Abstract
Bacterial cDNA expression libraries are made to reproduce protein sequences present in the mRNA source tissue. However, there is no control over which frame of the cDNA is translated, because translation of the cDNA must be initiated on vector sequence. In a library of nondirectionally cloned cDNAs, only some 8% of the protein sequences produced are expected to be correct. Directional cloning can increase this by a factor of two, but it does not solve the frame problem. We have therefore developed and tested a library construction methodology using a novel vector, pKE-1, with which translation in the correct reading frame confers kanamycin resistance on the host. Following kanamycin selection, the cDNA libraries contained 60–80% open, in-frame clones. These, compared with unselected libraries, showed a 10-fold increase in the number of matches between the cDNA-encoded proteins made by the bacteria and database protein sequences. cDNA sequencing programs will benefit from the enrichment for correct coding sequences, and screening methods requiring protein expression will benefit from the enrichment for authentic translation products.
Keywords: cloning vector, ORF, ORF vector, in-frame vector
Bacterial cDNA expression libraries are made to reproduce protein sequences from a source tissue and are typically screened for antibody-binding or functional domains. Our goal is to use pooled cDNA-encoded proteins as complex antigens for making panels of monoclonal antibodies, then to isolate the epitope-encoding cDNAs. It is of great importance in such screens that the expressed polypeptides be authentic, lest the isolated clones be useless. This requires cDNA expression libraries that produce mostly authentic proteins. But translation of library cDNAs is initiated by prokaryotic signals in the vector, so cloning cDNAs in an expression vector without respect to orientation or knowledge of the correct frame will yield a majority of clones in either the wrong orientation (1/2) or the wrong frame (2/3), or containing 5′ or 3′ untranslated sequences (≈1/2). Only ≈8% of clones can be expected to produce authentic protein sequences.
Methods have been devised that partially circumvent these problems. Directional cloning of oligo(dT)-primed cDNA fragments ensures that only forward reading frames are expressed in the library (1–4). The pORF vectors contain transcription and translation start sites, followed by a cDNA cloning site and an out-of-frame β-galactosidase coding sequence (5, 6). Recently, vectors have been constructed that translate cloned sequences in all three reading frames (pFlag·shift12, IBI), so that correct proteins are produced somewhere in the library. However, these vectors do not reduce the background of incorrect translation products, do not exclude 5′ and 3′ nontranslated sequences, and do not permit the selective production or purification of the correct protein sequences.
We have designed and tested a new vector, pKE-1 (for Kosher Expression-1; pronounced picky one). By its design, cDNAs cloned in ORFs should confer kanamycin resistance upon the host, whereas the vector alone and cDNA inserts containing stop codons in the frame translated by the bacteria should not. We describe here the construction of this vector and the testing of the selection process and show that it is now possible to make cDNA libraries in Escherichia coli that are highly enriched in clones that produce authentic polypeptides.
MATERIALS AND METHODS
Construction of pKE-1.
A DNA fragment bearing the coding region of the kanamycin resistance gene, flanked 5′ to the ATG translation initiation codon by the sequence 5′-GCGGATCCTGGCATCGACTAG and 3′ to the termination codon by the sequence 5′-TCAGAATTCGGC was constructed by PCR amplification of a kanamycin phosphotransferase (KanPT) cassette (Pharmacia). This introduced a BamHI site and a TAG stop codon 5′ to the KanPT start codon, ATG, and an EcoRI site at its 3′ end. The KanPT coding sequence begins with ATGA so there is a TGA stop codon in a second reading frame overlapping the KanPT start codon. After cleaving with BamHI and EcoRI, the fragment was ligated into the expression vector pGEX-2T (Amrad, Melbourne, Australia; ref. 7). For directional cDNA cloning, AscI and MluI restriction sites were inserted at the BamHI site by ligating in the short double-stranded DNA fragment 5′-GATCCGGCCGTATGGCGCGCC GCCGGCATACCGCGCGGCTAG-5′.
