Abstract
The green fluorescent protein (GFP) from the jellyfish Aequorea victoria is finding wide use as a genetic marker that can be directly visualized in the living cells of many heterologous organisms. We have sought to express GFP in the model plant Arabidopsis thaliana, but have found that proper expression of GFP is curtailed due to aberrant mRNA processing. An 84-nt cryptic intron is efficiently recognized and excised from transcripts of the GFP coding sequence. The cryptic intron contains sequences similar to those required for recognition of normal plant introns. We have modified the codon usage of the gfp gene to mutate the intron and to restore proper expression in Arabidopsis. GFP is mainly localized within the nucleoplasm and cytoplasm of transformed Arabidopsis cells and can give rise to high levels of fluorescence, but it proved difficult to efficiently regenerate transgenic plants from such highly fluorescent cells. However, when GFP is targeted to the endoplasmic reticulum, transformed cells regenerate routinely to give highly fluorescent plants. These modified forms of the gfp gene are useful for directly monitoring gene expression and protein localization and dynamics at high resolution, and as a simply scored genetic marker in living plants.
Keywords: plant transformation, confocal microscopy/cryptic splicing/genetic marker/endoplasmic reticulum
The β-glucuronidase (GUS) gene has been used extensively as a reporter for gene expression in plants (1). Transformed tissues or patterns of gene expression can be identified histochemically, but this is generally a destructive test and is not suitable for assaying primary transformants, for following the time course of gene expression in living plants, nor as a means of rapidly screening segregating populations of seedlings. The green fluorescent protein (GFP) from the jellyfish Aequorea victoria can be directly visualized, and therefore shares none of these problems. The protein undergoes posttranslational modification, and a tripeptide within the protein is cyclized and oxidized to form a covalently attached chromophore (2, 3). Mature GFP is intrinsically fluorescent, with a barrel-like fold that encases the chromophore and acts as a natural solvent shield (4, 5). GFP cDNA has been cloned (6) and successfully expressed in various heterologous organisms (2, 7–9). Use of the protein allows the direct visualization of gene expression and subcellular localization of fusion proteins, in living cells, without the need for invasive techniques or addition of cofactors.
GFP has been successfully expressed at high levels in tobacco plants using the cytoplasmic RNA viruses potato virus X (10) and tobacco mosaic virus (11). In these experiments, the gene was directly expressed as a viral mRNA in infected cells, and very high levels of GFP fluorescence were seen. However, poor or no fluorescence was seen when the gfp cDNA was transformed into isolated cells or transformed plants of Arabidopsis (12–14). We show here that expression of the gfp cDNA is curtailed by aberrant mRNA splicing in Arabidopsis. We have altered the codon usage of gfp to avoid recognition of a cryptic intron and have demonstrated that proper expression of the fluorescent protein is restored. We have also targeted the protein to localized compartments within the cell to obtain a gfp gene that can be used as a bright fluorescent marker for efficient transformation and improved regeneration of Arabidopsis plants. With these improvements we have been able to label cells and subcellular structures brightly within living plants to directly and noninvasively monitor vital processes at high resolution.
MATERIALS AND METHODS
Gene Construction and Testing.
Two oligonucleotides, GGC GGATCC AAGGAGATAT AACA ATG AGT AAA GGA GAA GAA CTT TTC ACT (optGFP5, the first codon of GFP, is underlined) and GGC GAGCTC TTA TTT GTA TAG TTC ATC CAT GCC (SacGFP3, sequence complementary to the termination codon for GFP, is underlined), complementary to the 5′ and 3′ ends of the GFP coding sequence, were synthesized and used for PCR amplification of pGFP10.1 template (6) with Thermococcus litoralis Vent DNA polymerase. The amplified fragment was cloned between the BamHI and SacI sites of pUC119 for expression in Escherichia coli, and pBI121 (1) for plant transformation experiments.
Plant Transformation.
Agrobacterium tumefaciens strain LBA4044 was transformed with the gfp-containing pBI121 plasmid by electroporation. Roots of Arabidopsis thaliana ecotype C24 were transformed using the protocol of Valvekens et al. (15). Transgenic callus and shoots were screened for GFP expression using a inverted fluorescence microscope (Leitz DM-IL) fitted with a filter set (Leitz-D excitation BP355–425, dichroic 455, emission LP460) suitable for the main 395-nm excitation and 509-nm emission peaks of GFP. The use of a 7-mm threaded extension tube with a 4× objective (EF 4/0.12) gave a greater working distance, and has allowed the convenient direct observation of tissue within inverted sealed Petri dishes. A hand-held 100 W long-wavelength UV lamp (B100AP; Ultraviolet Products) was also used for routine monitoring of transgenic shoots and plants.
Nucleic Acid Extraction.
