Abstract
Cytochrome P450 enzymes are monooxygenases that contain a functional heme b group linked to a conserved cysteine with a thiolate bond. In the native state, the central iron atom is hexacoordinated with a covalently bound water molecule. The exclusion of solvent molecules from the active site is essential for efficient enzymatic function. Upon substrate binding, water has to be displaced from the active site to prevent electron uncoupling that results in hydrogen peroxide or water. In contrast to typical hemoproteins, the protein surface is not directly accessible from the heme of cytochromes P450. We postulate a two-state model in which a conserved arginine, stabilizing the heme propionate in all known cytochrome P450 crystal structures, changes from the initial, stable side-chain conformation to another rotamer (metastable). In this new state, a functional water channel (aqueduct) is formed from the active site to a water cluster located on the thiolate side of the heme, close to the protein surface. This water cluster communicates with the surface in the closed state and is partly replaced by the flipping arginine side chain in the open state, allowing water molecules to exit to the surface or to reaccess the active site. This two-state model suggests the presence of an exit pathway for water between the active site and the protein surface.
Keywords: aromatase, potentials of mean force, protein cavities, protein dynamics/protein hydration
Cytochrome P450 enzymes are monooxygenases (1) that catalyze the incorporation of an oxygen atom into a large variety of endogenous (e.g., steroids and eicosanoids) and xenobiotic substrates. Mitochondrial/bacterial enzymes use a soluble iron-sulfur protein to transfer electrons between the flavoprotein (FAD reductase) and the P450 (class I enzymes), whereas microsomal P450 enzymes receive electrons directly from a FAD/FMN reductase (class II enzymes; ref. 2). Mitochondrial and microsomal P450 enzymes are coupled to NADPH as electron donors, whereas bacterial class I P450 enzymes are coupled to NADH.
These proteins contain a single heme b group, which plays a major role in activating molecular oxygen (3). In the absence of substrate, the Fe(III) atom is hexacoordinated with a cysteine-thiolate ligand from the protein and a covalently bound water (or hydroxide) trans from the sulfur atom (4). The Fe(III) atom oscillates between the low-spin state (S = 1/2) and the high-spin state (S = 5/2; ref. 5), with the low-spin state favored in the absence of substrate (hexacoordinated form). The incoming substrate displaces the covalently bound aqua ligand from the oxidized heme, followed by a transition to the high-spin state (6, 7).
Displacement of the active-site water molecules by the substrate shifts the Fe(II)/Fe(III) redox equilibrium potential (8) in camphor monooxygenase (P450cam) at pH 7.0 toward the ferro, Fe(II), state, from −300 to −173 mV, and is believed to decrease the polarity of the heme environment (9). This process favors reduction of the substrate-bound enzyme by putidaredoxin, the in vivo reductant of P450cam (1). The formal reduction potential ranges from −400 to −170 mV vs. normal hydrogen electrode for substrate-bound P450 enzymes (10).
The enzymatic cycle of cytochrome P450 proteins has been thoroughly investigated (11–19). The exclusion of solvent molecules from the active site is essential for efficient enzymatic function (19), but, in contrast to other hemoproteins, the protein surface is not directly accessible from the heme of cytochromes P450. Thus, the pathway followed by water from the active site to the bulk solvent is not obvious.
Some water molecules displaced by the natural substrate are suggested to occupy the internal solvent channel (14, 15, 20), described in the substrate-bound cytochrome P450 crystal structures (15, 21). This channel involves Thr-252cam and Glu-366cam, which are located opposite to heme propionates in the four known crystal structures. This channel involves three water molecules and a conserved glutamate side chain, all of which can act as a proton-relay system from the solvent toward the dioxygen–ferryl complex (16). In substrate-free cytochrome P450 (4, 22, 23), this channel is absent; only one or two water molecules are present in the vicinity of the glutamate side chain. The internal solvent channel does not account for the exclusion of all water molecules from the active site because it is not connected with the surface. This mechanism of excluding excess water molecules from the active site into bulk solvent is the subject of this paper.
Changes in volume measured during a low-spin- to high-spin-state transition induced by osmotic pressure indicated that about 19 water molecules are involved in this transition of the fenchone-bound P450cam (24). However, the actual number of water molecules might be affected by conformational changes associated with this process. Nevertheless, this number is surprisingly large compared with crystal structures of P450cam that show three to nine structural water molecules near the active site. Di Primo and coworkers (24) suggested that water molecules other than those located in the active site play a key role in modulating the spin equilibrium of P450cam.
We postulate a two-state model in which a functionally conserved arginine, which stabilizes the heme propionate in all known cytochrome P450 crystal structures (15, 21–23), changes from the initial (stable) side-chain conformation to another rotamer (metastable). This structural change may be one of many conformational substates sampled as a result of thermal fluctuations in proteins (25).
