Abstract
Generation of insect-resistant, transgenic crop plants by expression of the insecticidal crystal protein (ICP) gene of Bacillus thuringiensis (Bt) is a standard crop improvement approach. In such cases, adequate expression of the most appropriate ICP against the target insect pest of the crop species is desirable. It is also considered advantageous to generate Bt-transgenics with multiple toxin systems to control rapid development of pest resistance to the ICP. Larvae of yellow stem borer (YSB), Scirpophaga incertulas, a major lepidopteran insect pest of rice, cause massive losses of rice yield. Studies on insect feeding and on the binding properties of ICP to brush border membrane receptors in the midgut of YSB larvae revealed that cryIAb and cryIAc are two individually suitable candidate genes for developing YSB-resistant rice. Programs were undertaken to develop Bt-transgenic rice with these ICP genes independently in a single cultivar. A cryIAc gene was reconstructed and placed under control of the maize ubiquitin 1 promoter, along with the first intron of the maize ubiquitin 1 gene, and the nos terminator. The gene construct was delivered to embryogenic calli of IR64, an elite indica rice cultivar, using the particle bombardment method. Six highly expressive independent transgenic ICP lines were identified. Molecular analyses and insect-feeding assays of two such lines revealed that the transferred synthetic cryIAc gene was expressed stably in the T2 generation of these lines and that the transgenic rice plants were highly toxic to YSB larvae and lessened the damage caused by their feeding.
Keywords: insect resistance, resistance management, rice transformation, stable expression, synthetic cryIAc gene
Technologies of plant cell biology and molecular biology have attracted much attention because they provide a powerful and novel means to supplement as well as complement the traditional methods of crop improvement. The transgenic approach of plant genetic engineering provides access to an unlimited gene pool for the transfer of desirable genes between any two species of interest, irrespective of their evolutionary or taxonomic relation. The successful generation of transgenic insect-resistant crop plants by transferring the insecticidal crystal protein (ICP) gene of Bacillus thuringiensis (Bt; refs. 1–8), a Gram-positive, spore-forming bacterium, is one of the major breakthroughs in crop science that the new genetics has provided.
indica rice is one of the most important food crops (9) of the world. There are half a dozen different species of lepidopteran insects whose larvae bore into the tissue of rice plants, and the resultant damage causes massive yield losses (10). Yellow stem borer (YSB), Scirpophaga incertulas, is most certainly the major contributor to these losses. The first-stage YSB larvae cut small holes in the stem of the paddy plant and enter the plant tissue, remaining there for the rest of their lives as larvae and pupae. The larvae feed voraciously inside the stem, providing themselves complete protection against chemical pesticides. There are no known resistance genes in rice (11) that can be readily introduced through breeding methods into elite rice cultivars.
In this communication, we report the generation of transgenic indica rice IR64, an elite cultivar of the “new plant type” (12) with resistance to YSB larvae. We observed that the ICP of the CryIAc type was highly toxic to feeding larvae of YSB. We further discovered that the receptors in the brush border membrane of midgut epithelial cells of the YSB larvae showed differential binding to CryIAc toxin in relation to that of the most toxic CryIAb protein, thus offering a scope for development of a cultivar with two ICPs for resistance management performance (13–15). In the present program, we reconstructed the bacterial cryIAc gene and transferred it to indica rice IR64 to generate stable transgenic lines with high levels of the toxin. The presence of the CryIAc ICP in the transgenic rice plant successfully lessened the damage caused by feeding YSB larvae, and the trait was stably transmitted through seeds.
MATERIALS AND METHODS
Construction of the Synthetic cryIAc Gene.
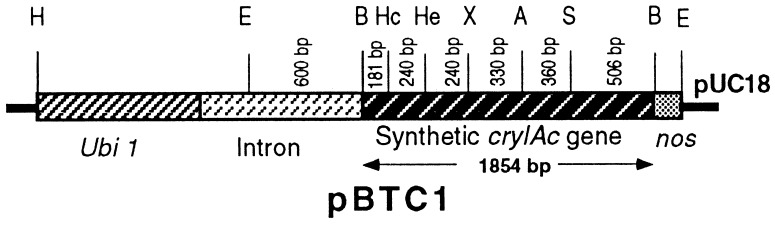
An engineered cryIAc gene was synthesized in six small fragments by using primers of 80–100 bp, following the “Recursive PCR” protocol (16). These six fragments were cloned in pUC18 vector and sequenced before ligation at the restriction sites, HincII, HaeII, XbaI, AvaI, and SstI, present in the gene to reconstruct the full-length cryIAc gene as a BamHI fragment. Additionally, during designing of the primer, the nucleotide context surrounding the initiation codon was changed to a plant-preferred one. The maize ubiquitin 1 promoter, the first intron of the maize ubiquitin 1 gene and the terminator of nopaline synthetase gene were fused to the modified cryIAc gene. The chimeric gene was cloned in a pUC18 vector to generate pBTC1 (Fig. 1).
Figure 1.
Chimeric construct of the synthetic cryIAc gene. A, AvaI; B, BamHI; E, EcoRI; H, HindIII; Hc, HincII; He, HaeII; nos, nopaline synthetase terminator; S, SstI; Ubi 1, ubiquitin 1 promoter; X, XbaI.
Rice Transformation.