Individual transgenic shoots (≈5 g) were immersed in liquid nitrogen and ground to a fine powder using a mortar and pestle. The frozen powder was treated with 10 ml of a buffer containing 200 mM Tris·HCl (pH 8.5), 300 mM LiCl, 10 mM Na3EDTA, 1.5% lithium dodecylsulfate, 1% sodium deoxycholate, and 1% Nonidet P-40, and extracted several times with phenol/chloroform (1:1). The resulting extract was ethanol-precipitated, and the resulting nucleic acid extract was divided and treated either with 100 μg/ml DNase 1, or with 100 μg/ml RNase A for 30 min at 37°C in 25 mM Tris·HCl, pH 7.5/10 mM MgCl2/1 mM DTT. After treatment, the RNA and DNA extracts were phenol/chloroform-extracted and ethanol-precipitated.
PCR Analysis and Sequencing.
Complementary DNA was synthesized by reverse transcription of 1 μg of extracted RNA using an oligo(dT)8 primer. The cDNA or 1 μg of extracted DNA was then used as the template for PCR amplification (35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C using Vent DNA polymerase) in the presence of 200 μCi/ml [α-32P]dCTP (1 Ci = 37 GBq) using flanking optGFP5 and SacGFP3 primers specific for the gfp expression cassette. The resulting radiolabeled fragments were probed for the presence of various restriction endonuclease recognition sites by enzyme digestion, electrophoresis in a native 5% polyacrylamide gel, and autoradiography. An RsaI-HincII fragment of the gfp cDNA that was found to contain a deletion was cloned into the phage vector M13mp18 and sequenced using a Sequenase Version 2.0 DNA sequencing kit (United States Biochemical) and the M13 (−40) oligonucleotide.
Modification of GFP Codon Usage.
Two mutagenic oligonucleotides, a 122-mer and a 126-mer, were synthesized and purified by PAGE. The oligonucleotides shared 17 nt of complementarity at their 3′ termini, and were annealed and elongated after three rounds of thermal cycling with Vent DNA polymerase. The extended product was cloned between the NdeI and AccI sites of gfp in pBluescript II KS(+), to give pBS-mgfp4. The mutant was subcloned into M13mp18 and sequenced using the dideoxynucleotide chain termination technique with T7 DNA polymerase.
Fusion of Endoplasmic Reticulum (ER) Targeting Sequences.
Two strands of a signal sequence were synthesized, annealed, end-filled using the Klenow fragment of DNA polymerase I, and digested with BamHI and EcoRI. PCR mutagenesis was used to fuse the amino acid sequence HDEL and an in-frame EcoRI site to the C and N termini, respectively, of the mgfp4 gene. Two oligonucleotides, GCC GAATTC AGT AAA GGA GAA GAA CTT TTC and GGC GAGCTC TTA AAG CTC ATT CAT GTT TGT ATA GTT CAT CCA TGC C, were synthesized and used for PCR amplification of the mgfp4 sequences, using Vent DNA polymerase (35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C). The product was digested with EcoRI and SacI (sites within the oligonucleotides are underlined), and cloned into pBI121 in a three-way ligation with the BamHI and EcoRI cut signal sequences.
Confocal Microscopy.
Transgenic Arabidopsis seedlings were grown in sterile agar culture for 5 days, and were mounted in water under glass coverslips for microscopy. The specimens were examined using a Bio-Rad MRC-600 laser-scanning confocal microscope equipped with a krypton-argon laser and filter sets suitable for the detection of fluorescein and Texas red dyes (Bio-Rad K1/K2), and a Nikon 60× PlanApo numerical aperture 1.2 water-immersion objective. Dual-channel confocal images and video montages of seedlings were composed using adobe photoshop.
RESULTS AND DISCUSSION
Construction of a gfp Expression Cassette.
A synthetic gfp gene was constructed using PCR. The plasmid pGFP10.1, which contains a cloned A. victoria gfp cDNA sequence (6), was used as template for amplification, and synthetic oligomer primers were used to modify the sequences flanking the GFP coding region. The amplified sequence differs from the published coding sequence of pGFP10.1, as it contains an arginine (CGG) positioned at residue 80, replacing a codon for glutamine (CAG). This difference has been noted by others (7). Recognition sites for the restriction endonucleases BamHI and SacI were placed at the 5′ and 3′ termini of the amplified fragment (Fig. 1). A Shine-Dalgarno sequence was positioned upstream of the initiation codon to ensure efficient translation of the transcribed gene in E. coli, and the sequence AACA was inserted between positions −4 and −1 for efficient translation in plants (16). Expression of the gene cassette in transformed E. coli and yeast cells gave rise to brilliant green fluorescence under long-wavelength UV illumination (17).