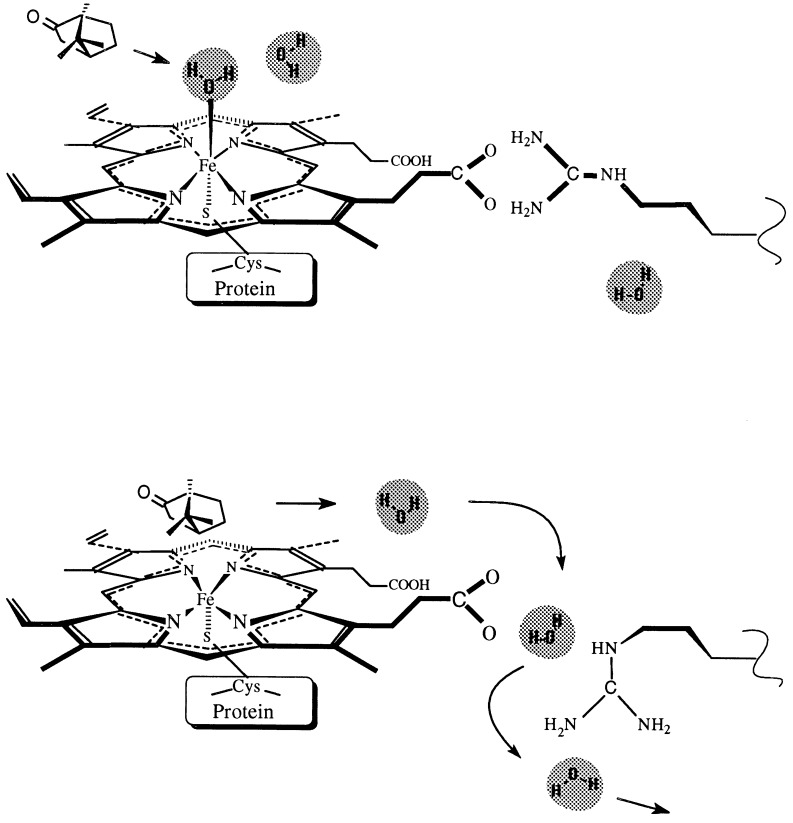
In this new state, a functional aqueduct is formed from the active site to a water cluster located on the thiolate side of the heme, close to the protein surface (see Fig. 1a). The term aqueduct is used to emphasize the active role of this channel in the transport of water and to distinguish it from the “internal water channel” (14, 15). The water cluster communicates with the surface in the closed state, and is partly replaced by the flipping arginine side chain in the open state, allowing water molecules to exit to the surface or to reaccess the active site.
Figure 1.
The two-state model of the functional aqueduct. Schematic illustration of the pathway followed by active-site water molecules upon binding of substrate (camphor; Upper), which forces the rotation of the arginine side chain, opening the functional aqueduct (Lower). The aqueduct closes after water is eliminated from the active site.
This two-state model may explain one exit pathway for water from the active site to the surface, as it is displaced by the incoming substrate, and the access of water from the surface to the active site as the catalytic product leaves the binding site. More important, to prevent uncoupling, this model allows displacement of excess water molecules from the active site toward the protein surface through use of a pathway different from the substrate access channel and the internal solvent channel. With this model, we can account for the other structural water molecules, which are not in the active site, that influence the spin equilibrium (24) in the osmotically perturbed P450cam. This model is supported by a detailed structural analysis of available high-resolution crystallographic data of cytochrome P450 enzymes and by the alignment of 200 sequences of P450 proteins. Theoretical calculations of cavities formed in the postulated two-state model and their hydration patterns are presented.
MATERIALS AND METHODS
Analysis of Structures Determined by Crystallography.
The structures of three class I (P450cam, P450terp, and P450eryF) and one class II (P450BM3) bacterial enzymes have been determined using x-ray crystallography. P450cam catalyzes the stereospecific hydroxylation of camphor at the 5-exo position and was studied in the substrate-bound (2cpp; ref. 4) and substrate-free (1phc; ref. 21) forms. P450terp catalyzes the hydroxylation of α-terpineol when this monoterpene is the sole carbon source for a Pseudomonas bacteria, and it was crystallized in its substrate-free form (1cpt; ref. 23). P450BM3 catalyzes the ω-end (nonpolar) hydroxylation of a variety of long-chain fatty acids, alcohols, and amides. The substrate was modeled (26) in the P450BM3 substrate-free (2hpd and 2bmh; refs. 22 and 26, respectively) crystal structures. P450eryF stereospecifically hydroxylates 6-DEB (6-deoxyerythronolide B) at the 6S position (27). Following hydroxylation, the 6-DEB macrolide ring is metabolized to erythromycin A. The crystal structure of substrate-bound P450eryF has been recently described (1oxa; ref. 15). Detailed analyses regarding these protein structures have been recently published (2, 15). Cartesian coordinates for 22 protein crystals of four different P450 proteins (18 for P450cam, 1 for P450terp, 2 for P450BM3, and 1 for P450eryF) have been retrieved from the Brookhaven Protein Data Bank (28, 29) and examined graphically (30, 31).