Embryogenic calli were initiated from scutellar tissues of mature seed embryo of indica rice IR64. Essentially the protocol of Cao et al. (17) was followed for this purpose except the medium used was as described earlier by Basu et al. (18). One-month-old calli were bombarded with plasmid DNA pBTC1 and pCH (19) using a Bio-Rad 1000/He Biolistic gun at 1100 psi according to the method described in the Bio-Rad manual. After 24 h, the bombarded calli were transferred to fresh medium (18) containing 50 μg/ml hygromycin B. After two subcultures, each lasting 2 weeks, rapidly growing calli were separated and cultured in the selection medium for another three subcultures. Surviving calli were transferred to the preregeneration medium [Murashige and Skoog (MS) salts and vitamins (20)/3 mg/l 6-benzylamino purine/1 mg/l α-naphthaleneacetic acid/3% sucrose/1% agar, pH-5.7] and incubated under a 16-h light/8-h dark photoperiod (3000 lux) at 24°C for 10 days, followed by transfer to MS medium without any growth regulators for plant regeneration. The regenerated plantlets were cultured for 2 weeks in liquid 1/2 MS medium (containing half strength of MS salts and vitamins, and 2% sucrose, pH 5.8) before being transferred to soil.
Analyses of Transformants.
Isolation of plant DNA was carried out following the method of Dellaporta et al. (21). PCR analysis employed the following primers (5′primer, 5′-ACCAGATCATGGCCTCTCCAGTT-3′; 3′primer, 5′-TTCAAGATTGTACTCAGCCTCAAG-3′) for amplification of a 905-bp fragment from the cryIAc gene. PCR analysis was carried out essentially as described by Basu et al. (18) with the addition of the hot-start step. Northern analysis followed the protocol of Sambrook et al. (22) using the total RNA extracted from the plant tissue following the protocol of Chomczynski and Sacchi (23).
Detection of CryIAc Protein in Transgenic Plants.
Isolation of protein from plant materials and Western blot analysis as well as Sandwich ELISA of the transgenic plant materials were performed following the methods described earlier (24).
Insect Bioassay. Insect-feeding tests for sensitivity of plant-expressed CryIAc toxin using Helicoverpa armigera neonates.
It is known (data not shown) that the CryIAc toxin is very sensitive to the feeding neonates of Helicoverpa armigera, the cotton bollworm. An insect-feeding bioassay for the sensitivity of this toxin to the feeding neonates when mixed with the synthetic medium is a standard method in the laboratory. Based on this, we designed an experiment to judge the entomocidal potentiality of the toxin peptide expressed in the transgenic rice plant tissues on these sensitive insects. Thus, transgenic plant tissues were crushed in liquid nitrogen, and the total protein was extracted in protein extraction buffer (24) excluding phenylmethylsulfonyl fluoride, leupeptin, and pepstatin. Appropriate volumes of this crude protein preparation containing plant-expressed toxin was added to freshly made diet (16 g of agar, 53 g of yeast, 1.7 g of sorbic acid, 3.3 g of methyl parabenzoate, 5.3 g of ascorbic acid, 0.25 g of streptomycin, and 13.5 ml of 10% formaldehyde in 800 ml distilled water) for the cotton boll worm neonates to offer 2.5 ng of plant-expressed toxin/250 μl of the diet. After solidification, 5 neonates were placed in each well in 10 replicates containing the diet. Crude protein extracted from the untransformed plant was used as the negative control. Mortality of larvae and reduction of body weight of the individual insects were recorded after 5 days.
Bioassays of transgenic rice plant tissue fed to YSB larvae.
The natural and preferred sites of feeding YSB larvae are the pith and nodal tissue of the stem. Young, freshly cut stems with nodal regions of transgenic rice plants were placed on moist tissue paper in Petri dishes. Ten first-instar YSB larvae were distributed in each plate. The plates were incubated in the dark at 28°C in 70% relative humidity, and a change of diet was provided after 48 h. Results were noted after the indicated periods of incubation. In another set of bioassays, plants were individually raised in pots in small netted containments. Each potted plant at the booting stage was infested with five first-instar larvae of YSB. The larvae were introduced into the young culm through incisions. The fate of the larvae was monitored after 5 days.
RESULTS
Toxicity of CryIAc ∂-Endotoxin to YSB Larvae.
Estimation of the sensitivity of activated CryIAc toxin was carried out through a feeding assay, using the first-instar larvae of YSB on semi-artificial medium. Since monophagous YSB larvae cannot be cultured in the laboratory on artificial diet (25), we standardized a semi-artificial diet for rearing of YSB larvae for a week in the laboratory. Different concentrations of the purified and activated toxin were mixed with the diet, and the sensitivity of the larvae to toxin was estimated. The analysis revealed that the LD50 and ID50 to CryIAc are ≈32 ng and ≈25 ng per ml of the diet, respectively.
The toxicity of CryIAc was correlated with specific binding to the receptors of midgut brush border membrane vesicles of YSB. Ligand blot analysis of the brush border membrane vesicles of first-instar larvae of YSB, revealed differences in the binding patterns of CryIAc and CryIAb toxins. CryIAb distinctly bound to a band of ≈30 kDa (Fig. 2) when there was no such binding exhibited by CryIAc. The latter bound more characteristically with 165-, 130-, 97-, and 68-kDa proteins, a trait shared to a lesser degree by CryIAb. Heterologous competition assays (data not included) showed a degree of binding of labeled CryIAb in the presence of excess CryIAc. The CryIAc toxin sensitivity to feeding YSB larvae possessing specific binding sites in the receptor was considered an important criterion for choosing CryIAc, along with CryIAb, as one of the two candidate entomocidal genes for transferring to rice.
Figure 2.
