Abstract
We have developed a new plant vector system for repeated transformation (called MAT for multi-auto-transformation) in which a chimeric ipt gene, inserted into the transposable element Ac, is used as a selectable marker for transformation. Selectable marker genes conferring antibiotic or herbicide resistance, used to introduce economically valuable genes into crop plants, have three major problems: (i) the selective agents have negative effects on proliferation and differentiation of plant cells; (ii) there is uncertainty regarding the environmental impact of many selectable marker genes; (iii) it is difficult to perform recurrent transformations using the same selectable marker to pyramid desirable genes. The MAT vector system containing the ipt gene and the Ac element is designed to overcome these difficulties. When tobacco leaf segments were transformed and selected, subsequent excision of the modified Ac produced marker-free transgenic tobacco plants without sexual crosses or seed production. In addition, the chimeric ipt gene could be visually used as a selectable marker for transformation of hybrid aspen (Populus sieboldii × Populus grandidentata). The chimeric ipt gene, therefore, is an attractive alternative to the most widely used selectable marker genes. The MAT vector system provides a promising way to shorten breeding time for genetically engineered crops. This method could be particularly valuable for fruit and forest trees, for which long generation times are a more significant barrier to breeding and genetic analysis.
Keywords: plant genetic engineering, DNA transformation, selectable marker, ipt gene, transposon Ac
Dominant genes encoding either antibiotic or herbicide resistance are widely used as selectable markers in plant transformation (1). The antibiotics and herbicides that select rare transgenic cells from nontransgenic cells generally have negative effects on proliferation and differentiation. These agents may retard differentiation of adventitious shoots during the transformation process. Some plant species are insensitive to or tolerant of the selective agents, and therefore, it is difficult to separate the transformed and untransformed cells or tissues. Therefore, it is difficult to find appropriate selectable markers and to establish optimal conditions for transformation of such difficult species (2–4). Selectable marker genes remain in transgenic plants, and their gene products need to be assessed for safety and environmental impact (5–7).
It is difficult to introduce a second gene of interest into a transgenic plant that already contains a resistance gene as a selectable marker. There are a large number of desirable traits and genes worth incorporating into plants, but only a limited number of selectable marker genes are available for practical use. The problem becomes even more difficult if one wants to introduce a number of genes, and it is impossible to introduce them simultaneously (1, 8). It is desirable, therefore, to develop a system for the removal of selectable marker genes to produce environmentally safe transgenic plants and pyramid a number of transgenes by repeated transformation. We developed the MAT (multi-auto-transformation) vector system, in which the selectable marker is composed of a chimeric ipt gene inserted into the maize transposable element Ac to overcome some of the difficulties of the current transformation methods.
The ipt gene encodes the enzyme isopentenyl transferase and is located on Ti-plasmids of Agrobacterium tumefaciens. This enzyme catalyzes the condensation of isopentenyl pyrophosphate with AMP to produce isopentenyl AMP, a precursor of several cytokinins (9, 10). Cytokinins stimulate organogenesis in many cultured plant tissues and are widely used to regenerate transgenic plants from cultured cells after transformation. Many studies have been performed to manipulate endogenous cytokinin levels using the ipt gene (11, 12). When a chimeric ipt gene under the control of the (CaMV) 35S promoter was introduced into cells of potato (13), cucumber (14), and several Nicotiana species (15), transgenic cells proliferated and adventitious shoots differentiated in hormone-free medium. These transgenic plants exhibited an (ESP) and loss of apical dominance. Therefore, it is easy to detect visually transgenic plants that carry a functional ipt gene. Chimeric ipt genes are not commonly used as selectable markers because the resulting transgenic plants lose apical dominance and are unable to root due to overproduction of cytokinins.
The maize transposable element Ac has the ability to move to new locations within a genome (16). We used Ac to remove the chimeric ipt gene from transgenic cells of ESP shoots after transformation. In the transposition process, about 10% of the Ac elements that excise do not reinsert and therefore disappear, or reinsert into a sister chromatid that is subsequently lost by somatic segregation (17). When the chimeric ipt gene is inserted into Ac in the MAT vector, the chimeric ipt gene may transpose or become lost along with Ac in transgenic cells. Consequently, we were able to obtain phenotypically normal transgenic plants that had lost the chimeric ipt gene. In this paper, we demonstrate that marker-free transgenic plants can be visually selected by using the chimeric ipt gene as a marker gene in tobacco plants and hybrid aspen.