The vector (see Fig. 1) was designated pKE-1.
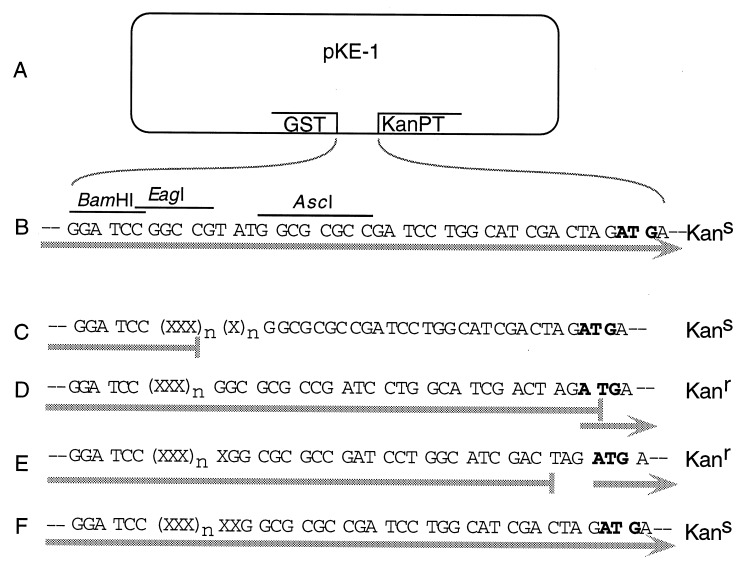
Figure 1.
Design and function of the pKE-1 vector. (A) pKE-1 was made by modifying pGEX-2T. It contains the glutathione-binding domain of GST under the regulation of the inducible tac promoter, directional cloning sites, a stop/start sequence, and the kanamycin phosphotransferase coding region (KanPT). (B) Sequence between GST and the KanPT start ATG codon. BamHI, EagI, and AscI are the cDNA cloning sites. Shaded arrow indicates the path of translation and nucleotide triplets show the reading frame. Bacteria containing pKE-1 alone are expected to be kanamycin-sensitive (Kans), as KanPT is translated in the wrong reading frame. (C–F) Insertion of cDNAs, represented by Xs, has four possible outcomes. (C) One possible outcome is that the cDNA fragment contains an in-frame stop codon. In this case, translation stops, the ribosomes disengage, KanPT is not translated, and the host bacterium is therefore expected to be Kans. If the cDNA is open, there are three possibilities. (D) One is that the length of the inserted cDNA is a multiple of three. Ribosomes traverse the entire length of the cDNA and stop at the TGA codon, but may reinitiate translation on the overlapping KanPT start codon. The host bacteria are therefore expected to be kanamycin-resistant (Kanr). (E) Another possibility is that the length is a multiple of three plus one. Ribosomes stop at the TAG stop codon, but can reinitiate translation on the adjacent KanPT start codon. The host bacteria are expected to be Kanr. (F) Finally, it is possible that the length is a multiple of three plus two. In this case, the reading frame is the same as in the vector alone. Although it is open, the host bacteria are expected to be Kans.
Preparation of cDNA.
Short, random-primed cDNAs were synthesized from 15-day embryonic mouse brain poly(A)+ RNA by the substitution method (8). For directional cloning, we made the following modifications to the procedure supplied with the Superscript reverse transcriptase (BRL). (i) Instead of random hexamers, primers with the sequence 5′-GCTCGCCCTCGCGGCGCGCCNNNNNT were used. The primers introduce an AscI site at the end of the cDNA, corresponding to the 3′ end of the mRNA, and so permit directional cloning. The sequence 5′ of the AscI site buffers the restriction site from the 5′ → 3′ exonuclease activity of the E. coli polymerase I used during the second strand synthesis. The terminal T residue prevents self-priming, since the 3′ portion of the primer is devoid of A residues. (ii) The first strand was synthesized at 15°C for 16 hr. Double-stranded linkers with the sequence 5′-CTGGCTCGCCCTCGCGGATCCG AGACCGAGCGGGAGCGCCTAGGC-5′
were ligated on at 16°C overnight. Since these cDNAs were also used for constructing self-subtracted libraries, they were subjected to 10 cycles of PCR amplification using the primer 5′-CTGGCTCGCCCTCGCGG. This has no bearing on the efficiency or use of the selection system described here, as shown by the results.