Cavity Analysis.
The presence and location of cavities in each protein were determined from a grid search. At each grid position, distances from a probe atom to the atoms of the molecule are calculated. If all distances are smaller than the sum of the van der Waals radii of each probe atom–molecule atom pair, the grid point is considered part of a cavity. The connectivity of cavities was determined using a clustering algorithm. The following settings were used for all 22 cytochrome P450 structures: probe atom radius, 1.0 Å; grid spacing, 0.3 Å; and atomic van der Waals radii, C = 1.8, N = 1.8, O = 1.5, and S = 1.85 Å. We use a small (1 Å) probe atom to compensate for the lack of structural fluctuations in the protein models. Crystal waters were not included in the cavity analysis.
Theoretical Calculations of Structural Hydration.
We used the Potentials of Mean Force (PMF) approach (32) to describe the organization of water inside protein cavities and at the surface of cytochrome P450 proteins. This approach is based on a statistical mechanical expression for the water-density distribution in terms of particle correlation functions. The local water density was calculated at Cartesian grid points (0.3 Å width). PMF calculations provide information on the equilibrium structural hydration, in addition to the crystallographic data. The intrinsic flexibility of the hydration network is reflected in the spatial extension and connectivity of calculated high water density regions. The accuracy of the local water density predictions have been verified against high resolution (<1 Å) DNA and RNA crystals (32).
The primary sequence alignment was used to determine the presence of conserved arginines in cytochrome P450 enzymes. The sequence alignment was based on the crystallographically determined alignments of P450cam with P450terp and P450BM3 (2) and with P450eryF (15), with preservation of structurally conserved motifs. The primary sequences of two microsomal P450 sequences, aromatase (estrogen synthetase, P450aro) and cholesterol side-chain cleavage (desmolase, P450scc) have been included in the analysis to represent mammalian P450 enzymes. Currently, 200 primary sequences of putative or known cytochromes P450 are deposited in the Swiss-Prot Protein Sequence Data Bank (33). These are part of the 481 genes and 22 pseudogenes of the 74 gene families (34) described so far. The 200 protein sequences have been aligned using the Swiss-Prot Data Bank and the blast algorithms (35).
RESULTS AND DISCUSSION
The Functional Aqueduct and the Two-State Model.
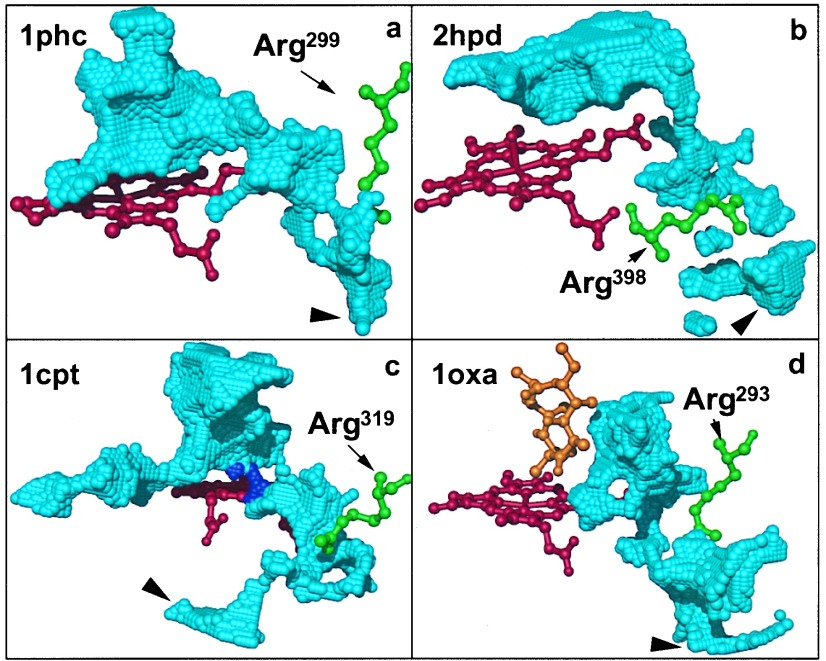
A possible pathway for the iron-bound water from the active site to the protein surface is located between the heme propionates, toward the thiolate side of the heme, into a conserved water cluster that communicates with the surface. This path is blocked by an arginine side chain (Arg-299cam, Arg-319terp, Arg-398BM3, and Arg-293eryF), which forms a salt bridge with one of the heme propionates. The arginine–propionate surroundings have been examined in the crystallographic structures of the four cytochrome P450 proteins, as summarized in Table 1. Hydrated salt bridge interactions are observed between propionate A and arginine in P450cam, P450terp, and P450eryF. In P450BM3, the arginine side chain interacts with the other (D) heme propionate. The arginine side chain is exposed to a water-occupied cavity (the conserved water cluster) in all four proteins, which allows the side chain to flip toward the protein surface (Fig. 1). This motion can be accomplished by modifying one or more torsional angles, as described for each protein in Table 2.