MATERIALS AND METHODS
Construction of MAT Vector Plasmids (pNPI106).
The 20.4-kb PstI fragment from the T-DNA (portion of the Ti plasmid that is transferred to plant cells) of A. tumefaciens P022 (18) was cloned into an unique PstI site of pUC 7. From this plasmid, the 1.9-kb BamHI–PstI fragment containing the entire ipt gene was excised and cloned into the BamHI–PstI site of pUC119. From this plasmid, the 1.3-kb RsaI fragment containing the coding sequence and terminator of the ipt gene was excised and cloned into the unique SmaI site of pUC119. The BamHI–SacI fragment was inserted to the BamHI–SacI site of pBI121 (CLONTECH), downstream of the CaMV 35S promoter. The HindIII–SacI fragment containing the chimeric ipt gene with the CaMV 35S promoter was converted to a blunt end fragment with T4 polymerase I and inserted to the unique blunted BamHI site of Ac in pCKR97 (19). Finally, PstI fragments containing the Ac and the inserted ipt gene with the CaMV 35S promoter were cut from this plasmid and ligated to the unique SseI site of the binary vector pBI121. This plasmid was called MAT vector pNPI106 (Fig. 1).
Figure 1.
Diagram of MAT vector pNPI106. Plasmid pNPI106 has a “hit and run” cassette in which the chimeric ipt gene with 35S promoter is inserted into Ac as a selectable marker. The gusA and nptII genes are unselected markers in these experiments. Arrows, PCR primers (see Fig. 3); 35S-P, CaMV 35S promoter; ipt, isopentenyl transferase gene; T, isopentenyl transferase terminator; N-P, nopaline synthase promoter; N-T, nopaline synthase terminator; nptII, neomycin phosphotransferase gene; gusA, β-glucuronidase gene.
Plant Transformation.
Plasmid pNPI106 was transformed into A. tumefaciens LBA4404 (20) using a freeze–thaw method (21). Leaves of Nicotiana tabacum cv. xanthi were sterilized with 1% sodium hypochlorite and cut into leaf segments of approximately 0.8 × 0.8 cm. These leaf segments were inoculated for 1 min with an overnight culture of bacteria diluted to OD630 = 0.25 and put on a sterilized filter paper to remove the bacteria in suspension. The infected leaf segments were cocultivated for 3 days on hormone-free Murashige-Skoog (MS) medium containing 2% sucrose, 0.8% agar, and 50 mg/liter acetosyringone and then transferred to hormone-free MS medium containing 500 mg/liter carbenicillin but no kanamycin (nonselective medium). When adventitious shoots were regenerated, they were separated from the leaf segments, transferred to fresh nonselective medium (with carbenicillin), and cultured under 3000 lux at 25°C.
A stem of an aseptically flask-grown hybrid aspen “Kitakami Hakuyo” (Populus sieboldii × Populus grandidentata) (22) was cut to obtain an internodal stem segment 5 mm in length, further cut lengthwise in two, and then inoculated with the same strain of A. tumefaciens (LBA4404 containing pNPI106) used for tobacco plants. These infected stem segments were transferred to nonselective medium for hybrid aspen (hormone-free modified MS medium with 800 mg/liter ammonium nitrate and 2 g/liter potassium nitrate). Regenerated shoots were cultured in the same medium. The normal shoots were transferred to root-inducing medium (2/3 MS medium/2% sucrose/0.25% Gelrite/0.05 mg/liter 3-indolebutyric acid).
Kanamycin and Histochemical Assays.
Leaf segments were placed on MS agar medium containing l mg/liter benzyladenine, 0.2 mg/liter 1-napthalene, and 200 mg/liter kanamycin. After 1 month in culture, the formation of callus and adventitious shoots was observed on the leaf segments. Histochemical assays for β-glucuronidase (GUS) activity were performed as described (23).
DNA Analysis.