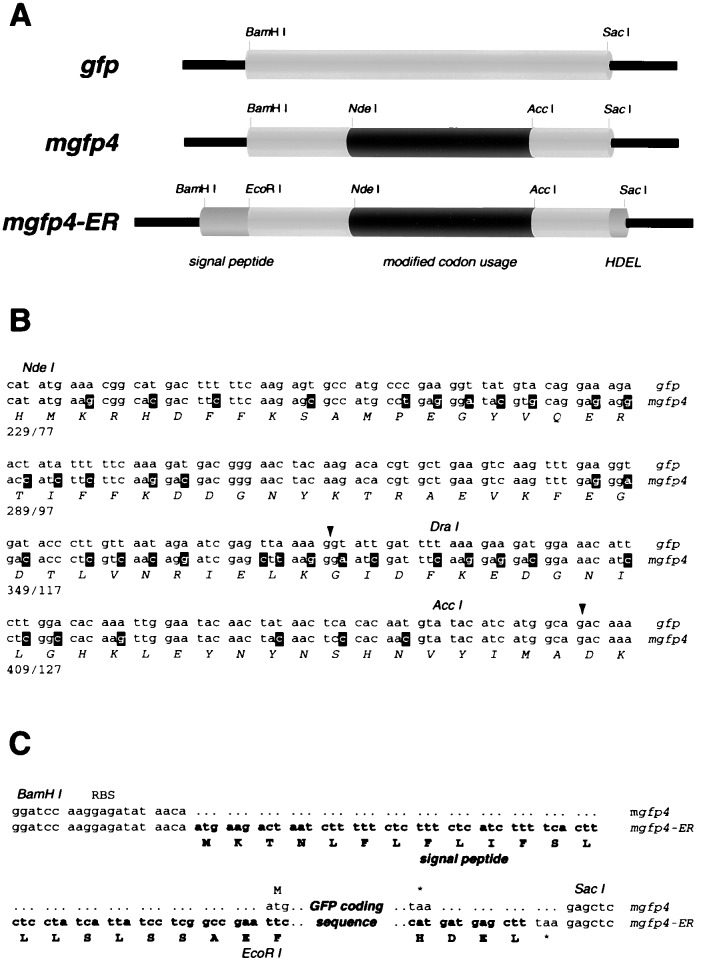
Figure 1.
Modified gfp sequences. (A) Schematic diagrams showing gene expression cassettes that contain the wild-type sequence (gfp) (6), modified codon usage (mgfp4), and additional peptide targeting sequences (mgfp4-ER). The sequences that have altered codon usage are indicated in black, and targeting sequences are shown in dark grey. (B) Altered codon usage in mgfp4. The cryptic intron is shown underlined with the 5′ and 3′ splice sites arrowed. The upper sequence corresponds to that of gfp, and the lower nucleotide sequence is that of mgfp4. Mutated nucleotides are shown in reverse type, and restriction endonuclease sites are shadowed. The amino acid sequence is the same for each gene and is shown below. (C) Sequences flanking the GFP coding region. Both gfp and mgfp4 are flanked by restriction endonuclease sites for BamHI and SacI, a ribosome binding site (RBS) for bacterial expression, and the sequence AACA upstream of the start codon for improved plant translation. The mgfp4-ER gene cassette contains additional sequences shown in boldface type, which comprise a 5′ terminal signal peptide and 3′ HDEL sequence. An EcoRI site was used to link the signal peptide and coding sequences.
After we determined that the PCR-amplified gfp cDNA cassette correctly produced fluorescent protein product in transformed microbial cells, the sequence was inserted into the plant transformation vector pBI121 (1), replacing the GUS gene behind the CaMV 35S promoter. An Agrobacterium-mediated root transformation procedure (15) was used to produce transgenic Arabidopsis tissue explants from which plantlets were regenerated. However, at no stage during the transformation procedure did we detect GFP-related fluorescence, using UV lamp illumination or epifluorescence microscopy. Subsequent reports have confirmed that expression of gfp results in poor fluorescence in a number of plant expression systems, including Arabidopsis. Although green fluorescence has been seen in gfp-transformed maize protoplasts (13, 14), Hu and Cheng (13) have reported that no signal was seen in transformed Arabidopsis protoplasts. Reichel and colleagues (18) also detected no fluorescence in gfp-transformed Arabidopsis, tobacco, or barley protoplasts. It has been claimed that gfp fluorescence can be detected in bombarded Arabidopsis tissues (14). However, in these experiments, leaf tissue was treated with methanol prior to microscopic examination, where methanol causes rapid and irreversible bleaching of GFP (19). Local wounding due to particle bombardment can cause punctate patterns of bright autofluorescence, and this type of experiment needs to be interpreted with care. The same authors saw no expression of a CAB2-driven gfp gene in transgenic Arabidopsis plants (14), and there is general agreement on the need for substantial improvement of the marker (12, 18, 20).
gfp mRNA Is Misspliced in Arabidopsis.
Useful expression of the gfp cDNA in plants requires that (i) the GFP apoprotein is produced in suitable amounts within plant cells, and (ii) the nonfluorescent apoprotein must undergo efficient posttranslational modification to produce the mature GFP. Early in the course of this work we were aware that gfp could be expressed at high levels in tobacco plants using cytoplasmic RNA virus vectors (10, 11). It was clear that efficient posttranslational maturation of the protein could take place in plants. As our own experiments relied on the expression of integrated copies of the gene, we used PCR-based methods to verify the correct insertion of the 35S promoter-driven gfp cDNA and to check mRNA transcription and processing in transformed plantlets. Nucleic acids were extracted from plantlets, and samples were either (i) treated with RNase or (ii) treated with DNase and reverse-transcribed using oligo(dT)8 primer. The gfp sequences in these extracts were therefore derived from genomic DNA or transcribed mRNAs, respectively. The gfp sequences were amplified via PCR from these separate extracts, and the products were analyzed for the presence of certain restriction endonuclease sites (Fig. 2A). While the expected full-length product was obtained after amplification of the integrated gfp gene, RT-PCR of gfp mRNA sequences gave rise to a truncated product. This product was 80–90 bp shorter than expected and was missing recognition sites for the restriction endonucleases DraI and AccI. This is consistent with a small deletion within the coding sequence of the gfp mRNA as shown in Fig. 2B. Because the amplified gfp gene sequences were of the expected size, we presumed that the mRNA was misprocessed.