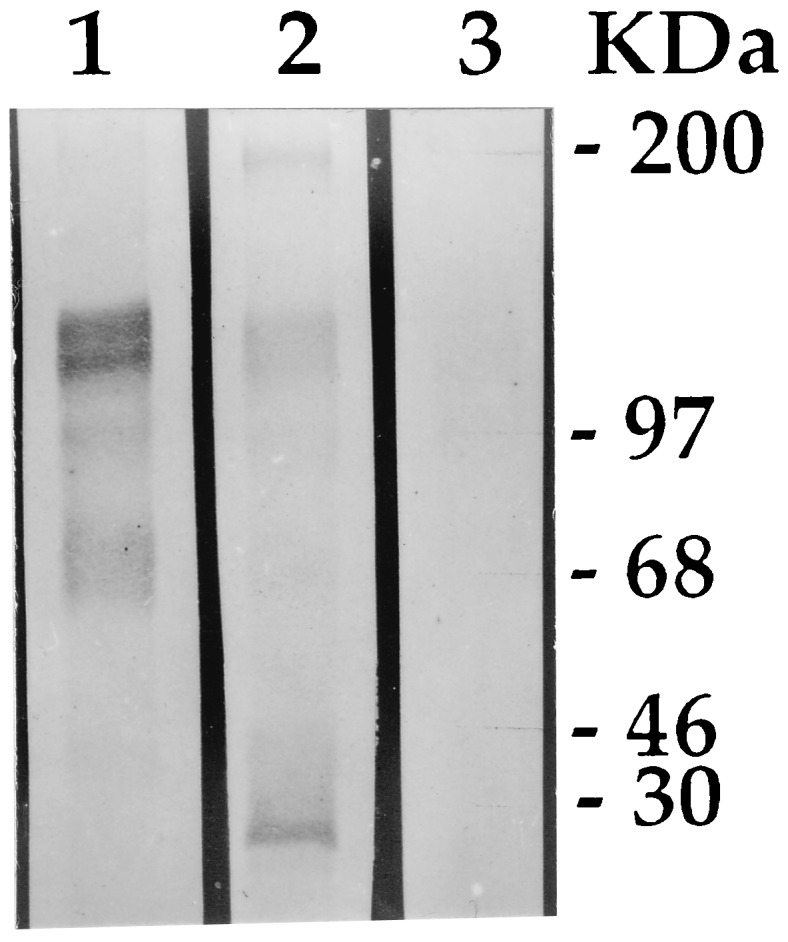
Ligand blot analysis of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate-extracted total brush border membrane vesicles proteins of first-instar YSB larvae. Equal amount of extracts in each lane were electrophoresed in an SDS/10% polyacrylamide gel and blotted onto nitrocellulose membrane; the membrane was cut into strips, and the strips were incubated in the respective toxins. The strips were probed further using anti-CryIA polyclonal rabbit IgG followed by anti-rabbit alkaline phosphatase (Sigma). Blots were processed using nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate mixture (Sigma). Lanes: 1, CryIAc; 2, CryIAb; 3, negative control (without toxin treatment).
Reconstruction of a Truncated cryIAc Gene.
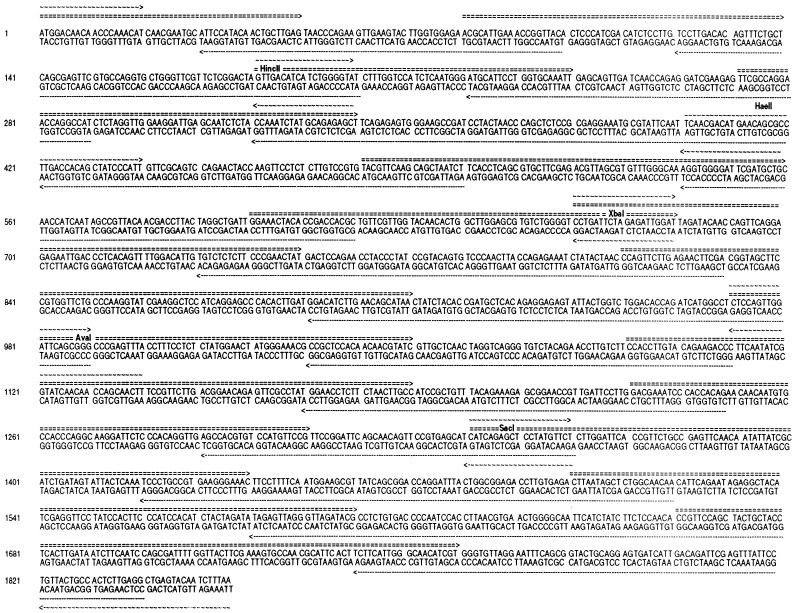
The coding sequences of the cryIAc gene were reconstructed to remove potential RNA processing sequences and polyadenylylation signals, and to optimize for more plant-preferred codon usage. Reconstruction was done essentially on the same line as that of Perlak et al. (7) with additional modifications. Care was taken to eliminate exon/intron junctions (donors) in the region between 1350 bp and 1854 bp that exist in the native gene. A complete account of the nucleotide sequences of the reconstructed cryIAc gene is provided in Fig. 3.
Figure 3.
Shown are the nucleotide sequences of the reconstructed cryIAc toxin gene with the restriction sites and the long primers (5′, ===; 3′, ······) and short primers (∼∼∼) that were used for reconstruction of the synthetic gene.
Expression of Synthetic cryIAc Gene in Transgenic Rice.