Table 1.
Interatomic distances in the arginine-propionate region of cytochrome P450 crystal structures
| Contact groups | P450cam Arg-299 | P450terp Arg-319 | P450BM3 Arg-398 | P450eryF Arg-293 |
|---|---|---|---|---|
| Heme propionate–arginine | 3.07 (O2A-Nη1) | 3.33 (O1A-Nη2) | 2.93 (O2D-Nη2) | 2.94 (O2A-Nη1) |
| 2.82 (O1A-Nη2) | 4.33 (O2A-Nη2) | 2.89 (O1A-Nη2) | ||
| Heme propionate–other | 2.74 (O2A-Asp-297Oδ2) | 5.49 (O1A-Asn-72Nδ2) | 2.97 (O2A-Lys-69Nζ) | 4.06 (CγA-His-349Cβ) |
| side chains | 3.20 (O2A-Asp-297Oδ1) | 5.78 (O1A-Asn-72Oδ1) | 2.64 (O1A-502) | |
| 3.06 (O2A-652) | 2.64 (O2A-547) | |||
| Arginine–other side chains | 4.23 (Nη1-Asp-297Oδ2) | 2.97 (Nη1-Asn-72Oδ1) | 3.03 (Nη2-Ser-89O) | 3.21 (Nη1-Thr-292O) |
| 4.00 (Nη2-Asp-297Oδ1) | 3.63 (Nη1-Asn-72Nδ2) | 3.06 (Nη1-Ser-89O) | 3.42 (Nɛ-Ser-59Oγ) | |
| 2.90 (Nη1-Thr-91Oγ) | ||||
| 2.97 (Nη2-Leu-86O) | ||||
| Water–arginine | 543, 558, and 582 (within 4.25 Å of Nɛ) | 520, 662, and 664 (within 3.8 Å of Nɛ) | 3.87 (Nη2-1035) | 3.16 (Nη1-547) |
| 1032 and 1036 (within 4.1 Å of Cγ) | 3.02 (Nη2-538) | |||
| 4.41 (Nη2-539) |
Distances (in angstroms) are given for the following structures: 1phc (P450cam), 1cpt (P450terp), 2hpd (P450BM3), and 1oxa (P450eryF). Water molecules are labeled with the numbers assigned in the corresponding Brookhaven Protein Data Bank file.
Table 2.
Torsional angles for the native-state (experimental) and “open-aqueduct” state
| Arginine | χ1 | χ2 | χ3 | χ4 | χ1′ | χ2′ | χ3′ | χ4′ |
|---|---|---|---|---|---|---|---|---|
| Arg-299cam | 178.0 | −79.1 | −176.3 | −88.5 | 178.0 | −160.7 | −176.3 | −160.0 |
| Arg-319terp | 170.5 | −51.3 | −173.1 | −101.8 | 180.0 | −32.3 | −60.0 | −101.8 |
| Arg-398BM3 | −177.1 | −48.3 | −58.7 | −83.9 | −177.1 | −179.2 | −58.7 | −80.3 |
| Arg-293eryF | 172.8 | −62.4 | −168.5 | −104.2 | 172.8 | −62.4 | −168.5 | 90.7 |
These changes in the side-chain conformation open a continuous pathway from the heme to the conserved water cluster. In the modified torsional state, the salt bridge is no longer present, and the arginine guanidinium group occupies the water cluster cavity and is partly exposed to the protein surface. Potentially positively charged residues (e.g., His-355cam, His-375terp, His-100BM3, and His-349eryF) are close to the heme propionate, providing a highly polar environment that eases the fluctuations of the Arg-293cam side to alternate torsional states. Surrounding water molecules can also lower the energy barrier for the transient breakage of the arginine–propionate ion pair. The energetics of an internal salt bridge interaction in proteins (36–38) suggest a free energy of ion-pair formation between 3.0 and 5.0 kcal/mol. The structural changes postulated in our two-state model include the breaking of a direct ion-pair contact and the formation of a water-mediated ion pair that should cost less free energy (39). We expect that most of the energy changes are related to the rotational barrier of the torsional side chain, of approximately 2 kcal/mol. The substrate binding is the limiting step for the reaction (1, 40). From the energetics of this fluctuation, we expect the arginine side-chain motion to be much faster than the time scale of substrate binding.