Construction of Sized cDNA Libraries.
cDNA was cleaved with BamHI and AscI and electrophoresed through a 2% low melting temperature agarose gel. Fragments in the size ranges 100–200, 200–300, and 300–500 bp were eluted from the gel and ligated into pKE-1, which had also been cleaved with BamHI and AscI. E. coli DH10B (BRL) were transformed with the ligation product by electroporation, and aliquots were plated onto agar containing 50 μg/ml ampicillin. Libraries in E. coli XL1-Blue (Stratagene) were constructed by purifying DNA from aliquots of the libraries in E. coli DH10B and then transforming XL1-Blue.
Response to Kanamycin Selection.
To assay the effect of kanamycin selection, ≈10,000 colonies were pooled from each library in E. coli DH10B and plated in quadruplicate on agar containing 0, 6.25, 12.5, 25, 37.5, 50, 62.5, 75, 87.5, or 100 μg/ml kanamycin. As a control, E. coli DH10B bearing pKE-1 alone were plated on agar containing 0, 12.5, 25, 50, or 100 μg/ml kanamycin. For the libraries in E. coli XL1-Blue, 10,000 colonies were pooled from each library and plated in duplicate on agar containing 0, 12.5, 25, 50, 100, or 150 μg/ml kanamycin. In all cases, survival fractions of visible colonies were determined by examination of the plates after ≈20 hr of incubation at 37°C.
Sequence Analysis of Cloned cDNA Fragments.
Ten clones from each DH10B library grown at 0, 12.5, or 37.5 μg/ml kanamycin, a total of 120 clones, were sequenced by the dideoxy technique (9) using the primer 5′-CTGGCAAGCCACGTTTGGTG, which binds within the glutathione S-transferase (GST) coding sequence, and a deazaG sequencing kit (United States Biochemical). Twenty clones from the 100- to 200-bp and 200- to 300-bp and 19 clones from the 300- to 500-bp library in XL1-Blue were completely sequenced using an Applied Biosystems automatic sequencer and dye terminator protocol. For each clone, conceptually translated proteins from all six reading frames were derived from the sequence. The lengths of the translated reading frames were calculated, and all protein sequences were used to search the combined protein sequence databases for homologous sequences, using the blast (10) server at the National Center for Biotechnology Information (NCBI). In the clones from the libraries in DH10B, sequence data were complete for all but 8 of the 60 clones in the 100- to 200-bp and 200- to 300-bp size ranges. Between 100 and 200 nt of sequence were collected for each of the 30 clones in the 300- to 500-bp size range. In cases where the sequence data were incomplete, the sizes of the cDNAs were determined by agarose gel electrophoresis, following cleavage with BamHI and AscI.
Western Blot Analysis.
To determine the sizes of polypeptide sequences encoded, individual clones were grown overnight in Luria–Bertani broth plus 50 μg/ml ampicillin, diluted 30-fold in fresh medium, and grown for 2.5 hr at 37°C. The cultures were induced with 0.05 mM isopropyl β-d-thiogalactoside (IPTG) for 5.5 hr at room temperature, then pelleted, lysed in an SDS/urea buffer, and subjected to 10% SDS/PAGE (11). The fusion proteins were detected by Western blot analysis using an anti-GST polyclonal antiserum (Pharmacia). The length in nucleotides of the translated cDNA sequence was calculated as [(MW − 26,500) × 3]/110, where MW is the molecular weight of the fusion protein, estimated from its gel mobility, 26,500 is the molecular weight of the GST fusion partner, 110 is the average molecular weight of an amino acid, and 3 is the number of nucleotides per codon. These lengths were compared with those of the cloned cDNAs as determined by sequencing or agarose gel mobility. It was not expected that the sequencing and Western data would precisely match, due to the variability in amino acid molecular weights and the effect of protein sequence on mobility, but if the ratio (estimated coding region length)/(estimated cDNA length) was >0.8 or if the two did not differ by >30 nt, the cDNA was considered to be cloned in an ORF.
