,6-dehydratase activity separately. Due to the chemical instability of the 4-keto-6-deoxy intermediate, we reduced the 4-keto group and assayed the resultant diagnostic monosaccharides, 6-deoxy-d-talose, and 6-deoxy-d-mannose (d-rhamnose). Protein from wild-type leaves was able to catalyze the formation of the 4-keto-6-deoxy intermediate (Fig. 2B, lane 3), whereas mur1-derived extracts were not (Fig. 2B, lane 4), indicating that no 4,6-dehydratase activity was present. Small amounts of [14C]rhamnose were formed during these in vitro assays even in the absence of chemical reduction, a result which is in agreement with previous reports (23). We confirmed that mur1 plants possessed functional 3,5-epimerase-4-reductase activity by using wild-type protein without NADPH to generate the 4-keto-6-deoxy intermediate and using this as the substrate for the epimerase-reductase. In the presence of NADPH, both mur1 and wild-type extracts were able to convert the intermediate into GDP-l-fucose (Fig. 2B, lanes 5 and 7).
Two Similar cDNAs Were Isolated from a cDNA Library Using an Arabidopsis EST with High Sequence Similarity to Putative Bacterial GDP-d-Mannose-4,6-Dehydratases.
An EST (clone 90A12T7) that showed high sequence similarity to putative bacterial GDP-d-mannose-4,6-dehydratases was identified in the database of ESTs (dbEST; ref. 20). This clone was provided by the Arabidopsis Biological Resource Center, and its entire nucleotide sequence was determined. Sequence comparisons with bacterial clones suggested that the cDNA was truncated at the 5′ end. To obtain a full-length cDNA, a lambda PRL-2 cDNA library (20) was screened using the EST as a probe. Twenty clones were isolated from this screen that could be divided into two separate classes based on restriction mapping of individual isolates. The nucleotide sequences from the longest cDNAs in each class were determined, indicating the identification of two different cDNAs with substantial sequence similarity to each other and to putative bacterial GDP-d-mannose-4,6-dehydratases. The two cDNAs were tentatively designated GMD1 and GMD2 (guanosine-diphosphod-mannose-4,6-dehydratase).
Southern Blot Analysis Reveals at Least Two Genes for Putative GDP-d-Mannose-4,6-Dehydratases Within the Arabidopsis Genome.
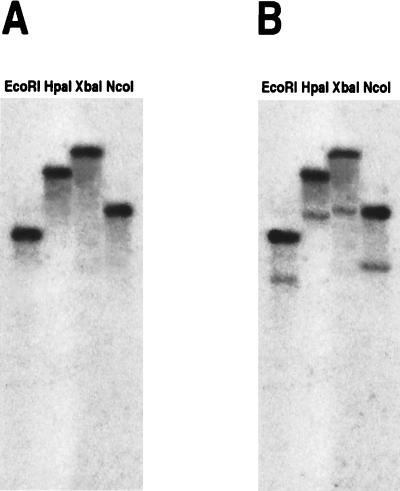
As shown in Fig. 3, Southern blots probed with GMD2 at high stringency revealed a single hybridizing fragment, while two fragments were observed at reduced stringency, providing further evidence that two genes for putative GDP-d-mannose-4,6-dehydratases were present in the Arabidopsis genome. Since both cDNAs represented candidate MUR1 genes, they were used to determine possible linkage to the mur1 locus.
Figure 3.
Southern blot analysis of total Arabidopsis DNA at high and low stringency using 32P-labeled GMD2 as a probe. (A) Digested Columbia DNA probed at high stringency (68°C hybridization, 65°C washes). (B) Digested Columbia DNA probed at low stringency (58°C hybridization, 55°C washes).
GMD2 Is Tightly Linked to mur1.
Restriction fragment length polymorphisms (RFLPs) were identified on Southern blots between HpaI-digested Columbia and Landsberg DNA when GMD1 was used as a probe and between ScrFI-digested Columbia and Landsberg DNA when GMD2 was used as a probe. These RFLPs were then used to determine the genotype (Columbia or Landsberg) of 50 mur1 plants obtained from a segregating F2 population (Landsberg (MUR1) × Columbia (mur1)). GMD1 segregated independently from mur1, while GMD2 showed complete cosegregation with the mur1 locus providing initial evidence that GMD2 was a MUR1-derived cDNA (data not shown).
All mur1 Alleles Harbor Mutations Within the Coding Region of GMD2.
After establishing linkage between GMD2 and mur1, we determined that three separate GMD2 cDNAs contained a 373-aa open reading frame. As shown in Fig. 4, the sequence surrounding the first codon showed a perfect match with the plant initiation codon consensus sequence AACAAUGGC (24). Northern blot analysis indicated a length of ≈1.4 kb for the GMD2 mRNA, matching the size of the longest cDNAs obtained (data not shown). These results indicate that cDNAs containing the entire GMD2 coding region had been cloned.