Genomic DNA samples were prepared from transgenic tobacco shoots and plants using cetyltrimethylammonium bromide (24), and used as templates for PCR amplification and Southern blot analysis. The PCR mixture contained 1 μg of genomic DNA, each primer at a concentration of 0.5 μM, 10 mM Tris·HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 1% Triton X-100, 0.1 mM dNTP, and 1.25 units of Taq DNA polymerase (TAKARA shuzo Otsu, Shiga, Japan) in a total volume of 50 μl, overlaid with 40 μl of mineral oil. After the mixtures were heated at 94°C for 1.5 min, the amplification occurred during 30 cycles of 1 min at 94°C, 2 min at 60°C, and 3 min extension at 72°C. Reaction products were resolved by electrophoresis through 1.8% agarose gel. The sequences of the two primers used to detect excision of the modified Ac element were 5′-TTGTCAAGACCGACCTGTCC-3′ (Fig. 1, primer a) and 5′-TGCATCGGCGAACTGATCGT-3′ (Fig. 1, primer b). The expected fragment size of the empty donor is approximately 3 kb. The sequences of the two primers to detect the ipt gene in were 5′-CTTGCACAGGAAAGACGTCG-3′ (Fig. 1, primer c) and 5′-AATGAAGACAGGTGTGACGC-3′ (Fig. 1, primer d). The expected fragment size of the amplified DNA segment is 800 bp. Genomic DNA (10 μg) was digested with HindIII and separated by 0.8% agarose gel electrophoresis, and then analyzed on Southern blots. The DNA probe, a fragment of the nptII gene, was labeled by PCR using DIG-DUTP (Boehringer Mannheim). Southern blots were carried out using DIG Easy Hyb (hybridization solution) and DIG Wash and Block Buffer Set (Boehringer Mannheim). The sequences of the two primers used to amplify a labeled fragment of the nptII gene were 5′-AGAGGCTATTCGGCTATGAC-3′ and 5′-CCATGATATTCGGCAAGCAG-3.
RESULTS
MAT Vector Constructs and Transformation of Tobacco Plants.
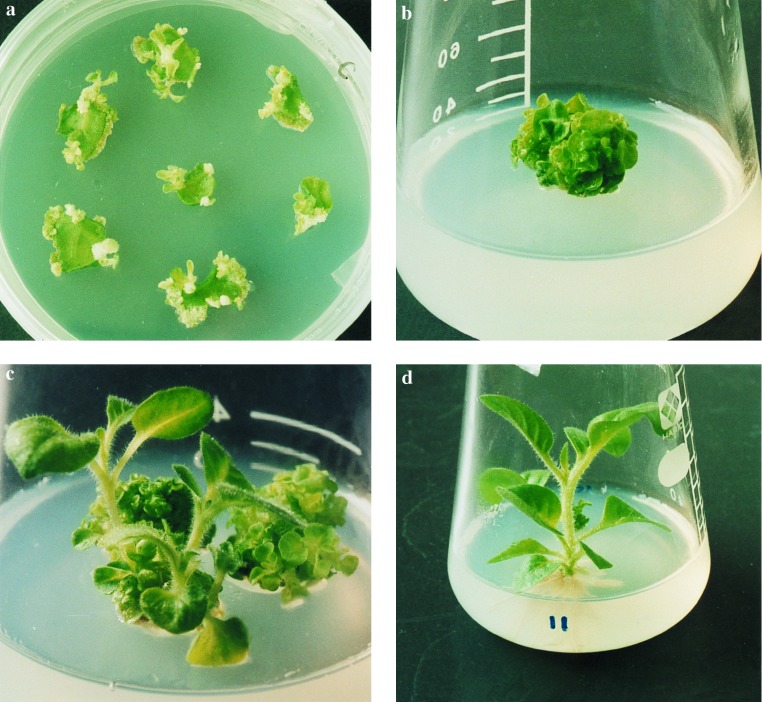
The chimeric ipt gene with a 35S promoter was inserted into Ac to create the vector pNPI106. The modified Ac cassette was used for selection and called the hit and run cassette of the MAT vector. Both gusA and nptII genes were ligated outside of the hit and run cassette and used as a model for desirable genes. The pNPI106 plasmid in A. tumefaciens LBA4404 was used to infect 50 tobacco leaf segments, which were subsequently cultivated on nonselective medium. Adventitious shoots differentiated on the leaf segments 3 weeks after infection (Fig. 2a). One hundred shoots were transferred to the same medium 2 weeks later. After 1 month of cultivation, we visually identified and selected 63 abnormal shoots for further cultivation that exhibited an ESP from the original 100 cultured shoots (Fig. 2b). Until 6 months after infection, several normal shoots that exhibited normal apical dominance appeared in three of the 63 ESP lines (Fig. 2c). We visually identified, selected, and transferred these normal shoots to the same medium. These shoots grew normally and rooted (Fig. 2d).