RESULTS
Design Rationale and Function of pKE-1.
The expression vector pKE-1 (Fig. 1A) was made from the commercially available vector pGEX-2T, in which the cDNAs are cloned 3′ to the glutathione-binding domain of GST. Transcription of the latter is under control of the inducible tac promoter. We added to this a synthetic DNA fragment containing directional cloning sites followed by a stop/start sequence and the coding region of kanamycin phosphotransferase (KanPT). Although coding sequences of eubacterial polycistronic messages are usually preceded by their own Shine–Dalgarno ribosome-binding sites, translation can reinitiate without a ribosome-binding site if an AUG codon is within a few nucleotides of the stop codon of a preceding translated sequence (12).
In the vector itself (Fig. 1B), translation proceeds through the GST sequence and into KanPT, but in the wrong frame, leaving the host kanamycin-sensitive (Kans). Nonrecombinants are therefore eliminated from the library. A bacterium containing a cDNA clone with a stop codon in the frame translated by the bacteria (Fig. 1C) will also be Kans, since KanPT is not translated. For cDNAs that do not contain any stop codons in the frame translated by the bacteria, there are three different outcomes. In two of these cases (Fig. 1 D and E), translation proceeds through the GST–cDNA fusion sequence and continues until the ribosome encounters one of the two stop codons, TGA (Fig. 1D) or TAG (Fig. 1E), which are near the ATG start codon of KanPT. Some ribosomes will then reinitiate translation of KanPT, conferring resistance to kanamycin (Kanr). In the third case (Fig. 1F), translation beyond the cDNA is the same as in the vector alone, and such a clone will be lost upon selection.
pKE-1 is designed such that cloned cDNA sequences containing stop codons in the frame translated by the bacteria will leave the host bacteria kanamycin-sensitive. cDNA sequences cloned in reading frames which do not contain stop codons in the frame translated by the bacteria should (with a 2/3 probability) confer kanamycin resistance. mRNA sequences are likely to have frequent stop codons in all reading frames except the correct one, so selecting for sequences cloned in an ORF enriches for those that are cloned in the correct reading frame and produce authentic protein sequences.
Response to Kanamycin Selection.
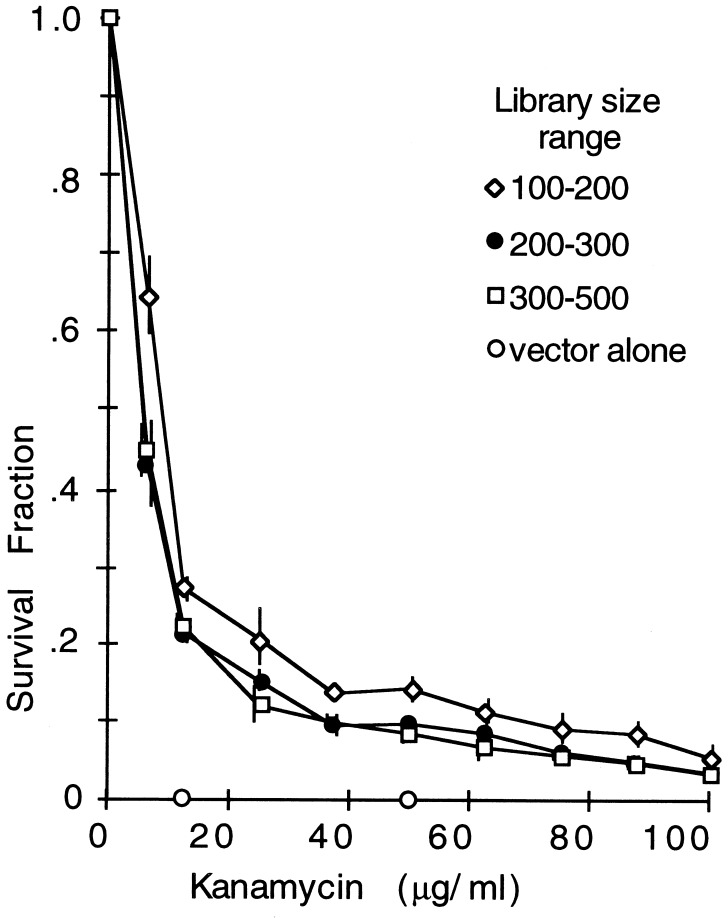