Figure 4.
Nucleotide sequence of the MUR1 cDNA and derived amino acid sequence of the MUR1 protein. Nucleotide changes and predicted amino acid substitutions in each of seven mur1 alleles are indicated. The consensus sequence for recognition of the initiation codon and a putative polyadenylylation signal are underlined.
The coding regions of the genes corresponding to the GMD2 cDNA were isolated via PCR from seven mur1 alleles and wild-type Columbia. The nucleotide sequences of the PCR products were then determined to identify possible sites of mutations. The sequences of the wild-type PCR product and the GMD2 cDNA were identical, indicating the absence of introns within the coding region. The PCR products from all seven mur1 alleles investigated showed single point mutations as indicated in Fig. 4. Based on these data we conclude that GMD2 represents a MUR1-derived cDNA.
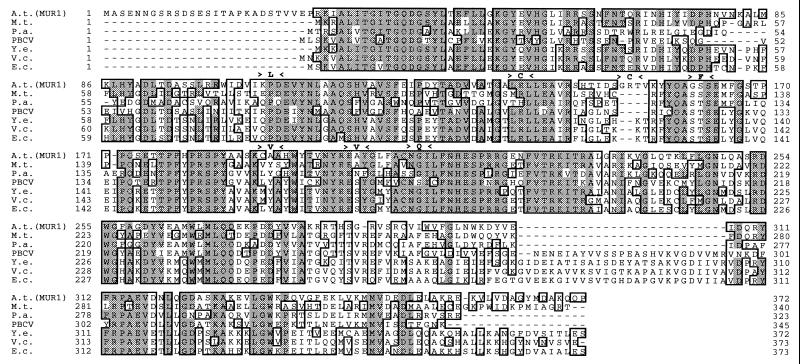
The MUR1 Protein Is Similar in Amino Acid Sequence to Putative Bacterial GDP-d-Mannose-4,6-Dehydratases.
A comparison between the derived amino acid sequence of the MUR1 gene product from Arabidopsis and putative bacterial GDP-d-mannose-4,6-dehydratases is shown in Fig. 5. The first 25 aa of the MUR1 protein are absent from the bacterial sequences, suggesting a function as a signal sequence. However, hydropathy plots (data not shown) and the presence of GDP-d-mannose-4,6-dehydratase activity in the cytosolic fraction (see Materials and Methods) suggest that the MUR1 protein is soluble and cytosolic. Furthermore, the amino acid sequence of the N-terminal extension does not fit the criteria for chloroplast or endoplasmic reticulum transit peptides (25, 26).
Figure 5.
Alignment between the derived MUR1 amino acid sequence and putative GDP-d-mannose-4,6-dehydratases. Identical residues are shaded in gray and conserved residues are boxed. Positions of the amino acid substitutions from seven mur1 alleles are indicated above the Arabidopsis sequence. A.t., A. thaliana; M.t., Mycobacterium tuberculosis; P.a., Pseudomonas aeruginosa; PBCV, Paramecium bursaria Chlorella virus; Y.e., Yersinia enterocolitica; V.c., Vibrio cholerae; and E.c., E. coli. Sequence alignments were done with clustal w and viewed with seqvu.
MUR1 Protein Expressed in E. coli Has GDP-d-Mannose-4,6-Dehydratase Activity That Complements the Biochemical Defect of mur1 Protein Extracts in Vitro.
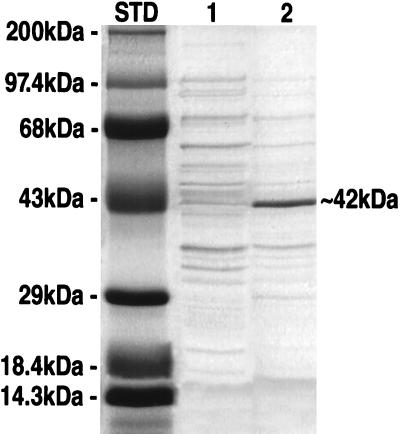
To verify the predicted function of the MUR1 protein, we expressed the MUR1 coding region in E. coli, determined the size of the protein, and subjected it to the 4,6-dehydratase assay. Fig. 6 shows an SDS/PAGE analysis of protein extracts from induced cells, indicating that the MUR1 gene encodes a protein ≈42 kDa in size, which is in agreement with the size predicted from the amino acid sequence (41.9 kDa). As shown in Fig. 7, lanes 1 and 2, extracts from the MUR1-expressing cells converted GDP-d-[14C]mannose into GDP-4-keto-6-deoxy-d-[14C]mannose, while extracts from control cells exhibited no 4,6-dehydratase activity. Moreover, when the bacterial extract was combined with a protein extract from mur1 plants, GDP-l-[14C]fucose was produced (Fig. 7, lane 3). We also noted that the E. coli strain possessed an endogenous 4-reductase activity that converted some of the GDP-4-keto-6-deoxy-d-[14C]mannose intermediate to GDP-d-[14C]rhamnose; however, in the absence of the MUR1 gene, bacterial extracts were unable to convert GDP-d-[14C]mannose into any detectable products.