Cavity Analysis.
The cavities forming the aqueduct are shown in Fig. 2 for four representative P450 structures (1phc, 1cpt, 2hpd, and 1oxa). The arginine is rotated with the guanidinium group pointing into the water cluster (Table 2). The distal (upper) side of the heme faces the large substrate cavity. With the arginine rotated, the substrate cavity is connected to a second solvent cavity on the proximal (under) side of the heme plane. This second cavity is present in all 22 crystal structures and is occupied by water. A channel leads from the solvent cavity to the surface. Thus, rotating the arginine side chain is sufficient to open a connection from the active site to the exterior, even when the rest of the protein matrix is kept rigid.
Figure 2.
Cavities forming the aqueduct for four representative P450 structures (1phc, 1cpt, 2hpd, and 1oxa). The grid points comprising the cavities are shown in blue. Large arrowheads indicate where the cavities reach the protein surface. The substrate-access channel is not shown. (a) In P450cam (1phc), rotation of the Arg-299 side chain opens a wide channel directly connecting the substrate cavity to the protein surface. (b) In the Arg-398-modified P450BM3 (2hpd), the substrate cavity extends into the region previously occupied by water. However, the guanidinium group of the rotated Arg-398 blocks a direct path from the substrate cavity to the protein exterior. (c) In P450terp, substrate and solvent cavities are only connected if the probe radius is reduced to 0.82 Å in the cavity calculations (purple). Dynamic fluctuations are expected to widen the narrow channel formed between Phe-317 and Thr-103, which separate the two cavities. The second cavity, extending to the left, does not connect with the surface. (d) In P450eryF (1oxa), the solvent channel extends from the substrate cavity to the surface, with the narrowest part between the heme propionate A and Thr-291.
Structural Hydration.
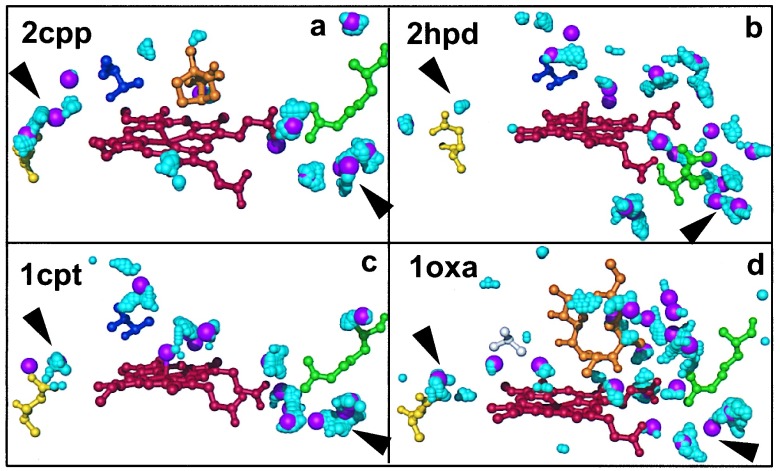
PMF calculations (32) were used to analyze the P450 hydration pattern. Regions of interest were the salt bridge between arginine and the heme propionate A (D for P450BM3) and the internal solvent channel formed between a threonine (alanine for P450eryF and the Thr-252 → Ala P450cam mutant; ref. 17) and a glutamate residue on the opposite side of the heme. Results are shown in Fig. 3. The PMF calculations of the local water density (probability of finding a water molecule) agree with the crystallographic findings; regions of high water density are observed near the heme–arginine salt bridge in all four different crystal structures, overlapping with waters Wat-652 in P450cam, Wat-522 in P450terp, Wat-1038 in P450BM3, and Wat-547 in P450eryF. Large, connected high-density clusters are found in the solvent cavity below the heme near the conserved arginine, overlapping with several water molecules (e.g., Wat-537 in P450cam, Wat-590 in P450terp, Wat-1036 in P450BM3, and Wat-503 in P450eryF). These solvent clusters are structurally conserved, suggesting a functional role. The spatial extension of the high-density regions connecting individual crystal water sites suggests a significant degree of flexibility in the hydration network. Fluctuations in the water cluster can facilitate a partial opening of the heme–arginine salt bridge.
Figure 3.