Two bacterial host strains were used, E. coli DH10B and E. coli XL1-Blue. In the initial experiments, E. coli DH10B bearing pKE-1 alone were grown on plates containing 0, 12.5, 25, 50, or 100 μg/ml kanamycin (Fig. 2). The survival fractions were 2 × 10−4 at 12.5 μg/ml and <5 × 10−6 at 50 or 100 μg/ml. Three different libraries of cDNAs in the size ranges 100–200, 200–300, and 300–500 bp were constructed in pKE-1 from 15-day embryonic mouse brain mRNA using DH10B. These libraries were grown on plates containing different concentrations of kanamycin (Fig. 2). As expected, insertion of cDNAs into pKE-1 resulted in kanamycin-resistant colonies, whereas the vector alone did not. There was no way to judge, a priori, what selection stringency would work best, but the slight survival plateau observed between 37.5 and 50 μg/ml kanamycin was suggestive.
Figure 2.
Survival of sized cDNA libraries after selection with kanamycin. Four different libraries in E. coli DH10B were made from cDNA fragments in the size ranges 50–100, 100–200, 200–300, and 300–500 bp. Each was grown on agar containing 6.25, 12.5, 25, 50, and 100 μg/ml kanamycin. Bacteria containing vector alone were grown on agar containing 0, 12.5, 25, 50, or 100 μg/ml kanamycin. The surviving colonies were counted. The survival values for the vector alone at 12.5 and 50 μg/ml kanamycin were 10−4 and 10−6, respectively. Bars indicate 95% confidence intervals.
Selection for Cloning in ORFs.
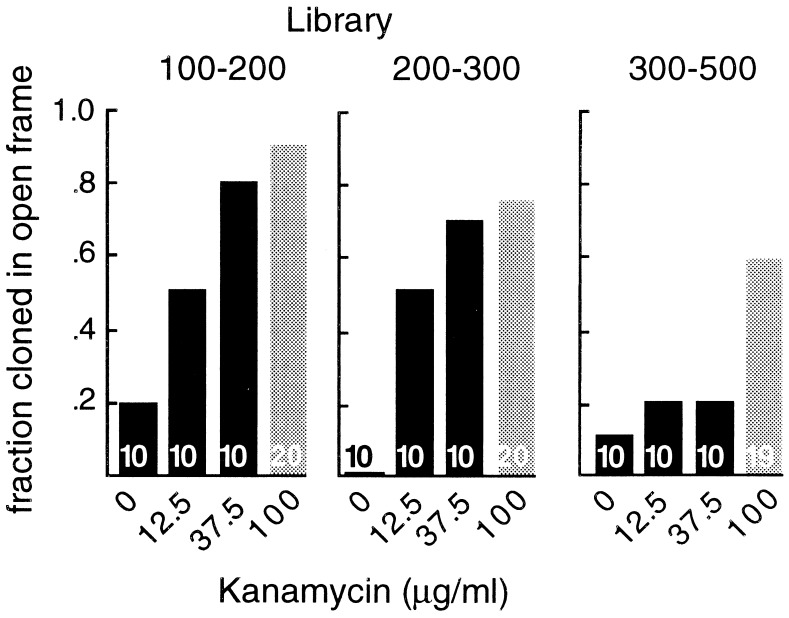
To determine whether the libraries were enriched for sequences cloned in ORFs, clones were isolated and sequenced. Ten clones were chosen at random from the different sized libraries in E. coli DH10B, each grown at 0, 12.5, and 37.5 μg/ml kanamycin, for a total of 120 clones. We also estimated the lengths of the translated sequences by Western blot analysis of the fusion proteins produced and compared them to the lengths of the cDNA inserts. Similar lengths indicate that the entire cDNA was translated and so was cloned in an ORF (see Materials and Methods). The two independent data sets were in agreement, and the combined results are shown in Fig. 3 (black bars). In the 100- to 200-bp library, 8 of 10 clones selected at 37.5 μg/ml kanamycin were cloned in ORFs. In the 200- to 300-bp library, 7 of 10 clones selected at 37.5 μg/ml kanamycin were cloned in ORFs. However, in the 300- to 500-bp library, only 4 of the 20 kanamycin-selected clones were judged to be cloned in ORFs. So selection enriched considerably for sequences cloned in ORFs, but worked poorly for the larger, 300- to 500-bp cDNAs.
Figure 3.
Fraction of sequences cloned in ORFs. Clones were picked at random from the different sized cDNA libraries that had been subjected to selection with 0, 12.5, 37.5, or 100 μg/ml kanamycin. Two independent methods, DNA sequencing and Western blotting, were used to determine whether they were cloned in ORFs. Black and gray bars represent groups of clones from libraries in E. coli DH10B and E. coli XL1-Blue, respectively. The numbers of clones analyzed in each of the four sets is shown in the corresponding bar.