Figure 6.
SDS/PAGE analysis of crude extracts from E. coli harboring the pET11d vector with or without the MUR1 coding region. Lane 1, protein extracted from E. coli containing a pET11d vector without an insert; and lane 2, protein extracted from E. coli containing a pET11d vector with the MUR1 coding region. STD, prestained protein molecular weight standards.
Figure 7.
Assay of the MUR1 protein for GDP-d-mannose-4,6-dehydratase activity and reconstitution of the l-fucose biosynthetic pathway in mur1-derived protein extracts. Lane 1, protein from E. coli expressing the MUR1 gene, incubated with GDP-d-[14C]mannose; lane 2, protein from E. coli transformed with pET11d vector minus the MUR1 insert; lane 3, same as lane 1 but with mur1-derived protein extract added; and lane 4, same as lane 2 but with mur1-derived protein extract added. All final products were hydrolyzed and reduced as described earlier.
DISCUSSION
The mur1 mutant of A. thaliana has allowed us to study the effects of l-fucose deficiency in the cell wall of higher plants and provided us with a means of cloning a plant gene involved in the de novo synthesis of an important cell wall component. The work reported in this paper provides biochemical evidence that a cloned gene encodes a functional GDP-d-mannose-4,6-dehydratase.
Enzymatic assays of protein from mur1 and wild-type plants led us to conclude that the mur1 mutation eliminated GDP-d-mannose-4,6-dehydratase activity in the mutant plants. A candidate EST was identified through sequence similarity searches and used to isolate two similar yet distinct cDNAs. One of the cDNAs showed complete linkage to the mur1 locus, and the corresponding gene was sequenced from wild-type as well as seven mur1 alleles. Nucleotide sequence comparisons indicated sequence identity between the cDNA and the wild-type gene but revealed single point mutations in all seven mur1 alleles investigated. We then verified the suspected function of the cDNA-encoded protein by expressing it in E. coli and assaying for 4,6-dehydratase activity. This analysis indicated that the cDNA encoded a functional GDP-d-mannose-4,6-dehydratase, which was able to restore l-fucose synthesis to mur1-derived protein extracts in vitro. This combination of genetic and biochemical data indicates that the MUR1 gene has been cloned.
Even with the information we have gained on the MUR1 gene, we are still unable to account for the presence of l-fucose in root tissue. The simplest explanation for this observation is the existence of additional gene(s) encoding GDP-d-mannose-4,6-dehydratase activity in the root. GMD1 may encode another isoform of this enzyme based on its sequence similarity to MUR1 and putative bacterial GDP-d-mannose-4,6-dehydratases. Further characterization of this cDNA clone will reveal the function of this putative isoform and its role within A. thaliana.
Our sequence data of the seven mur1 alleles indicate that all of the mutations are caused by transitions leading to amino acid substitutions. The mutations are clustered within a region of ≈300 nucleotides, and six out of seven nucleotide changes represent C → T transitions on the coding strand. Some of the substitutions are within conserved regions of the protein as compared with the bacterial sequences, but without any knowledge of the protein structure it is difficult to determine how these substitutions may affect protein folding or function. These point mutations are characteristic of ethyl methanesulfonate mutagenesis, but it is noteworthy that nonsense mutations were not identified.
Another intriguing aspect of the results from the cell wall mutant screen is that all mutations leading to l-fucose deficiency mapped to the mur1 locus. If GMD1 does in fact encode an isoform of GDP-d-mannose-4,6-dehydratase with a root-specific expression pattern, mutations in this gene could not have been identified in a mutant screen based on leaf material (12). Since the GDP-4-keto-6-deoxy-d-mannose-3,5-epimerase-4-reductase necessary for the second step in the de novo pathway of GDP-l-fucose synthesis must be encoded by a second gene, we wonder why no l-fucose deficient mutants affecting this enzyme were isolated. Mutations eliminating this activity may be lethal. Alternatively, genetic redundancy in this enzymatic step may defeat any attempts to identify mutants.