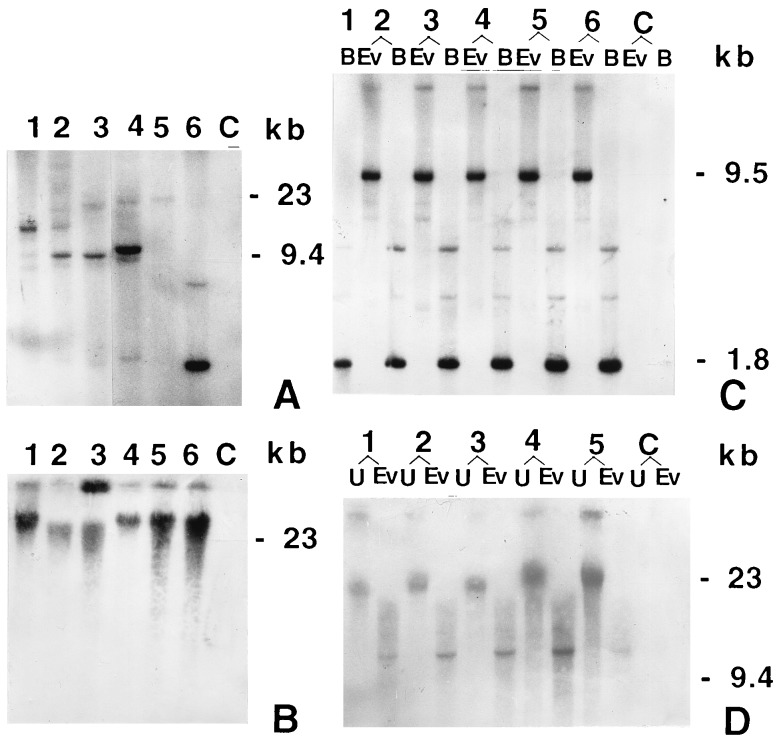
The combination of maize ubiquitin 1 promoter and the first intron of the coding region is capable of expressing foreign genes (26) at a high level in transgenic rice plants (27). DNA isolated from leaves of all the hygromycin-resistant plants was individually analyzed by PCR for the presence of the cryIAc gene. The PCR-positive plants were further tested through ELISA for the presence of the toxin peptide and were either discarded or grown to maturity inside the growth house in peat pots. Table 1 summarizes the results of our transformation experiments. On the basis of ELISA and Southern analysis (Fig. 4A), six primary independent transformants expressing high levels of the Bt toxin were identified (Table 1). All of them were fully fertile. Two independent transformants (events 3112 and 4017) were critically analyzed.
Table 1.
Summary of transformation experiments
| Calli mass bombarded, n | Hyg-resistant plants, n | T0crylAc+ plants, n | Transformants showing high ICP expression, n |
|---|---|---|---|
| 462 | 11 | 8 | — |
| 384 | 7 | 3 | — |
| 457 | 17 | 12 | 2 |
| 387 | 14 | 11 | — |
| 456 | 9 | 6 | 1 |
| 389 | — | — | — |
| 417 | 16 | 12 | 1 |
| 433 | 8 | 6 | — |
| 446 | 24 | 18 | 2 |
Hyg, hygromycin.
Figure 4.
Southern blot analysis results. The [32P]dCTP-labeled 660-bp DNA fragment from the N-terminal end of the gene was used as the probe for Southern blot analysis. Digested or undigested genomic DNA from each sample was fractionated through an 0.8% agarose gel and blotted to Hybond-N+ nylon membrane (Amersham). After hybridization, the membrane was exposed to Kodak X- Omat film at −70°C. B, BamHI; C, untransformed plant; Ev, EcoRV; U, undigested. (A) Genomic DNA (6 μg per lane) from T0 plants was digested with EcoRV (which has no restriction site in the transforming DNA). Six independent T0 transgenic plants are shown in lanes 1–6, events 3112, 3421, 4017, 756, 1321, and 1804, respectively. (B) Undigested genomic DNA (4 μg per lane) of T0 event 4017 (lane 1) and its five T1 progenies (lanes 2–6). (C) Genomic DNA (6 μg per lane) of the same plants as in B digested with BamHI and with EcoRV. Lanes: 1, T0 event 4017; 2–6, T1 progenies, events 4017-62, 4017-14, 4017-23, 4017-51, and 4017-39, respectively. The EcoRV fragments indicate that event 4017 has two linked integration sites. The fainter high molecular weight band probably represents a single copy, whereas the 9.4-kb band represents multiple copies of the gene in a single site. (D) Genomic DNA (6 μg per lane) of T0 event 3112 (lane 1) and four T1 progenies (lanes 2–5, events 3112-20, 3112-27, 3112-38, and 3112-42, respectively) were digested with EcoRV or not digested. Among the transgenic lines, the presence of a single band in the EcoRV-digested lanes is evident.
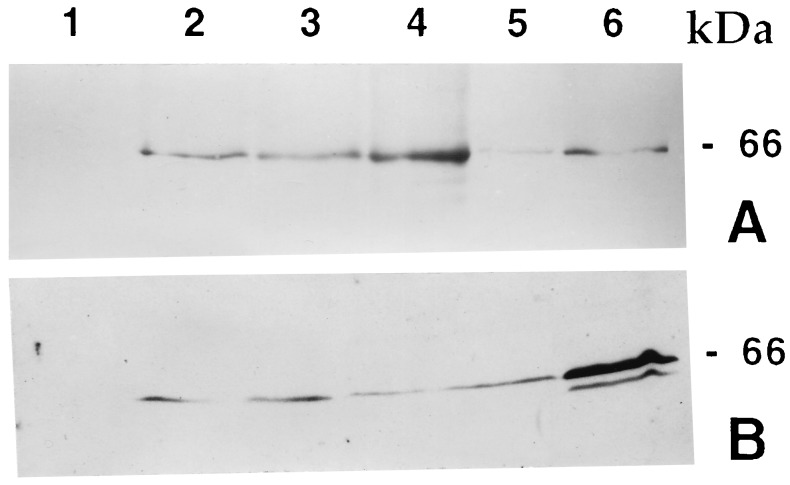
Northern analysis revealed that high levels of intact full-length cryIAc transcripts were present, and no degradation of mRNA was detected (Fig. 5). An ELISA was performed to estimate the levels of the CryIAc protein present in leaves and nodal tissues of the R0 plants of events 3112 and 4017. Western blot analysis indicated the presence of single band of ≈66 kDa (Fig. 6A). From the band intensity, we estimated that the toxin peptide constituted 0.02–0.025% of the total soluble protein of the plant tissue.