Figure 2.
Visible selection of marker-free transgenic tobacco plants. (a) Regeneration of adventitious shoots from leaf segments on nonselective medium. (b) Differentiation of ESP from adventitious shoots. (c) Appearance of “normal” morphological shoots from ESP. (d) Normal rooted plant.
DNA Analysis of Transgenic Tobacco Plants.
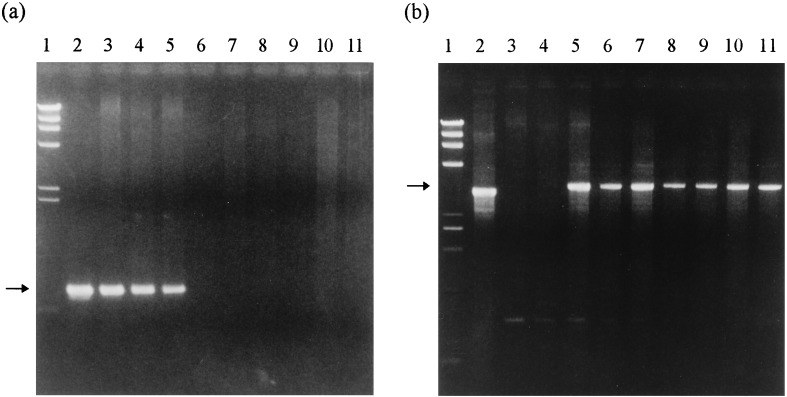
We isolated genomic DNA from 11 ESP shoots and 6 normal shoots, 2 each from 3 lines of 63 ESP shoots, and subjected them to PCR analysis. The specific primers a and b (Fig. 1) flanking the position of the hit and run cassette were able to amplify a DNA fragment when the excision of the cassette had occurred. If no excision occurred, the primers were too far apart to amplify a fragment. The expected fragment size of approximately 3 kb was observed in all normal shoots (Fig. 3b). Meanwhile, the amplification of the DNA fragment of approximately 800 bp was observed in all ESP shoots using the primers c and d (Fig. 1) flanking the ipt gene (Fig. 3a). These results show that the chimeric ipt gene is present in the chromosomal DNA of the ESP shoots but was excised from that of the normal shoots along with the modified Ac. In addition, we investigated the GUS activity and kanamycin resistance and confirmed the expression of these in all 11 ESP shoots and 6 normal shoots. These results indicate that the normal shoots separated from the ESP shoots are ipt marker-free transgenic plants that have retained gusA and nptII genes but not the selectable marker gene (the ipt gene). We observed both the 3-kb and the 800-bp DNA fragments, which indicate the excision and presence of the ipt gene in only one ESP shoot (lane 5 in Fig. 3). This ESP exceptional shoot appears to be composed of two kinds of transgenic cells, with and without the ipt gene, or two kinds of integrated copies with and without the ipt gene.
Figure 3.
PCR analysis of ESP shoots and normal plants obtained from ESP shoots. (a) Amplification of the DNA fragments using primers c and d flanking the ipt gene (Fig. 1). The arrow indicates a fragment of approximately 800 bp. (b) Amplification of the fragments using primers a and b (Fig. 1) flanking the position of hit and run cassette. The arrow indicates amplified fragments of 3 kb that result from empty donor sites. Lanes: 1, HindIII size marker (TAKARA shuzo); 2, plasmid pNPI106 in a and plasmid pBI121 in b; 3 and 4, DNA from two independent ESP shoot-derived clones, in which normal shoots did not reappear; 5, DNA from a ESP shoot-derived clone, in which normal shoots reappeared; 6–11, DNA from two independent normal shoots from each of three ESP shoot-derived clones. Lanes 6 and 7 represent 2 shoots from line 2, lanes 8 and 9 represent 2 shoots from line 3, and lanes 10 and 11 represent 2 shoots from line 4.