The dependence of selection on cDNA size suggested that interactions between the cDNA sequences and the bacterial transcription/translation machinery, rather than a feature of the pKE-1 vector, might be responsible for the reduced efficiency. We therefore tried a different E. coli host strain. The initial host, DH10B, contains a mutation in the S12 small ribosomal subunit that confers resistance to streptomycin (RpsL) and has been shown to alter the translation process (13, 14). The libraries were therefore transferred into a non-RpsL host, E. coli XL1-Blue, and the analyses were repeated. The survival curves in this host were essentially similar, although with plateaus at 50–100 μg/ml kanamycin. Twenty clones from each size range, selected at 100 μg/ml were analyzed as before to determine the efficiency of selection for sequences cloned in ORFs. Switching the host strain indeed increased the efficiency of the selection process (Fig. 3, gray bars). The fraction of sequences cloned in ORFs in the 100- to 200-bp, 200- to 300-bp, and 300- to 500-bp libraries were 0.90, 0.75, and 0.58, respectively.
Selection for Cloning in the Correct Reading Frame.
Are the open frames in which the cDNA are cloned also the authentic coding frames? Calculations based on sequences chosen at random by computer from GenBank murine cDNA sequences indicated that the expected frequency of open but incorrect reading frames in the 100- to 200-bp range would be ≈15% and <5% for the larger sequences (calculations not shown). We also searched the National Center for Biotechnology Information (NCBI) combined protein databases for sequences matching closely (P < e−11) to the clones that had been sequenced after selection. Kanamycin selection resulted in an ≈2-fold increase in sequences matching protein database entries (Table 1, [kan] = 100 μg/ml vs. [kan] = 0 μg/ml), probably due to the unique ability of the cloning system to discard 5′ and 3′ untranslated sequences from the cDNA libraries. Since only correct reading frames encode naturally occurring proteins, a close match also identifies the correct reading frame of the cloned cDNA. In the selected libraries, we observed an ≈10-fold increase in matches to the frames of the cloned cDNAs translated by the bacterial host (Table 2, [kan] = 100 μg/ml vs. [kan] = 0 μg/ml). Details of the set of 300- to 500-bp clones selected at 100 μg/ml kanamycin are in Table 3. This shows that pKE-1 does, indeed, strongly select for sequences that are cloned in the correct reading frame and therefore produce authentic polypeptides.