Figure 5.
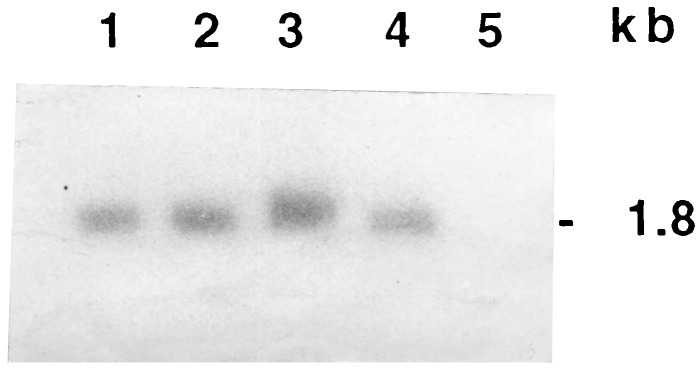
Northern blot analysis of total RNA of transgenic plants. RNA from T0 plants of events 3112 (lane 1) and 4017 (lane 2), two T1 progenies of event 3112 (lanes 3 and 4), and untransformed IR64 plant (lane 5). One of the T1 transgenics (lane 3) shows a higher level of transcription than the others. The full-length synthetic cryIAc gene was used as a probe.
Figure 6.
Immunoblot analysis of protein. Total protein from tissue extracts (50 μg) was boiled with SDS gel loading buffer and loaded onto an SDS/10% polyacrylamide gel. For a positive control, a known amount of activated (trypsin-digested) CryIAc protein was loaded in one lane; untransformed rice tissue extract was used as negative control. (A) Four T0 lines and untransformed rice plant. Lanes: 1, untransformed; 2–4, protein from events 756, 4017, and 3112, respectively; 5, protein from event 1146, a low expressing transgenic line; 6, positive control. (B) Four T1 progenies of event 3112. Lanes: 1, untransformed plant; 2–5, T1 progenies; 6, positive control.
Inheritance of the cryIAc Gene in Transgenic Plants.
Inheritance of the transgene through seed was studied by monitoring T1 individuals for the presence of the cryIAc gene (PCR analysis) and for expression of the gene (ELISA). The inheritance of T1 plants of events 3112 and 4017 showed a 1:1 segregation ratio for cryIAc gene expression when crossed with untransformed IR64 plants, whereas a 3:1 segregation ratio was observed when selfed. This observation and Southern blot analyses data (Fig. 4 B–D) indicated that the transferred cryIAc gene in event 3112 had integrated into a single site of the genome and in event 4017 into two sites of a single chromosome. Levels of expression of T1 progenies of event 3112 varied from 58 to 240 ng per mg of total soluble protein depending upon the developmental growth stages of the plant, season of growth, and plant parts. ELISA analyses of 33 T2 plants indicated that the expression remained at par with that of T1 plants (data not shown). Forty-two T1 plants of event 4017 were analyzed for levels of expression by ELISA and Western blot analysis (Fig. 6B). Levels of expression ranged from 0.018 to 0.34% of total soluble protein (data not shown). Progenies of T1 plants showed the presence of the transgene in the T2 population.
Evaluation of Biological Activity of the Toxin Peptide in Transgenic Rice Plants.
Entomocidal activity of the toxin peptide in the tissues of three expressive T1 transgenic plants of each of the two primary transgenic lines was assayed after 5 days (Table 2) through a feeding assay of neonates of H. armigera. Adverse developmental response to the toxin present in the transgenic rice tissue was evident within 60 h of the feeding assay (Fig. 7A).
Table 2.
Feeding assay of H. armigera neonates when fed with CryIAc protein (2.5 ng/250 μl of diet) present in transgenic rice plant tissue
| ICP line | Weight of the young stem tissue, g | Expressed toxin in % of the total soluble protein | Mortality, % | Body weight reduction, % |
|---|---|---|---|---|
| T0 3112 | 0.21 | 0.01 | 44 | 79.3 |
| T1 3112-20 | 0.17 | 0.0095 | 46 | 80.2 |
| T1 3112-27 | 0.24 | 0.012 | 42 | 78.8 |
| T1 3112-38 | 0.18 | 0.024 | 40 | 82.2 |
| T0 4017 | 0.2 | 0.012 | 42 | 82.2 |
| T1 4017-6 | 0.18 | 0.01 | 40 | 79.7 |
| T1 4017-14 | 0.19 | 0.013 | 38 | 84.2 |
| T1 4017-23 | 0.192 | 0.018 | 44 | 79.6 |
| Negative control | 0.18 | 0 | 0 | 0 |
| Positive control | 0.2 | — | 50 | 82.1 |
Figure 7.
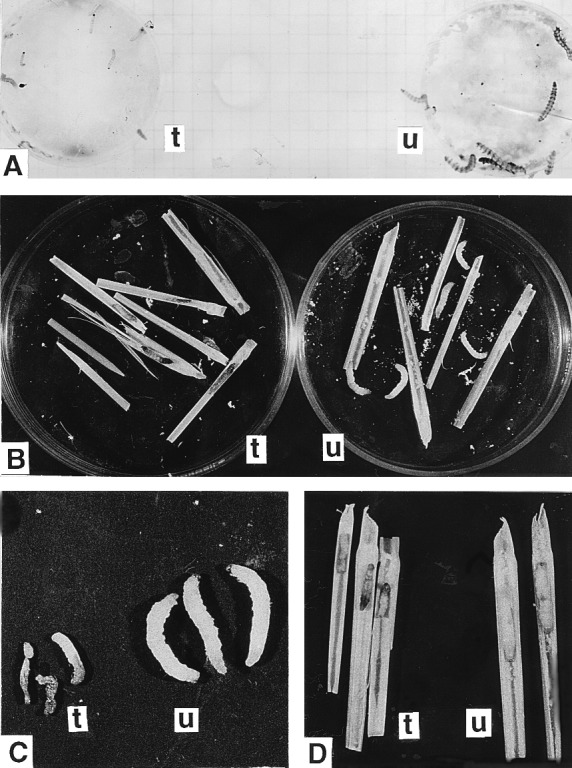
Insect bioassay. (A) H. armigera neonates fed on an artificial diet containing toxin extracted from transgenic rice plant tissue (t) and untransformed rice plant tissue (u) after 60 h. (B and C) YSB larvae fed on transgenic plant parts showing browning and severe growth retardation (t) in comparison to larvae fed on untransformed plant (u) parts (B), and mortality resulting from transgenic-plant feeding (C). (D) Larvae fed on T2 plants showing mortality (t) and larvae fed on untransformed plant (u) revealing no such effect.