Structural hydration of the cytochrome P450 crystals. Hydration analysis using crystallographic data and PMF calculations of the regions near the heme propionate–arginine salt bridge and the internal solvent channel. The heme is shown in red, arginine in green, glutamate in yellow, threonine in blue, and alanine (1oxa) in gray. The large magenta spheres indicate crystallographic water positions. The cyan spheres correspond to grid points with high water density in the PMF calculations. Medium and small cyan spheres correlate to densities larger than five, and between three and five, respectively, in units of the bulk density of water. Results are shown for P450cam (a), P450BM3 (b), P450terp (c), and P450eryF (d). An extended water cluster is found near the arginine on the proximal side of the heme plane (lower right corners of a–d, indicated by arrowheads). The internal solvent channel is found in the P450cam (a) and P450eryF (d) structures (upper left corners of a–d, indicated by arrowheads). In P450cam, the solvent channel appears as a continuous high-water density wire in the PMF calculations. In P450eryF, water density regions are strongly localized.
Aspects of the internal hydration of P450cam were studied by Wade (41), using the grid program (42), and by molecular dynamics simulations (43). In agreement with the present study and the crystal structure, the heme–arginine salt bridge was found to be hydrated in both substrate-free and substrate-bound P450cam (Wat-652). The region of the conserved water cluster, critical to our postulate, was not specifically studied by Wade and coworkers (41).
Sequence Alignment.
The primary sequence alignment (adapted from Hasemann and coworkers, ref. 2, and Cupp-Vickery and Poulos, ref. 15) shows two distinct patterns; bacterial class I P450 sequences have a conserved arginine (Arg-299cam, Arg-319terp, and Arg-293eryF) that forms a salt bridge with the heme propionate A. The corresponding aligned residues from the class II P450 sequences are Leu-333BM3, Arg-375aro, and Arg-357scc. However, in the P450BM3 crystal structure, Arg-398 is observed to interact with the heme propionate D, whereas Lys-69 interacts with the propionate A (see Table 1). In the primary sequence alignment, the corresponding residues are histidines for class I sequences (His-355cam, His-375terp, and His-349eryF), and arginines for the class II proteins (Arg-435aro and Arg-421scc).
The 504-residue sequence of aromatase was used to search for the presence of other proteins with similar sequence in the Swiss-Prot Data Bank, using the blast algorithm. All of these proteins are biochemically characterized as cytochrome P450 enzymes. Among these proteins, 35 contain an arginine, and 1 contains a lysine, aligned to Arg-375aro (4–8 residues from the K helix, containing the Glu-X-X-Arg conserved residues; ref. 2); 15 other sequences have an arginine and 8 have a histidine aligned within 1 residue of Arg-375aro. Of these proteins, 44% have a positively charged residue (37% have an arginine) at the Arg-375aro location. Of the 134 sequences, 133 have an arginine located 2 residues before the thiolate cysteine, and 1 has a histidine.
The presence of arginine in the proximity of the thiolate ligand was studied using the heme–thiolate signature (44), F-[SNGH]-X-[GD]-X-[+]-X-C-[LIVMFAP]-[GAD], where + can be arginine, histidine, proline, or threonine, and X is any residue. This signature is characteristic for cytochrome P450 enzymes. The search identified 200 sequences in the Swiss-Prot Data Bank. Among the 198 biochemically characterized cytochromes P450, the + position was predominantly occupied by arginine (173 times). Histidine occurred in 19 sequences, whereas threonine and proline occurred rarely (3 and 1 sequences, respectively). The remaining two cytochrome-like sequences had a histidine and a threonine in that position. Thus, more than 85% of these sequences have an arginine at the Arg-398BM3 location.
The presence of Lys-69 in P450BM3 is unique, in both sequence alignment and crystal structures, in its stabilizing role for the heme propionate. A sequence probe of 100 residues from P450BM3 (with Lys-69 as the central residue) was used to look for the presence of other proteins with similar sequence in the Swiss-Prot Data Bank, using the blast algorithm. Among the most similar 50 sequences retrieved, no cytochrome P450 (except BM3 itself) was retrieved, suggesting an anomalous sequence in this region of P450BM3.
In summary, the primary sequence analyses suggest a high probability of finding an arginine that interacts with the heme propionate, which is involved in the functional aqueduct. The position of the residue is, however, not absolutely conserved in the primary sequence.
Comparison with the Internal Solvent Channel.