Changes in cell wall polysaccharides in the mur1 mutant have been recently investigated, leading to the discovery that l-fucose is partly replaced by l-galactose in mutant-derived xyloglucan (27). This apparent switch from one monosaccharide to another closely related sugar could be explained based on our knowledge of the remaining steps in the pathway for the de novo synthesis of GDP-l-fucose. In the absence of a functional GDP-d-mannose-4,6-dehydratase, the 3,5-epimerase-4-reductase normally involved in l-fucose synthesis may be able to use GDP-d-mannose directly forming GDP-l-galactose. If this hypothesis were true, it would be interesting both biochemically and genetically to study organisms such as jojoba (28) and the unicellular green alga, Chlorella (29), that produce significant amounts of l-galactose.
Acknowledgments
We thank the Arabidopsis Biological Resource Center at Ohio State University for providing EST clones and the lambda PRL-2 cDNA library. This work was supported, in parts, by grants from the U.S. Department of Agriculture National Research Initiative (93-37304-9510), the U.S. Department of Energy (DE-FG02-95ER20203), and the National Science Foundation (MCB-9405739).
ABBREVIATION
- EST
expressed sequence tag
Footnotes
References
- 1.Levy S, York W S, Stuike-Prill R, Meyer B, Staehelin L A. Plant J. 1991;1:195–215. [PubMed] [Google Scholar]
- 2.York W S, Darvill A G, Albersheim P. Plant Physiol. 1984;75:295–297. doi: 10.1104/pp.75.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginsburg V. J Biol Chem. 1961;236:2389–2393. [PubMed] [Google Scholar]
- 4.Markovitz A. J Biol Chem. 1964;239:2091–2098. [PubMed] [Google Scholar]
- 5.Feingold D S, Avigad G. In: The Biochemistry of Plants: A Comprehensive Treatise. Stumpf P K, Conn E E, editors. Vol. 3. New York: Academic; 1980. pp. 101–170. [Google Scholar]
- 6.Liao T H, Barber G A. Biochim Biophys Acta. 1971;230:64–71. doi: 10.1016/0304-4165(71)90054-7. [DOI] [PubMed] [Google Scholar]
- 7.Liao T H, Barber G A. Biochim Biophys Acta. 1972;276:85–93. doi: 10.1016/0005-2744(72)90010-1. [DOI] [PubMed] [Google Scholar]
- 8.Reitman M L, Trowbridge I S, Kornfield S. J Biol Chem. 1980;255:9900–9906. [PubMed] [Google Scholar]
- 9.Overton K, Serif G S. Biochim Biophys Acta. 1981;675:281–284. doi: 10.1016/0304-4165(81)90238-5. [DOI] [PubMed] [Google Scholar]
- 10.Ripka J, Adamany A, Stanley P. Arch Biochem Biophys. 1986;249:533–545. doi: 10.1016/0003-9861(86)90031-7. [DOI] [PubMed] [Google Scholar]
- 11.Chang S, Duerr B, Serif G. J Biol Chem. 1988;263:1693–1697. [PubMed] [Google Scholar]
- 12.Reiter W-D, Chapple C C S, Somerville C R. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- 13.Dörmann P, Benning C. Arch Biochem Biophys. 1996;327:27–34. doi: 10.1006/abbi.1996.0088. [DOI] [PubMed] [Google Scholar]
- 14.Tenhaken R, Thulke O. Plant Physiol. 1996;112:1127–1134. doi: 10.1104/pp.112.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoyama K, Haase A M, Reeves P R. Mol Biol Evol. 1994;11:829–838. doi: 10.1093/oxfordjournals.molbev.a040166. [DOI] [PubMed] [Google Scholar]
- 16.Bastin D A, Reeves P R. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 17.Fry S C. The Growing Plant Cell Wall: Chemical and Metabolic Analysis. New York: Wiley; 1988. [Google Scholar]
- 18.Dellaporta S. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 19.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman T, DeBruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, Retzel E, Somerville C. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatas T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Barber G A. Biochim Biophys Acta. 1968;165:68–75. doi: 10.1016/0304-4165(68)90189-x. [DOI] [PubMed] [Google Scholar]
- 24.Lütcke H A, Chow K C, Mickel F S, Moss K A, Kern H F, Scheele G A. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keegstra K, Olson L J. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- 26.VonHeijne G. Eur J Biochem. 1983;133:17–21. [Google Scholar]
- 27.Zablackis E, York W S, Pauly M, Hantus S, Reiter W-D, Chapple C C S, Albersheim P, Darvill A. Science. 1996;272:1808–1810. doi: 10.1126/science.272.5269.1808. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe T, Takahashi K, Matsuda K. Agric Biol Chem. 1980;44:791–797. [Google Scholar]
- 29.Barber G A. Arch Biochem Biophys. 1971;147:619–623. doi: 10.1016/0003-9861(71)90420-6. [DOI] [PubMed] [Google Scholar]