Young nodal stem pieces of three T1 ICP lines each of events 3112 and 4017 were offered to first instar larvae of YSB. Larvae fed on untransformed tissue grew well with slight mortality caused by handling (Table 3). However, larvae fed on transgenic plant tissues suffered from severe deleterious effects (Fig. 7 B and C). Mortality ranged between 75.9% and 92.4%, and surviving insects suffered reduction in body weight ranging from 70.4% to 83.4% and failed to pupate. When fed directly on the plant, the larvae did not survive or were severely affected as indicated by the typical symptom of browning (Fig. 7D) before ultimate death.
Table 3.
Feeding assay of first-instar larvae of YSB on young stem tissue (0.2 g) of transgenic plants
| T1 ICP line | Toxin content in total of soluble protein, % | Mortality, % | Body weight reduction, % |
|---|---|---|---|
| 3112-24 | 0.015 | 75.9 | 79.2 |
| 3112-17 | 0.018 | 85.8 | 70.4 |
| 3112-38 | 0.021 | 79.2 | 80.6 |
| 4017-39 | 0.014 | 82.5 | 81.5 |
| 4017-51 | 0.013 | 89.1 | 79.2 |
| 4017-62 | 0.024 | 92.4 | 83.4 |
| C | 0 | 6.6 | 0 |
The fate of the larvae after 5 days was recorded in three sets of experiments allowing 10 larvae in each sets. Larvae fed on untransformed nodal stem pieces (C) served as the control.
DISCUSSION
The initial feeding assay revealed that CryIAc ICP is toxic to the larvae of YSB. The ligand blotting experiments further established that distinct differences exist in the binding pattern between CryIAc and CryIAb to YSB brush border membrane vesicles proteins. Binding-site detection by blotting experiments has been demonstrated to be quite specific (28) and, hence, informative. Binding site specificity is not a simple system in which each toxin binds to a unique receptor. A high degree of heterogeneity among binding sites exists in some species, suggesting that some sites may bind a single toxin, whereas sites others may bind two or more toxins. Similarly, specific toxins may bind to more than one site in some insect species.
It has been observed (29, 30) that significant differences exist between the binding patterns of CryIAc and CryIAb to the lepidopteran insect midgut brush border membrane. That this difference can be exploited for the development of Bt-resistant strategy has been indicated (13, 14, 31). This approach has been used in this study. Attempts to develop independent transgenic IR64 plants with cryIAc as well as cryIAb genes were thus undertaken. This report concerns development of IR64 transgenics with the cryIAc gene. The significance of identifying the most effective ICP has been pointed out (32). Since native cry genes have poor coding capacity in plants (33), the cryIAc gene was duly reconstructed.
Past efforts (34, 35) to enhance the level of expression of the Bt-toxin gene in plants with the help of 5′ untranslated region/enhancer or strong plant promoter elements were not successful, unlike similar efforts for other bacterial genes (36). Northern blot analysis of poly(A)+ mRNA or total RNA of the cyclohexamide-treated plant cells of cryIAc-transformed plants often showed the presence of small transcripts (37). On the other hand, half-lives of mRNA of in vitro synthesized cryIAb and bar genes in electroporated tobacco protoplasts showed that low accumulation of Bt RNA is due to some mechanisms that operate between transcription initiation and mRNA degradation and is not due to the instability of the Bt-toxin RNA (37). This result was further confirmed (38) by a nuclear run-on analysis and reverse transcriptase–PCR. Reverse transcriptase–PCR with the total RNA isolated from the electroporated protoplast indicated that, in the case of unmodified cryIAb, there are two exon/intron donors (gt) and two intron/exon acceptors (ag) lying beyond 1350 bp in the cryIAb gene that result in generation of shorter transcripts when compared with the modified cryIAb gene, where the sequence around gt has been altered. Both the native as well as the modified cryIAc gene of Perlak et al. (7) in the same region contained four such intron/exon acceptors (ag) and eight and seven exon/intron donors (gt), respectively. Site detection analysis was carried out with the help of the pcgene program, where the cutoff values for exon/intron (donor) junctions and intron/exon (acceptor) junctions were set at −15 and −22, respectively. In the present reconstructed cryIAc gene, all the intron/exon (ag) junctions in the region beyond 1350 bp were eliminated, and the number of exon/intron (gt) junctions were reduced to 3. Another important area that called for modification to bring in mRNA stability has been motifs such as the ATTTA sequences contained in cry genes (39). They are responsible for acting as a signal for mRNA decay, although the minimal domain appeared to be longer than ATTTA alone (37). Polyadenylylation signal sequences that have resemblances within the A+T-rich Bt gene sequences are suspected to be limiting transcription beyond those points. Reverse transcriptase–PCR analysis identified two such sites within both cryIAc and cryIAb genes that resulted in the generation of short transcripts of ≈600 and ≈900 bp (37). Attempts to reduce such process-limiting possibilities were important when the cryIAc gene was reconstructed. Finally, codon usage of the bacterial cryIAc gene in plant creates a distinct possibility for stalling ribosomes at the rare codon, thereby making the mRNA more vulnerable to degradation. Thus, all such expression-limiting elements present in the native cry gene in plant that can destabilize the Bt-toxin mRNA and limit expression of the cry gene in plant were mostly replaced by the use of translationally neutral plant preferred codons. As the result of the reconstruction, the new cryIAc gene in the region beyond 1350 bp maintained 78% homology with the native cryIAc gene and, the GC content was raised to 44.95%. Furthermore, this reconstructed gene in the same region had 52% of the 165 codons altered, whereas in the earlier reconstructed cryIAc gene (7), only 28% codons were altered. The reconstructed cryIAc gene contained only the first 1854 bp of the 3537-bp-long cryIAc bacterial gene and has 59% translationally neutral codons compared with the native cryIAc gene.