Our PMF calculation results agree with the crystallographic hydration pattern of the internal solvent channel (Fig. 4). We did not find high water density patterns connecting glutamate and threonine in the substrate-free P450cam, P450BM3, and P450terp structures. Interestingly, in the case of P450eryF, we found three strongly localized water-density peaks at the crystal–water positions. A gap of 5.4 Å between two water molecules is bridged with the carbonyl oxygen of Gly-242. In contrast to P450eryF, a wire of high water density connects Thr-252 and Glu-366 in most P450cam structures with a substrate or inhibitor bound (1pha, 1phb, 1phd, 1phf, 1phg, 1cp4, 2cpp, 3cpp, 4cpp, 5cpp, 6cpp, 7cpp, and 8cpp), indicative of flexibility in the water structure. In these crystal structures, two of the three water molecules forming the internal solvent channel are always present. The water molecule close to the threonine is absent in three structures (3cp4, 4cpp, and 6cpp). For two of these (4cpp and 6cpp), however, the calculations show high water density extending toward the threonine, suggesting that the position close to the threonine could in part be occupied by another water molecule. Interestingly, the water molecule close to the threonine appears to be tightly packed in the P450cam structures. This is most apparent in 1phe, where Wat-687 (occupancy 0.45) is within 2.9 Å and 2.42 Å of three carbon atoms and an oxygen atom, respectively.
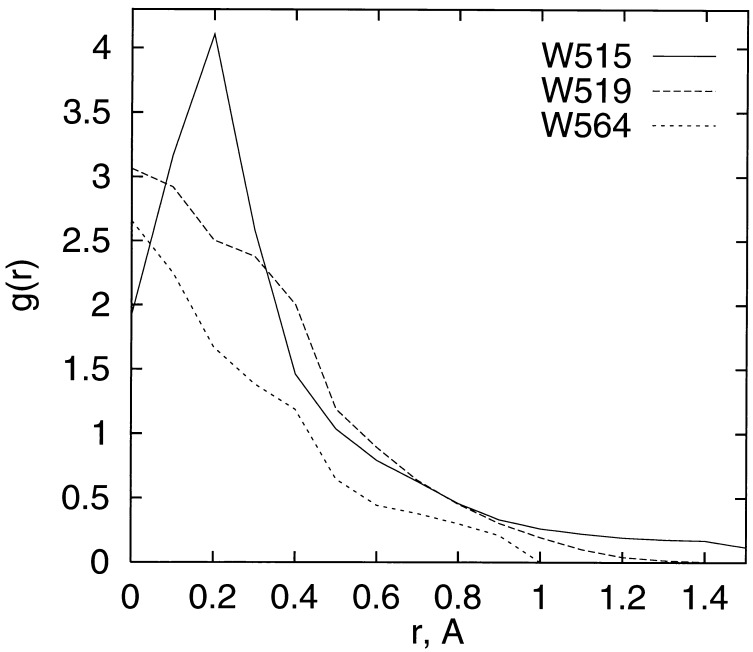
Figure 4.
Calculated radial distribution function g(r) of water around the water sites Wat-515, Wat-519, and Wat-564, comprising the internal solvent channel in P450eryF (1oxa). The function g(r) is the calculated average density of water in units of the bulk-water density on a spherical shell of radius r around a water site in the crystal. Values of g(r) larger than 1 indicate water localization. Around each of the three water sites in the crystal, the water density was calculated using the PMF method by averaging over 100,000 random points on each of the shells. The estimated statistical error of the g(r) values is 0.02. All three g(r) values show a peak at or near the crystal–water site (r = 0), indicative of high water density at the crystal position, in agreement with the crystallographic results.
The absence of three water molecules linking Thr-252 to Glu-366 in the internal solvent channel of substrate-free structures is indicative of the way the enzyme avoids hydrogen peroxide production in the absence of the substrate. The presence of water in the active site of these structures suggests two possible roles for water during enzyme activation: (i) removal of the aqua ligand and of the active-site structural water molecules reduces the redox potential and induces the transition from low to high spin; and (ii) water entering the internal solvent channel activates the charge relay system. The remaining water molecules, not involved in this process, can be readily eliminated through the functional aqueduct.
Osmotic pressure-induced volume changes during low- to high-spin transitions (24) in fenchone-bound P450cam suggest that approximately 19 water molecules are involved in this process. The quantitative aspect of this estimate is limited by the nature of the experimental (thermodynamic) measurement, and could be affected by conformational changes of the complex. Of the 19 structural waters, crystal structures account for three to nine water molecules near the active site. In our model, the open-aqueduct state may provide access to the active site for the seven structural water molecules located in the water cluster of substrate-bound (2cpp) and substrate-free (1phc) P450cam. This water cluster is open to bulk solvent, which can supply the remaining water molecules. Thus, the two-state model offers a possible explanation for the involvement of structural water molecules, in addition to those present in the active site, in modulating the spin equilibrium of P450cam.