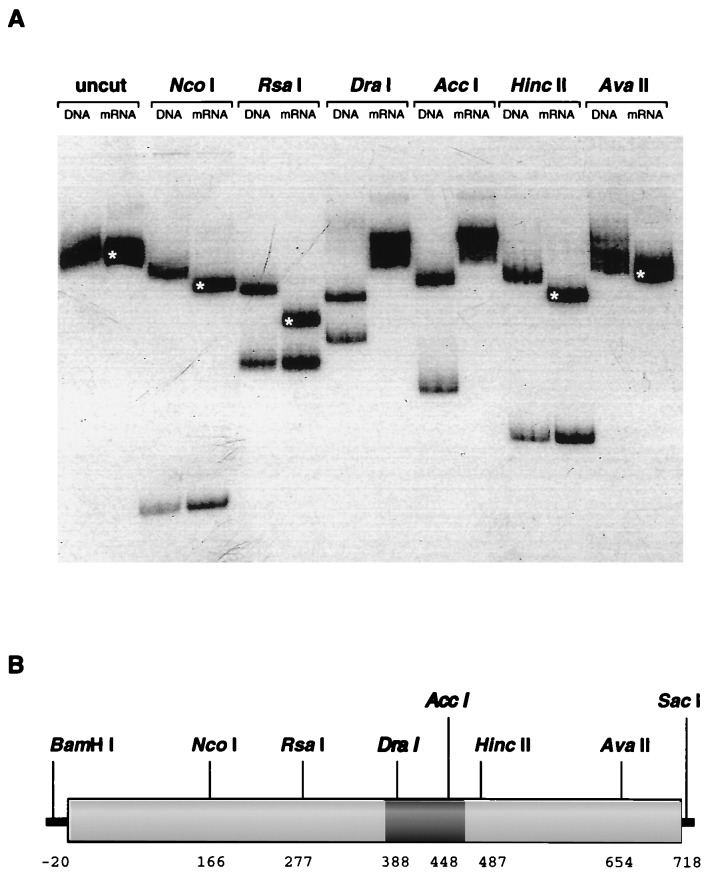
Figure 2.
Aberrant posttranscriptional processing of gfp mRNA. (A) Restriction endonuclease digestion of PCR fragments derived from gfp DNA and mRNA sequences. Sequences corresponding to the integrated gfp gene and to mRNA transcripts were isolated and separately amplified using PCR techniques and incubated with various restriction endonucleases. The radiolabeled fragments were fractionated by electrophoresis in a 5% polyacrylamide gel, and are shown labeled with the source of the amplified sequences (DNA or mRNA) and the name of the restriction endonuclease used for digestion, or not (uncut). The mRNA-derived sequences appeared to lack sites for DraI and AccI and to contain deleted sequences. Fragments that are smaller than expected have been indicated with a white asterisk. (B) Schematic diagram of the gfp coding sequence. The positions of the restriction endonuclease cleavage sites that were used for analysis of PCR products are indicated, and these are numbered according to the coding region of gfp. The cleavage pattern of the amplified gfp mRNA sequence corresponds to a deletion of 80–90 nt, and this is indicated by a dark shaded region.
The shortened RT-PCR product was cloned and sequenced, and a deletion of 84 nt was located between residues 380–463 of the GFP coding sequence (Fig. 3A). The sequences bordering the deletion are shown in Fig. 3B and demonstrate similarity to known plant introns. Matches were found for sequences that are conserved at the 5′ and 3′ splice sites of plant introns (reviewed in ref. 21) and for conserved branchpoint nucleotides in plant introns (22, 23). The excised gfp sequence also contains a high AU content (68%) that has been shown to be important for recognition of plant introns (24–27). It is likely that this 84-nt region of the jellyfish gfp cDNA sequence is efficiently recognized as an intron when transcribed in Arabidopsis, resulting in an in-frame deletion and the production of a defective protein product, which is predicted to be 28 aa shorter. This explanation would also account for the efficient expression of gfp from RNA virus vectors that replicate in the cytoplasm, and thus evade splicing. It should be noted that the borders of the cryptic intron do not coincide with any of the natural spliced junctions found after processing of the gfp mRNA in A. victoria (6). No full-length gfp mRNA is detectable by RT-PCR, and therefore misprocessing must be close to complete in transformed Arabidopsis plantlets.