DNA samples of these ESP and normal shoots were analyzed by Southern blotting to determine the copy number of the nptII genes inserted. We found one hybridizing band in two of the normal shoots and two bands in another normal one when the nptII gene was used as the probe. A single band hybridizing to nptII was observed in only one ESP shoot, and more than two bands were found in nine other ESP shoots. To investigate the inheritance of the kanamycin-resistance trait, three independently transformed normal shoots (1 × 3 lines) were selfed, and the number of seedlings resistant to kanamycin was determined. Segregation of resistant and sensitive progeny were consistent with either a 3:1 ratio (observed ratio, 138:38 or 87:21) or a 15:1 ratio (89:9). GUS activity was also detected in the progeny carrying the nptII gene (10 positive of 10 plants tested). These results indicate that one or two independently segregating copies of the T-DNA were integrated in ipt marker-free transgenic plants. Accordingly, the presence or absence of the chimeric ipt gene is sufficient to identify visually both primary transgenic plants and subsequent marker-free transgenic plants after transformation.
Transformation of Hybrid Aspens.
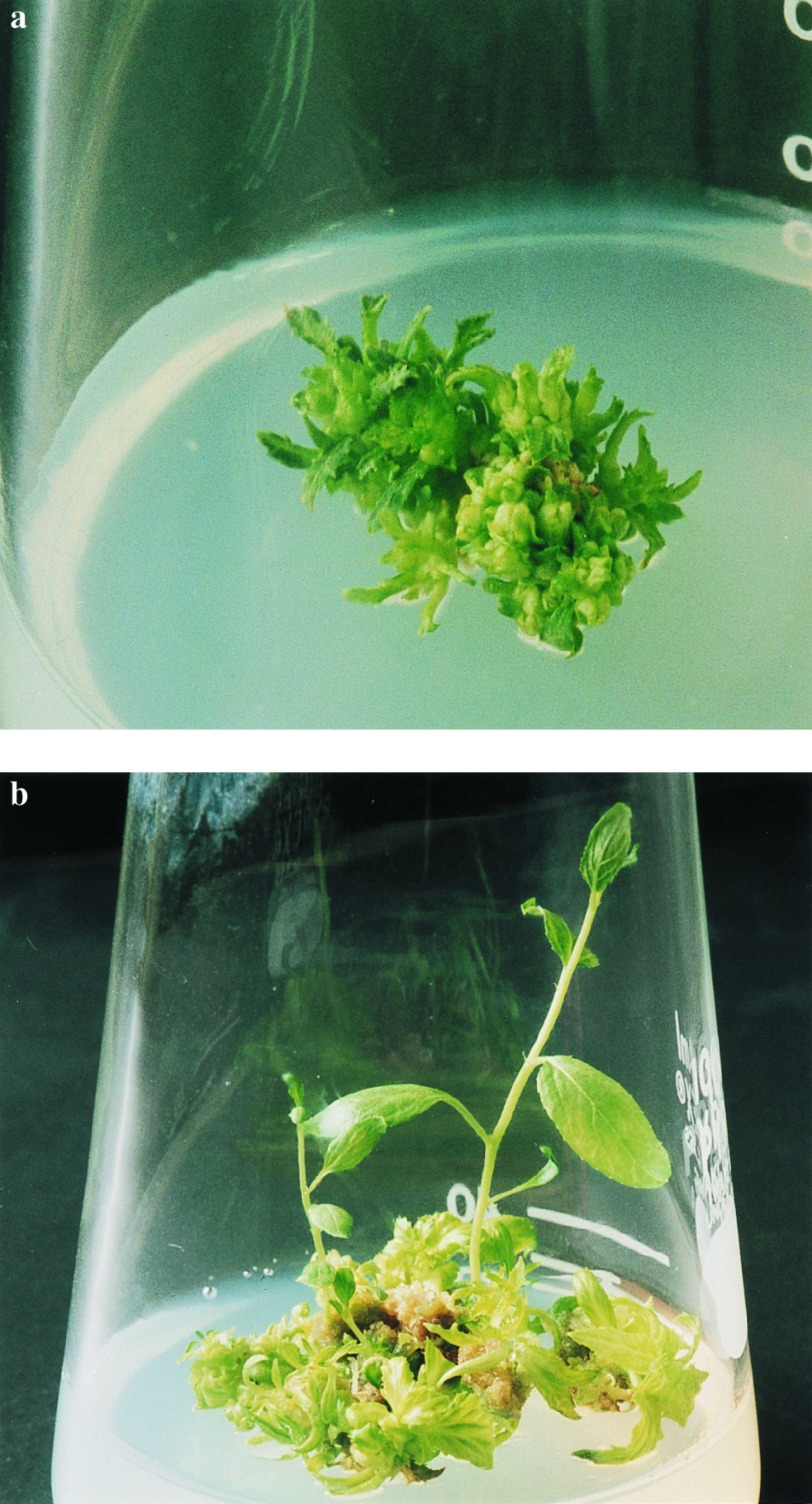
We inoculated A. tumefaciens containing pNPI106 into 50 stem segments from hybrid aspens. These infected stem segments were transferred to nonselective medium. After cultivation for 1 month, adventitious shoots appeared and were transferred to fresh medium. We separated 20 ESP shoots and cultured them (Fig. 4a). Normal shoots appeared in three ESP shoot clones by 8 months after infection (Fig. 4b). We separated the normal shoots and transferred them to root-inducing medium. These shoots grew normally and rooted. By PCR analysis, the ipt gene was detected in 20 ESP shoots but not 6 normal shoots (2 × 3 lines). Although only shoots from one clone showed GUS activity, these results demonstrate that a chimeric ipt gene can be selected visibly in hybrid aspens also. These results confirm work showing that Ac can function in poplar (R. Ahuja, personal communication).
Figure 4.