Table 1.
Fraction of clones scoring database matches in any frame
| Host | [kan], μg/ml | Library
|
||
|---|---|---|---|---|
| 100- to 200-bp | 200- to 300-bp | 300- to 500-bp | ||
| E. coli DH10B | 0 | 0.4 | 0.1 | 0.2 |
| 12.5 | 0.2 | 0.6 | 0.4 | |
| 37.5 | 0.2 | 0.6 | 0.3 | |
| E. coli XL1-Blue | 100 | 0.3 | 0.7 | 0.6 |
Comparison of the sequences of the isolated clones with those in the combined databases at the NCBI, using the blast algorithm. To ensure correct assignment of the reading frame, only those similarities with a random match probability of less than e−11 were scored. The number of clones in each group is indicated in Fig. 3. The fraction of clones in each group scoring a match in any of six possible reading frames is shown. [kan], Kanamycin concentration.
Table 2.
Fraction of clones scoring database matches in the host-translated frame
| Host | [kan], μg/ml | Library
|
||
|---|---|---|---|---|
| 100- to 200-bp | 200- to 300-bp | 300- to 500-bp | ||
| E. coli DH10B | 0 | 0.3 | 0 | 0 |
| 12.5 | 1 | 0.7 | 0.5 | |
| 37.5 | 1 | 0.7 | 0.7 | |
| E. coli XL1-Blue | 100 | 0.7 | 0.7 | 0.8 |
The fraction of the scored matches (Table 1) that are in the reading frame actually translated by the host bacterium are shown. [kan], Kanamycin concentration.
Table 3.
Details of the 300- to 500-bp clones in XL1-Blue host selected at 100 μg/ml kanamycin
| Parameter | Value |
|---|---|
| Total no. clones analyzed | 19 |
| No. cloned in an open frame | 11 |
| No. of clones scoring database matches in any frame | 11 |
| No. of clones scoring database matches in the host-translated frame | 9 |
Database match entries and descriptions are as follows: spQ01149, mouse procollagen; spQ09163, mouse Δ-like protein; pirΔ27671, rat spectrin; spP15303, yeast sec23; pirA33313, goose malonyl-CoA decarboxylase; spP41123, rat 60S ribosomal protein L13; spP37397, rat calponin; gi1228982, Xenopus C4SR protein; and gi554476, myosin heavy chain.
DISCUSSION
The reason for making bacterial cDNA expression libraries is to reproduce the source tissue protein repertoire in the host. However, since translation must be initiated on vector-encoded sequence and since there is no control over the frame in which the cDNA is cloned, most translation products in a cDNA expression library are irrelevant. We have described here the development and testing of a new methodology for making cDNA expression libraries that contain a majority of sequences cloned in the correct reading frame and so produce only authentic protein sequences. The system uses a novel expression vector, pKE-1, which selectively eliminates nonrecombinants and clones containing stop codons in the cDNA reading frame translated by the bacteria.
The vector was tested by constructing and analyzing sized cDNA libraries. The frequency of sequences cloned in ORFs depended upon selection and upon the sizes of the cDNAs. (In χ2 tests, the null hypotheses—no effect of either size or selection—are rejected with P < 0.01.) Efficiency was also better in E. coli XL1-Blue than in DH10B for the larger library (P < 0.05). Selection increased the proportion of in-frame clones from ≈8% to 70%. This is a major change; it decreases the number of different protein sequences that must be screened by almost 10-fold. A perfect library would only decrease the number by another 30%.