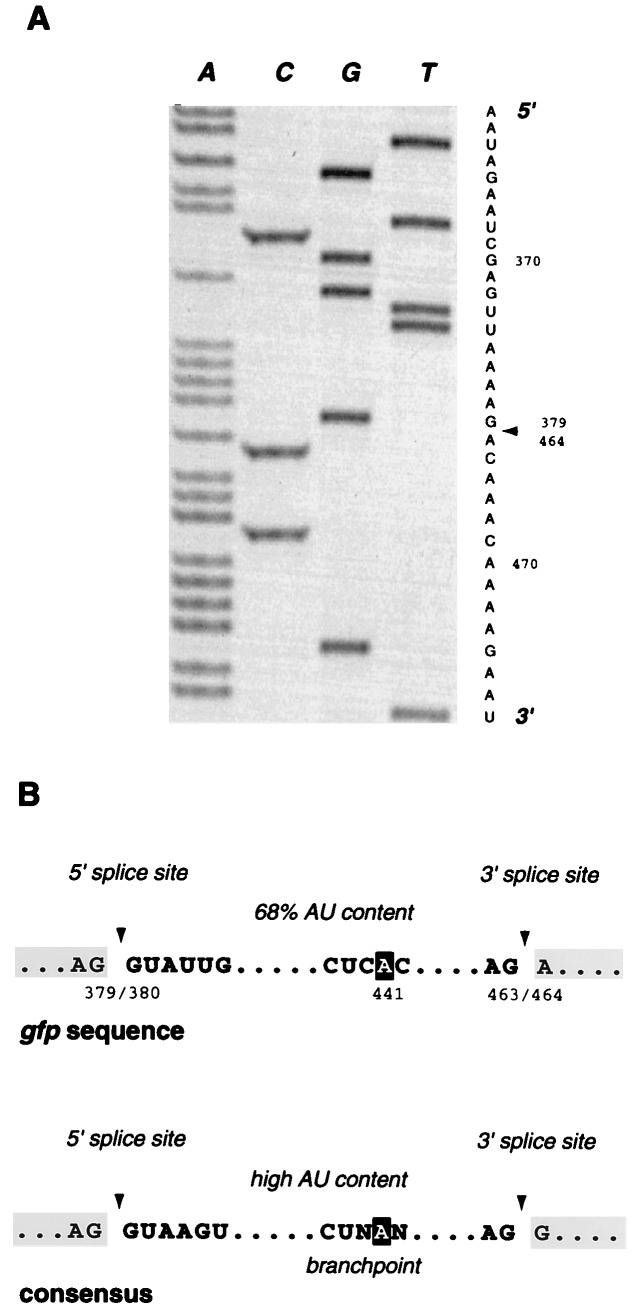
Figure 3.
Sequence analysis of cloned gfp mRNAs. (A) Autoradiograph and sequence of the amplified gfp mRNA sequence. Nucleotides 380–463 are absent from the transcribed sequence, and the site of this 84-nt deletion is indicated by an arrowhead. (B) Comparison of gfp cryptic intron sequences with the conserved sequences that are found at the splice sites and branchpoints of plant introns (21, 22). Splice sites are indicated by arrowheads, and the nucleotide involved in branchpoint formation is shown in reverse type.
Modification of gfp Codon Usage.
To destroy this cryptic intron we have mutated the sequences involved in splice-site and branchpoint recognition and decreased the AU content of the putative intron, as shown in Fig. 1B. All modifications affected only codon usage, and this modified gene, mgfp4, encodes a protein product that is identical to that of the jellyfish. The mgfp4 sequence was inserted behind the 35S promoter in pBI121, and introduced into Arabidopsis using the root transformation technique (15). There was little expression of the 35S-driven gene in A. tumefaciens, and bacterial fluorescence did not interfere with the detection of transformed plant cells, which were brightly green fluorescent within 2–3 days of cocultivation. As cell proliferation continued, the brightest clumps of callus and developing shoot tissue were so intensely fluorescent that they were clearly visible by eye, using a hand-held long-wavelength UV lamp. We have also adapted the microscope objective of an inverted fluorescence microscope to allow more sensitive, higher-magnification observation of cells in sterile culture during transformation and regeneration (see Materials and Methods). During regeneration experiments, we observed a wide range of GFP fluorescence intensities in 35S-mgfp4 transformed plantlets, which we expect arose from position-dependent modulation of gene expression in different transformants. It proved difficult to regenerate fertile plants from the brightest transformants, with cells remaining as a highly fluorescent callus or mass of shoots after several months of culture. It is possible that high levels of GFP expression were mildly toxic or interfered with differentiation. This is of special concern with a fluorescent molecule such as GFP, which would be expected to generate free radicals upon excitation, and which undergoes oxidative modification and could possess catalytic properties. The conditions that we have used for plant regeneration should provide a stringent test for any deleterious effect due to GFP. The 35S promoter was used to drive expression of the protein at high levels throughout the plant, including meristematic cells (Fig. 4), and regeneration took place under continual illumination, allowing the possibility of GFP-mediated phototoxicity. Despite poor regeneration of the brightest transformants, we managed to obtain more than 50 separate transgenic Arabidopsis lines, most of which contained levels of GFP that were easily detectable by microscopy.