Transformation of hybrid aspens by the MAT vector system. (a) Differentiation of the ESP from adventitious shoots. (b) Appearance of morphologically normal shoots from ESP.
DISCUSSION
In this study, we demonstrate that a chimeric ipt gene with a 35S promoter can be a visually selectable marker for transformation of tobacco plants and hybrid aspens. When the ipt gene fused individually to several different promoters was introduced into potato (13), cucumber (14), tobacco (15, 25–29), Arabidopsis (27), peach (30), and poplar (31), the cytokinin level of transgenic plants was elevated, and the same effect as exogenously supplied cytokinin was observed (11, 12). It is well established that an elevated cytokinin-to-auxin ratio can promote shoot formation from plant tissue cultures in many species, and it may be expected that an ipt gene introduced by transformation would work in a similar way in many other species also. Some species may not respond to exogenously supplied hormones because of low hormone uptake, compartmentalization, or metabolism (14, 15). The MAT vector system, however, may provide an alternate approach to regenerate some plant species that have been difficult to transform, through internal manipulation of the cytokinin-to-auxin ratio.
We obtained marker-free transgenic tobacco plants from which Ac had disappeared from 4.8% of transgenic clones by 6 months after infection. When the ipt gene was introduced into tobacco plants, transgenic plants developed a great many shoots and lost apical dominance. We cultured about 150 shoots per transgenic clone for 6 months. The frequency of marker-free plants was 0.032%. The frequency of transposition events that occur in tobacco plants (32) during the transformation process and are transmitted to the progeny is in the range of 1–5%. About 10% of Ac elements disappear from transgenic cells during the transposition events because Ac cannot reinsert or because it has transposed into a sister chromatid that is lost by somatic segregation (17). Therefore, we estimate the frequency of somatic elimination of Ac, involving cells in the apical meristem that can give rise to a shoot, is also about 0.1–0.5%.