The reconstructed cryIAc gene was transferred to IR64, an elite breeding line of indica rice. As the result of the gene transfer, six independent Bt-transgenic rice lines expressing high levels of ICP could be generated. Analysis of the T2 population of two of the independent Bt-transgenic lines segregating in a Mendelian pattern of inheritance revealed production of stable toxin peptide. The level of toxin present is distinctly higher than the lethal dose of the toxin required for mortality of the YSB larvae. Insect bioassays conducted with YSB larvae showed that the toxin present in the transgenic plant tissue indeed offers plant protection when compared with nontransgenic plants. Thus, so long as no clear, practical utility of a very high level of expression (3, 40) of Bt toxin in transgenics emerges, the level of expression obtained in this study appears adequate for the purpose of protecting plants. It is interesting to note in this context that Wunn et al. (5), using the synthetic cryIAb gene construct of Koziel et al. (3), achieved lepidopteran insect resistance in rice even though the level of expression was considerably low.
To date, there is insufficient field data on the absolute potential for transgenic Bt plants on their own. It seems appropriate not to use the Bt plants in a stand-alone mode but to incorporate them in pest management strategies. A cautious approach to the use of Bt genes in transgenic crops and of strategies adapted to extend the useful life of Bt-transgenic plants merits consideration. Thus, resistance management is considered to be an important aspect of Bt plants in the integrated pest management concept as it can contribute significantly toward the desired robustness of insect resistance based on genes for insect management.
The strategy for resistance management to delay or prevent adaptation of YSB to the IR64 Bt transgenics has been the hallmark of the present transgenic approach. Two of the four strategies as earlier suggested (13), based upon which resistance management within integrated pest management should be effected, can be attained from the approach reported here. Emphasis has been placed on more than one source of a mortality mechanism and, concurrently, on the reduction of selection pressure for each mortality mechanism. YSB is not known to have any hosts other than rice. Transgenic lines with high levels of Bt toxin could be potentially effective when grown in conjunction with untransformed lines to serve as refuges (41).
To our knowledge, the work presented here is the first report of success in developing insect-resistant transgenics with a synthetic cryIAc gene in any cereal crop to date.
Acknowledgments
We thank Dr. G. S. Khush (International Rice Research Institute, Manila, The Philippines) for providing the IR64 seed material and Dr. H. Uchimiya (University of Tokyo, Tokyo) for the gift of plasmid pCH. We also thank Dr. John C. O’Toole (Rockefeller Foundation, New York) for providing relevant scientific documents and the scientists of the Entomology Division of the Central Rice Research Institute, Cuttack, India, and the Directorate of Rice Research, Hyderabad, India, for technical discussions. Technical support from Arup K. Dey, Sugata Munshi, Manoj Aditya, and Bishnupada Pradhan of our laboratory and Khudiram Bhuin of Seva-Bharati, West Bengal, are gratefully acknowledged. Financial support from the Rockefeller Foundation and the Department of Biotechnology, Government of India, to S.K.S. and from the Council for Scientific & Industrial Research, India. to P.N. are acknowledged gratefully.
ABBREVIATIONS
- ICP
insecticidal crystal protein
- Bt
Bacillus thuringiensis
- YSB
yellow stem borer
Footnotes
References
- 1.Perlak F J, Fuchs R L, Dean D A, McPherson S L, Fischhoff D A. Proc Natl Acad Sci USA. 1991;88:3324–3328. doi: 10.1073/pnas.88.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton D W, Havstad P K, Kemp J D. Transgenic Res. 1992;1:228–236. doi: 10.1007/BF02524753. [DOI] [PubMed] [Google Scholar]
- 3.Koziel M G, Beland G L, Bowman C, Carozzi N B, Crenshaw R, Crossland L, Dawson J, Desai N, Hill M, Kadwell S, Launis K, Lewis K, Maddox D, McPherson K, Meghji M R, Merlin E, Rhodes R, Warren G W, Wright M, Evola S V. Bio/Technology. 1993;11:194–200. [Google Scholar]
- 4.Fujimoto H, Itoh K, Yamamoto M, Kyojuka J, Shimamoto K. Bio/Technology. 1993;11:1151–1155. doi: 10.1038/nbt1093-1151. [DOI] [PubMed] [Google Scholar]
- 5.Wunn J, Kloti A, Burkhardt P K, Ghosh-Biswas G C, Launis K, Iglesias V A, Potrykus I. Bio/Technology. 1996;14:171–176. doi: 10.1038/nbt0296-171. [DOI] [PubMed] [Google Scholar]
- 6.Nayak, P., Basu, D., Das, S., Ghosh, D., Basu, A. & Sen, S. K. (1997) Proceedings of the International Rice Genetics Symposium in press.