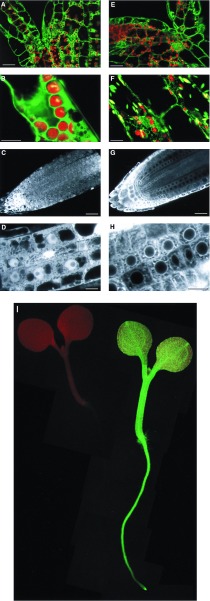
Figure 4.
Confocal images of 35S-mgfp4 and 35S-mgfp4-ER transformed Arabidopsis plants. (A–D) Images of 35S-mgfp4 transformed seedlings. (E–H) Images of 35S-mgfp4-ER transformed seedlings. A and E show optical sections through the shoot apex of the seedlings, at the junction of cotyledons and hypocotyl (bars = 25 microns). Cells from the hypocotyl are shown in B (cortex) and F (epidermal; bars = 10 microns). Median longitudinal optical sections of root tips are shown in C and G (bars = 25 microns). Cells within the root tips are shown at higher magnification in D and H (bars = 10 microns). The distribution of the different forms of GFP is clear, with GFP accumulating within the nucleoplasm, but being excluded from the nucleolus (D), and with the ER-targeted form of GFP being excluded from the nucleus and forming a distinct perinuclear pattern. Similar partitioning of the two GFP forms can be seen in the shoot (A and E). (I) 5-day-old wild-type (Left) and 35S-mgfp4-ER transgenic (Right) seedlings were mounted in water on a Leitz DM-IL inverted fluorescence microscope and illuminated with long-wavelength UV light.
It is likely that the mgfp4 gene and its derivatives will be useful for expression studies in other plants, particularly dicots, which share similar features involved in intron recognition (21). Some transient gfp expression has been seen in plant protoplasts of Citrus sinensis (28) and maize (13, 14), and therefore aberrant splicing of gfp mRNA may not always be as efficient as in transgenic Arabidopsis tissues. However, experiments with tobacco and barley protoplasts (18) have demonstrated that mgfp4-derived sequences are expressed at much higher levels than the wild-type gene in these species. Recently, it has been shown that alteration of gfp codon usage (which also alters the putative cryptic intron sequence) leads to 20-fold increased expression in maize protoplasts (20). It is possible that aberrant RNA processing may also interfere with gfp expression in other organisms. There have been recent reports of improved GFP expression in mammalian cells after alteration of gene codon usage (29, 30). Increased levels of expression have been attributed to improved rates of translation due to optimized codon usage, but it is also possible that altered mRNA sequences affect posttranscriptional processing in animal cells. However, introns found in animals, including A. victoria (6), share a conserved polypyrimidine tract adjacent to the 3′ splice site (reviewed in ref. 31), and introns in yeast cells possess a requirement for additional conserved sequences (UACUAAC) located at the branch point (32). The lack of these additional features may help to minimize aberrant processing of gfp mRNA in fungal and animal cells.
Confocal Microscopy of Living Plants.
GFP expression and localization can be directly visualized at high resolution in living plants, using laser-scanning confocal microscopy. Confocal imaging allows precise visualization of fluorescent signals within a narrow plane of focus, with exclusion of out-of-focus blur, and the technique permits the reconstruction of three-dimensional structures from serial optical sections. Intact plant tissue proves a difficult subject for fluorescence microscopy because it consists of deep, alternating layers of refractile cell walls and aqueous cytosol and contains various autofluorescent and light-scattering components. There are two approaches to the difficulties imposed by these conditions: (i) to fix and to clear the tissue with a high refractive index mounting medium, or (ii) to directly image living tissue using suitably corrected microscope optics. It has proved difficult to effectively clear Arabidopsis wholemounts without causing artifacts or losing GFP fluorescence, and there are considerable advantages to working with living tissues, so we have pursued the second approach. Arabidopsis seedlings can simply be mounted in water for microscopy and examined using a long-working distance water-immersion objective to minimize the effects of spherical aberration when focusing deep into an aqueous sample (12). Even with the use of such a specialized objective (Nikon 60× planapochromat, numerical aperture 1.2, working distance 220 μm), image quality degrades rapidly for optical sections deeper than 60–80 μM within the tissue. However, the small size of Arabidopsis seedlings allows very useful imaging despite this limitation (e.g., median longitudinal optical sections can be obtained from intact roots) (Fig. 4 C and G).
In transgenic Arabidopsis cells, GFP is found throughout the cytoplasm but appears to accumulate within the nucleoplasm. It is excluded from vacuoles, organelles, and other bodies in the cytoplasm, and is excluded from the nucleolus (Fig. 4 A–D). A similar subcellular distribution of GFP was seen in all Arabidopsis cell types examined, and red autofluorescent chloroplasts provide an effective counter-fluor for GFP in the upper parts of the plant. Cytoplasmic streaming and the movement of organelles could be observed in these living cells. In addition to cell ultrastructure, the architecture of the intact tissue was also clearly discernible, and the arrangement of different cell types could be seen in longitudinal optical sections of root tips and cotyledons. For example, cells within the epidermis of the cotyledon contain few mature chloroplasts and could be distinguished from layers of neighboring mesophyll cells (Fig. 4A), and files of developing cells around the primary root meristem are clearly evident (Fig. 4C).