If more than one expressed copy of the vector were inserted into the plant genome, elimination of one copy would not cause loss of the ipt function. This inference leads to the expectation that marker-free transgenic plants will be derived from low copy number transformants. Almost all transgenic plants with ESP have more than one copy of the insert. The observed frequency of marker-free transgenic plants is 10 times lower than the expected elimination frequency of Ac, perhaps due to the effect of multiple copies. The MAT vector system, therefore, might make it possible to select and identify transgenic plants with a single functional copy of the inserted gene of interest as a result of selecting marker-free transgenic plants. Nevertheless, the frequency of marker-free transgenic plants is relatively low. Therefore, we have recently constructed an improved vector (pNPI132) with a site-specific recombination system R/Rs that can remove the ipt gene with a 10-fold increase in frequency (unpublished work). We are also now carrying out a second cycle of transformation in both tobacco and poplar to verify this method.
Recently, transformation methods that use the transposable element Ac (33), the Cre/lox (34, 35) site-specific recombination system, and cotransformation (36) are reported to remove selectable marker genes from transgenic plants (1). The selectable marker genes can be removed from the first generation of transgenic plants (Ro) by further selfing or outcrossing for at least one additional generation. Three kinds of transgenic plants (marker, marker-free, and somatic mosaic) may coexist during the process. However, marker-free transgenic plants cannot survive on culture medium containing selective agents. Marker-free transgenic plants may be recovered, allowing growth in the absence of selection followed by testing of individual plants. Alternatively, F2 plants could be screened and distinguished by PCR. However, the MAT vector system enables us to use the ipt gene to select visually marker-free transgenic plants at the Ro generation without sexual crossing. Furthermore, the ipt gene acts as a dominant and is not cell autonomous relative to the marker-free cells, so that chimeric plants would be likely to show the ipt shooty phenotype. The MAT vector system, therefore, selects against chimeric transformants.
“Kitakami Hakuyo” are elite clones of hybrid aspen produced by vegetative propagation for paper making (22). We believe that this is the first report of the removal of a selectable marker gene without crossing from a vegetatively propagated crop. A great many important crops, including potato, apple, grapevine, strawberries, cassava, and banana, as well as hybrids of poplar and eucalyptus, are hybrids and must be vegetatively propagated to maintain the elite genome. The previous transformation systems for eliminating selectable marker genes cannot be applied to those crops because they need sexual crosses to produce marker-free plants and to be able to carry out successive transformation (1, 6, 33–36). Moreover, perennial horticultural crops such as fruit trees and forest tree species such as hybrid aspen have long reproductive cycles. A great deal of time would be required to pyramid several valuable genes into trees by conventional breeding. Therefore, development of transformation systems that enable us to repeat transformation and introduce many genes is a most promising way to bypass the difficulties imposed by long generation times and reduce the time required to improve trees through genetic engineering.
Acknowledgments
We thank Prof. Ronald Sederoff for critical review and helpful comments on the manuscript, and Prof. Shigeru Iida for supply of pCKR97.
ABBREVIATIONS
- CaMV
cauliflower mosaic virus
- ESP
extreme shooty phenotype
- GUS
β-glucuronidase
References
- 1.Yoder J I, Goldsbrough A P. Bio/Technology. 1994;12:263–267. [Google Scholar]
- 2.Vasil V, Brown S M, Re D, Fromm M E, Vasil I K. Bio/Technology. 1991;9:743–747. [Google Scholar]
- 3.Perl A, Galili S, Shaul O, Ben-Tzvi I, Galili G. Bio/Technology. 1993;11:715–718. [Google Scholar]