We do not understand why selection efficiency depends on the cDNA size. Nevertheless, the current functional size range is large enough to encode antibody epitopes and most protein domains. The increased efficiency obtained by simply changing the library host suggests that further improvements may be possible. Perhaps the very different codon usage in E. coli compared with eukaryotes is involved. Heterologous sequences containing a large number of codons that are rare in E. coli cause translation problems, including decreased protein production (15–17) and frame shifts (14, 17), which could result in both false positives and false negatives. Using the vector in a host optimized for the translation of eukaryotic sequences could possibly improve the selection efficiency for larger size inserts. Initial steps toward constructing such host E. coli strains have already been made by adding extra tRNAs for some of the rarely used codons (18, 19).
The cloning system described should be useful in several respects. Genome projects amassing large numbers of relatively short cDNA fragments (20) will benefit from the exclusion of nontranslated sequences from the libraries. One of the primary sources of information from such projects is via the identification of homologous sequences in the protein and cDNA sequence databases. Since significant homologies are most likely to be found in coding sequences, it is advantageous to exclude noncoding sequences from these programs (21). Screens of cDNA expression libraries that rely on antibody or ligand binding will benefit from the reduction in the background of irrelevant protein sequences that must be screened. Most importantly, pKE-1 will make feasible screening methods that depend on selectively producing authentic cDNA-encoded proteins. Our goal of using these proteins to generate antibody panels is one such example.
Acknowledgments
We thank Lynette Dowling and Viveca Sapin for expert technical assistance. This work was supported by fellowships to C.A.D. from the Human Frontiers in Science Program and the Canadian Medical Research Council and by grants to S.B. from the National Science Foundation (MCB-9408718), the National Institutes of Health (AG12289 and EY09278), the McKnight Foundation, and the James G. Boswell Foundation.
ABBREVIATION
- GST
glutathione S-transferase
References
- 1.Meissner P S, Sisk W P, Berman M L. Proc Natl Acad Sci USA. 1987;84:4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleclough C, Erlitz F L. Gene. 1985;34:305–314. doi: 10.1016/0378-1119(85)90139-8. [DOI] [PubMed] [Google Scholar]
- 3.Helfman D M, Fiddes J C, Hanahan D. Methods Enzymol. 1987;152:349–359. doi: 10.1016/0076-6879(87)52042-0. [DOI] [PubMed] [Google Scholar]
- 4.Miki T, Matsui T, Heidaran M A, Aaronson S A. Gene. 1989;83:137–146. doi: 10.1016/0378-1119(89)90411-3. [DOI] [PubMed] [Google Scholar]
- 5.Weinstock G M, Rhys C A, Berman M L, Hampar B, Jackson D, Silhavy T J, Weisemann J, Zweig M. Proc Natl Acad Sci USA. 1983;80:4432–4436. doi: 10.1073/pnas.80.14.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstock G M. Methods Enzymol. 1987;154:157–163. doi: 10.1016/0076-6879(87)54075-7. [DOI] [PubMed] [Google Scholar]
- 7.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 8.Gubler U, Hoffman B J. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 9.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli E K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Spanjaard R A, van Duin J. Nucleic Acids Res. 1989;17:5501–5507. doi: 10.1093/nar/17.14.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sipley J, Goldman E. Proc Natl Acad Sci USA. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spanjaard R A, Vanduin J. Proc Natl Acad Sci USA. 1988;85:7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane J F. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 16.Robinson M, Lilley R, Little S, Emtage J S, Yarranton G, Stephens P, Millican A, Eaton M, Humphreys G. Nucleic Acids Res. 1984;12:6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg A H, Goldman E, Dunn J J, Studier F W, Zubay G. J Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spanjaard R A, Chen K, Walker J R, Vanduin J. Nucleic Acids Res. 1990;18:5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltito B J D, Ward J M, Hodgson J, Gershater C J L, Edwards H, Wysocki L A, Watson F A, Sathe G, Kane J F. J Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelly J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 21.Adams M D, Soares M B, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:373–386. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]