Regeneration of Brightly Fluorescent Plants with ER-Targeted GFP.
We found it difficult to regenerate fertile plants from the brightest 35S-mgfp4 transformants. In jellyfish photocytes, where natural high levels of GFP are well tolerated, the protein is found sequestered in cytoplasmic granules (33). In contrast, the mature protein is found throughout the cytoplasm and nucleoplasm of transformed Arabidopsis cells. If GFP is a source of fluorescence-related free radicals, for example, it might be advisable to target the protein to a more localized compartment within the plant cell. We have fused several targeting peptides to GFP and expressed these variants in transgenic Arabidopsis plants. One, targeted to the ER, showed a substantial improvement over the unmodified GFP. The targeted form of GFP contains an N-terminal signal peptide derived from an Arabidopsis vacuolar basic chitinase and the C-terminal amino acid sequence HDEL (Fig. 1C) to ensure secretion and retention of the protein within the lumen of the ER. Using this modified gene (mgfp4-ER), it has been possible consistently to regenerate intensely fluorescent and fertile plantlets (Fig. 4I). Fluorescence within these plants could be readily observed by eye using a long-wavelength UV lamp. The mgfp4-ER-expressing plants were also examined by confocal microscopy, and fluorescent protein was found mainly within the endomembrane system. The protein is excluded from the nucleus, shows a perinuclear distribution, and is found associated with the ER, which forms a characteristic reticulate network in highly vacuolate cells (Fig. 4 E–H). In highly cytoplasmic meristematic cells, the nuclei and orientation of cell divisions can be clearly distinguished (Fig. 4H). Localization of the modified protein to cytoplasmic organelles was also evident, to what appear to be leucoplasts or proplastids. For example, an optical section of a hypocotyl epidermal cell is shown in Fig. 4F, and this includes a thin portion of cytosol that is pressed between the cell wall and vacuole. Such hypocotyl cells in mgfp4-ER-transformed seedlings appear to contain a spectrum of developing plastids that range from the brightly green fluorescent to those that take on a yellow, orange, or red appearance in dual-channel confocal micrographs. We presume that this is due to increasing chlorophyll synthesis, and that the green fluorescent plastids may be the maturing precursors of chloroplasts in these cells. These green fluorescent plastids are also found within the chloroplast-free epidermal cells of leaves and cotyledons, but are not found within the underlying mesophyll cells that are packed with mature chloroplasts (Fig. 4E). It seems likely that these organelles are proplastids and are capable of developing into chloroplasts, but we cannot exclude the possibility that they are some specialized form of leucoplast.
The accumulation of mgfp4-ER protein within leucoplasts or developing proplastids, in addition to its entry into the secretory pathway and retention in the ER, may indicate misrecognition of the N-terminal signal peptide. Proplastid accumulation of GFP is not seen in the 35S-mgfp4 transformed plants. If the mgfp4-ER encoded-signal peptide is inefficiently recognized before docking and cotranslational transport of the protein into the lumen of the ER, a proportion of GFP-bearing fused terminal sequences may be produced in the cytoplasm. If so, it is possible that the neglected signal peptide may act as a transit sequence for plastid entry. Alternatively, there may be some direct exchange between developing plastids and the endomembrane system. We see no free cytoplasmic fluorescence, and the protein is sorted very efficiently to the ER or to plastids.
It is unclear whether the beneficial effects of targeting GFP to the ER are due to increased levels or safer accumulation of mature GFP within cells. For example, if accumulation of fluorescent protein leads to the generation of free radicals in illuminated cells, it is conceivable that removing GFP from the nucleus could protect cells from DNA damage due to such short-range, highly reactive species. However, it is also possible that the fusion of peptide targeting sequences may improve the properties of the protein itself, or that the localization of GFP to the lumen of the ER may improve its maturation and accumulation. We have shown previously that maturation of the GFP apoprotein is sensitive to temperature, and that the apoprotein readily misfolds under certain conditions (17). The lumen of the ER is known to contain components such as chaperones and peptidyl prolyl isomerases that aid protein folding (34), and secretion and retention of GFP within the ER may allow improved formation and accumulation of the mature fluorescent protein.
Conclusions.
A major use for GFP will be as a replacement for the GUS gene, which is widely used as a reporter for promoter and gene fusions in transformed plants. The GUS gene product can be localized or quantified using histochemical techniques, but these are generally destructive tests (1). In contrast, GFP can be directly seen in living tissues. For example, high levels of fluorescence intensity are obtained in GFP-transformed microbial colonies, allowing simple screening for GFP expression with the use of a hand-held UV lamp. Such an assay for gene expression in living plants will be a very useful tool for plant transformation and genetic experiments. Many transformation techniques give rise to regenerating tissues that are variable or chimeric and that require testing of the progeny of the primary transformants. Potentially, GFP-expressing tissues could be monitored using in vivo fluorescence, avoiding any need for destructive testing, and the appropriate transformants could be rescued and directly grown to seed. Similarly, in vivo fluorescence will be an easily scored marker for field testing in plant breeding, allowing transgenes linked to the GFP gene to be easily followed.