- 4.Stomp A-M, Loopstra C, Sederoff R, Chilton S, Fillatti J, Dupper G, Tedeschi P, Kinlaw C. In: The Genetic Manipulation of Woody Plants. Hanover J W, Keathley D E, editors. New York: Plenum; 1988. pp. 231–241. [Google Scholar]
- 5.Bryant J, Leather S. Trends Biotechnol. 1992;10:274–275. [Google Scholar]
- 6.Gressel J. Trends Biotechnol. 1992;10:382. [Google Scholar]
- 7.Flavell R B, Dart E, Fuchs R L, Fraley R B. Bio/Technology. 1992;10:141–144. doi: 10.1038/nbt0292-141. [DOI] [PubMed] [Google Scholar]
- 8.Goldsbrough A P. Trends Biotechnol. 1992;10:417. doi: 10.1016/0167-7799(92)90289-8. [DOI] [PubMed] [Google Scholar]
- 9.Akiyoshi D, Klee H, Amasino R, Nester E W, Gordon M P. Proc Natl Acad Sci USA. 1984;81:5994–5998. doi: 10.1073/pnas.81.19.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barry G F, Rogers S G, Fraley R T, Brand L. Proc Natl Acad Sci USA. 1984;81:4776–4780. doi: 10.1073/pnas.81.15.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klee H J, Horsch R B, Rogers S G. Annu Rev Plant Physiol. 1987;38:467–486. [Google Scholar]
- 12.Hobbie L, Timpte C, Estelle M. Plant Mol Biol. 1994;26:1499–1519. doi: 10.1007/BF00016487. [DOI] [PubMed] [Google Scholar]
- 13.Ooms G, Kaup A, Roberts J. Theor Appl Genet. 1983;66:169–172. doi: 10.1007/BF00265193. [DOI] [PubMed] [Google Scholar]
- 14.Smigocki A C, Owens L D. Plant Physiol. 1989;91:808–811. doi: 10.1104/pp.91.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smigocki A C, Owens L D. Proc Natl Acad Sci USA. 1988;85:5131–5135. doi: 10.1073/pnas.85.14.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedoroff N V. In: Mobile DNA. Douglas E B, Martha M H, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 375–411. [Google Scholar]
- 17.Belzile F, Lassner M W, Tong Y, Khush R, Yoder J I. Genetics. 1989;123:181–189. doi: 10.1093/genetics/123.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wabiko H, Kagaya M, Kodama I, Masuda K, Kodama Y, Yamamoto H, Shibano Y, Sano H. Arch Microbiol. 1989;152:119–124. [Google Scholar]
- 19.Izawa T, Miyazaki C, Yamamoto M, Terada R, Iida S, Shimamoto K. Mol Gen Genet. 1991;227:391–396. doi: 10.1007/BF00273928. [DOI] [PubMed] [Google Scholar]
- 20.Huekema A, Hirsch P R, Hooykaas P J J, Schilperoort R A. Nature (London) 1983;303:179–180. [Google Scholar]
- 21.Höfgen R, Willmitzer L. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takayama Y. Nichirin-shi. 1968;50:267–273. [Google Scholar]
- 23.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–14. [Google Scholar]
- 25.Estruch J J, Prinsen E, Onckelen H V, Shell J, Spena A. Science. 1991;254:1364–1367. doi: 10.1126/science.254.5036.1364. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Hagen G, Guilfoyle T J. Dev Biol. 1992;153:386–395. doi: 10.1016/0012-1606(92)90123-x. [DOI] [PubMed] [Google Scholar]
- 27.Medford J I, Horgan R, El-Sawi Z, Klee H J. Plant Cell. 1989;1:403–413. doi: 10.1105/tpc.1.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmulling, T., Beinsberger, J., Greef, J. D., Schell, J., Onckelen, H. V. & Spena, A. FEBS Lett. 249, 401–406.
- 29.Smart M C, Scofield S R, Bevan M W, Dyer T A. Plant Cell. 1991;3:647–656. doi: 10.1105/tpc.3.7.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smigocki A C, Hammerschlag F A. J Am Soc Hortic Sci. 1991;116:1092–1097. [Google Scholar]
- 31.Schwartzenberg K V, Doumas P, Jouanin L, Pilate G. Tree Physiol. 1994;14:27–35. doi: 10.1093/treephys/14.1.27. [DOI] [PubMed] [Google Scholar]
- 32.Jones J D G, Carland F, Lim E, Ralston E, Dooner H K. Plant Cell. 1990;2:701–707. doi: 10.1105/tpc.2.8.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldsbrough A P, Lastrella C N, Yoder J I. Bio/Technology. 1993;11:1286–1292. [Google Scholar]
- 34.Dale E C, Ow D W. Proc Natl Acad Sci USA. 1991;88:10558–10562. doi: 10.1073/pnas.88.23.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russel S H, Hoopes J L, Odell J T. Mol Gen Genet. 1992;234:49–59. doi: 10.1007/BF00272344. [DOI] [PubMed] [Google Scholar]
- 36.Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. Plant J. 1996;10:165–174. doi: 10.1046/j.1365-313x.1996.10010165.x. [DOI] [PubMed] [Google Scholar]