- 7.Perlak F J, Deaton R W, Armstrong T A, Fuchs R L. Bio/Technology. 1990;8:939–943. doi: 10.1038/nbt1090-939. [DOI] [PubMed] [Google Scholar]
- 8.Adang M J, Brody M S, Cardineau G, Eagan N, Roush R T, Shewmaker C K, Jones A, Oakes J V, McBride K E. Plant Mol Biol. 1993;21:1131–1145. doi: 10.1007/BF00023609. [DOI] [PubMed] [Google Scholar]
- 9.Ayres N M, Park W D. Crit Rev Plant Sci. 1994;13(3):219–239. [Google Scholar]
- 10.Herdt R W. In: Rice Biotechnology. Khush G S, Toenniessen G H, editors. Wallingford, U.K.: CAB International; 1991. pp. 19–54. [Google Scholar]
- 11.Khan Z R, Litsinger J A, Barrion A T, Villanenva F F D, Fernandez N J, Taylo L D. World Bibliography of Rice Stem Borers. Manila, The Philippines: IRRI; 1991. pp. 1794–1990. [Google Scholar]
- 12.IRRI—International Rice Research Institute. IRRI Toward 2000 and Beyond. Manila, The Philippines: IRRI; 1989. pp. 36–37. [Google Scholar]
- 13.McGaughey W H, Whalon M E. Science. 1992;258:1451–1455. doi: 10.1126/science.258.5087.1451. [DOI] [PubMed] [Google Scholar]
- 14.Gould F. Trends Biotechnol. 1988;6:515. [Google Scholar]
- 15.Van Mellaert, H., Botterman, J., Van Rie, J. & Joos, H. (1991) Eur. Patent Appl. 19401427.1.
- 16.Chrisostomos P, Pearl L H. Protein Eng. 1992;5:827–829. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 17.Cao J, Duan X, McElroy D, Wu R. Plant Cell Rep. 1993;11:586–591. doi: 10.1007/BF00233098. [DOI] [PubMed] [Google Scholar]
- 18.Basu D, Das S, Nayak P, Sen S K. Curr Sci. 1995;68:1140–1144. [Google Scholar]
- 19.Goto F, Toki S, Uchimiya H. Transgenic Res. 1993;2:300–305. [Google Scholar]
- 20.Murashige T, Skoog S. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 21.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Kar, S., Basu, D., Das, S., Ramakrishnan, N. A., Mukherjee, P., Nayak, P. & Sen, S. K. (1997) Transgenic Res., in press.
- 25.Theunis, W., Aguda, R. M., Lambert, B. & Peferoen, M. IRRI/PGS B. T. Project: An Overview (IRRI, Manila, The Philippines).
- 26.Christensen A H, Quail P H. Trans Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 27.Toki S, Takamatsu S, Nojiri C, Ooba S, Anzai H, Iwata M, Christensen A H, Quail P, Uchimiya H. Plant Physiol. 1992;100:1503–1507. doi: 10.1104/pp.100.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vadlamudi R K, Ji T H, Bulla L A., Jr J Biol Chem. 1993;268:12334–12340. [PubMed] [Google Scholar]
- 29.Oddou P, Hartmann H, Geiser M. Eur J Biochem. 1991;202:673–680. doi: 10.1111/j.1432-1033.1991.tb16422.x. [DOI] [PubMed] [Google Scholar]
- 30.Garczynski S F, Crim J W, Adang M J. Appl Environ Microbiol. 1991;56:1378–1385. doi: 10.1128/aem.57.10.2816-2820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani G S. Genetics. 1985;109:761–774. doi: 10.1093/genetics/109.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacIntosh S C, Stone T B, Sims S R, Hunst P L, Greenplate J T, Marrone P G, Perlak F J, Fischhoff D A, Fuchs R L. J Invertebr Pathol. 1990;56:258–266. doi: 10.1016/0022-2011(90)90109-j. [DOI] [PubMed] [Google Scholar]
- 33.Murray E E, Rochlean T, Eberle M, Stock C, Sekar V, Adang M. Plant Mol Biol. 1991;16:1035–1050. doi: 10.1007/BF00016075. [DOI] [PubMed] [Google Scholar]
- 34.Carozzi N B, Warren G W, Desai N, Jayne S M, Lotstein R, Rice D A, Evola S, Koziel M G. Plant Mol Biol. 1992;20:539–548. doi: 10.1007/BF00040612. [DOI] [PubMed] [Google Scholar]
- 35.Parrott W A, All J N, Adang M J, Bailey M A, Boerma H R, Stewart C N., Jr In Vitro Cell Dev Biol. 1994;30:144–149. [Google Scholar]
- 36.Walbot V, Gallie D. In: Rice Biotechnology. Khush G S, Toenniessen G H, editors. Wallingford, U.K.: CAB International; 1991. pp. 225–251. [Google Scholar]
- 37.Dien S H, De Rocher E J, Green P J. In: Genetic Engineering. Setlow J K, editor. Vol. 18. New York: Plenum; 1996. pp. 83–99. [DOI] [PubMed] [Google Scholar]
- 38.Van Aarsen R, Soetaert, Stam M, Dockx J, Gosseli V, Seurinck J, Reynaerts A, Cornelissen M. Plant Mol Biol. 1995;28:513–524. doi: 10.1007/BF00020398. [DOI] [PubMed] [Google Scholar]
- 39.Ohme-Takagi M, Taylor C B, Newman T C, Green P J. Proc Natl Acad Sci USA. 1993;90:11811–11815. doi: 10.1073/pnas.90.24.11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride K E, Svab Z, Schaff D J, Patrick S H, Stalker D M, Maliga P. Bio/Technology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- 41.Denholm I, Rowland M W. Annu Rev Entomol. 1992;37:91. doi: 10.1146/annurev.en.37.010192.000515. [DOI] [PubMed] [Google Scholar]