Unlike enzyme markers, GFP can be visualized at high resolution in living cells using confocal microscopy. The images are not prone to fixation or staining artifacts, and can be of exceptional clarity. Moreover, the activities of living cells, such as cytoplasmic streaming, are clearly evident during microscopy. Genetic systems such as that of Arabidopsis provide a large resource of potentially informative mutants, and there has been much recent improvement in techniques for determining the molecular basis of a particular phenotype. The use of fluorescent proteins will provide a further tool for examining the biology of mutant cells. The ability to monitor simply and precisely both particular cells and subcellular structures that have been highlighted with a fluorescent signal will improve both the screening for particular abnormal phenotypes and the characterization of dynamic processes.
Acknowledgments
We wish to thank Brad Amos for generous and invaluable help with confocal microscopy, Peter York of Leica U.K. for help and the gift of objective adapters, and Andrea Brand, Dave Baulcombe, John Brown, and Mary Schuler for their advice and help. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council Plant and Animal Genome Analysis program (PAG/01518).
ABBREVIATIONS
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- RT-PCR
reverse transcription–PCR
- ER
endoplasmic reticulum
Footnotes
References
- 1.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heim R, Prasher D C, Tsien R Y. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cody C W, Prasher D C, Westler W M, Prendergast F G, Ward W W. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 4.Yang F, Moss L G, Phillips G N. Nat Biotechnol. 1996;14:1246–1220. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 5.Ormo M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 6.Prasher D C, Eckenrode V K, Ward W W, Prendergast F G, Cormier M J. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 8.Inouye S, Tsuji F I. FEBS Lett. 1994;341:277–280. doi: 10.1016/0014-5793(94)80472-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang S X, Hazelrigg T. Nature (London) 1994;369:400–403. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- 10.Baulcombe D C, Chapman S, Cruz S S. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 11.Heinlein M, Epel B L, Padgett H S, Beachy R N. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 12.Haseloff J, Amos B. Trends Genet. 1995;11:328–329. doi: 10.1016/0168-9525(95)90186-8. [DOI] [PubMed] [Google Scholar]
- 13.Hu W, Cheng C L. FEBS Lett. 1995;369:331–334. doi: 10.1016/0014-5793(95)00776-6. [DOI] [PubMed] [Google Scholar]
- 14.Sheen J, Hwang S B, Niwa Y, Kobayashi H, Galbraith D W. Plant J. 1995;8:777–784. doi: 10.1046/j.1365-313x.1995.08050777.x. [DOI] [PubMed] [Google Scholar]
- 15.Valvekens D, Van Montagu M, Van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutcke H A, Chow K C, Mickel F C, Moss K A, Kern H F, Scheele G A. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siemering K R, Golbik R, Sever R, Haseloff J. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 18.Reichel C, Mathur J, Eckes P, Langenkemper K, Reiss B, Koncz C, Schell J, Maas C. Proc Natl Acad Sci USA. 1996;93:5888–5893. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward W W, Cody C W, Hart R C, Cormier M J. Photochem Photobiol. 1980;31:611–615. [Google Scholar]
- 20.Chiu W L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 21.Luehrsen K R, Taha S, Walbot V. Prog Nucleic Acid Res Mol Biol. 1994;47:149–193. doi: 10.1016/s0079-6603(08)60252-4. [DOI] [PubMed] [Google Scholar]
- 22.Simpson C G, Clark G, Davidson D, Smith P, Brown J W S. Plant J. 1996;9:369–380. doi: 10.1046/j.1365-313x.1996.09030369.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu H X, Filipowicz W. Plant J. 1996;9:381–389. doi: 10.1046/j.1365-313x.1996.09030381.x. [DOI] [PubMed] [Google Scholar]
- 24.Wiebauer K, Herrero J-J, Filipowicz W. Mol Cell Biol. 1988;8:2042–2051. doi: 10.1128/mcb.8.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodall G J, Filipowicz W. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- 26.Goodall G J, Filipowicz W. EMBO J. 1991;10:2635–2644. doi: 10.1002/j.1460-2075.1991.tb07806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanley B A, Schuler M A. Nucleic Acids Res. 1988;16:7159–7176. doi: 10.1093/nar/16.14.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niedz R P, Sussman M R, Satterlee J S. Plant Cell Rep. 1995;14:403–406. doi: 10.1007/BF00234043. [DOI] [PubMed] [Google Scholar]
- 29.Zolotukhin S, Potter M, Hauswirth W, Guy J, Muzyczka N. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas J, Park E C, Seed B. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 31.Green M R. Ann Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 32.Langford C J, Klinz F-J, Donath C, Gallwitz D. Cell. 1984;36:645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- 33.Davenport D, Nichol J A C. Proc R Soc Ser B. 1955;144:399–411. [Google Scholar]
- 34.Fischer G. Angew Chem Int Ed Engl. 1994;33:1415–1436. [Google Scholar]