CONCLUSION
Elimination of water from the active site has been previously shown to be essential for efficient electron coupling with the monooxygenation reaction. We have proposed a functional aqueduct that allows simultaneous displacement of excess water molecules from the active site toward the protein surface, using a different pathway than the substrate access channel and the internal solvent channel. In our model, a conserved arginine that stabilizes the heme propionate in all known cytochrome P450 crystal structures can change from the crystal side-chain conformation to another rotamer. The new conformation opens an aqueduct that connects the active site to a water cluster located close to the protein surface, on the proximal side of the heme. In the crystal structure, the aqueduct is closed and the water cluster communicates with the surface. In the open state, the arginine side chain occupies in part this cluster, allowing water molecules to exit to the surface, or to re-enter the active site. Changes between the stable closed state and the metastable open state occur continuously, independent of the substrate binding state. During a substrate binding event, we expect the water flow to be outward. With this two-state model, we can explain how other structural water molecules, in addition to those present in the active site, may be involved in modulating the spin equilibrium of the osmotically perturbed P450cam (24). Hereby we do not exclude the possibility for structural water to exit the catalytic site using other pathways, such as a substrate access channel.
By combining site-directed mutagenesis and molecular dynamics (45), further insight could be gained in the functional role of the proposed aqueduct. Site-directed mutagenesis could identify residues that are involved in the proposed water transport. Designing a mechanism and/or ligand that interferes with this aqueduct could provide a general pharmacological tool to block cytochromes P450. Demonstration of this mechanism could prove an invaluable addition to the experimental study of metabolic pathways and to the understanding of protein hydration and dynamics.
Acknowledgments
This work was supported by the U.S. Department of Energy through a Los Alamos National Laboratory LDRD-CD grant in Bioremediation.
ABBREVIATION
- PMF
Potentials of Mean Force
References
- 1.Gunsalus I C, Sligar S G. In: Advances in Enzymology and Related Areas in Molecular Biology. Meister A, editor. Vol. 47. New York: Wiley–Interscience; 1978. pp. 1–44. [DOI] [PubMed] [Google Scholar]
- 2.Hasemann C A, Kurumbail R G, Boddupalli S S, Peterson J A, Deisenhofer J. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 3.Erman J E, Hager L P, Sligar S G. Adv Inorg Biochem. 1994;10:71–118. [PubMed] [Google Scholar]
- 4.Poulos T L, Finzel B C, Howard A J. Biochemistry. 1986;25:5314–5322. doi: 10.1021/bi00366a049. [DOI] [PubMed] [Google Scholar]
- 5.Sligar S G, Gunsalus I C. Proc Natl Acad Sci USA. 1976;73:1078–1082. doi: 10.1073/pnas.73.4.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deprez E, Gerber N C, Di Primo C, Douzou P, Sligar S G, Hoa G H-B. Biochemistry. 1994;33:14464–14468. doi: 10.1021/bi00252a012. [DOI] [PubMed] [Google Scholar]
- 7.Harris D, Loew G. J Am Chem Soc. 1993;115:8775–8779. [Google Scholar]
- 8.Sligar S G, Gunsalus I C. Biochemistry. 1979;18:2290–2295. doi: 10.1021/bi00578a024. [DOI] [PubMed] [Google Scholar]
- 9.Raag R, Poulos T L. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 10.Krainev A G, Shimizu T, Hiroya K, Hatano M. Arch Biochem Biophys. 1992;298:198–203. doi: 10.1016/0003-9861(92)90113-b. [DOI] [PubMed] [Google Scholar]
- 11.Andersson L A, Dawson J H. In: Metal Complexes with Tetrapyrrole Ligands II. Buchler J W, editor. Berlin: Springer; 1991. pp. 2–40. [Google Scholar]
- 12.Atkins W M, Sligar S G. Biochemistry. 1988;27:1610–1616. doi: 10.1021/bi00405a033. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Narasimhulu S, Havran L M, Winkler J D, Poulos T L. J Am Chem Soc. 1995;117:6297–6299. [Google Scholar]
- 14.Raag R, Poulos T L. Biochemistry. 1991;30:2674–2684. doi: 10.1021/bi00224a016. [DOI] [PubMed] [Google Scholar]
- 15.Cupp-Vickery J R, Poulos T L. Nat Struct Biol. 1995;2:144–153. doi: 10.1038/nsb0295-144. [DOI] [PubMed] [Google Scholar]
- 16.Gerber N C, Sligar S G. J Am Chem Soc. 1992;114:8742–8743. [Google Scholar]
- 17.Raag R, Martinis S A, Sligar S G, Poulos T L. Biochemistry. 1991;30:11420–11429. doi: 10.1021/bi00112a008. [DOI] [PubMed] [Google Scholar]
- 18.Atkins W M, Sligar S G. J Am Chem Soc. 1989;111:2715–2717. [Google Scholar]
- 19.Loida P J, Sligar S G. Biochemistry. 1993;32:11530–11538. doi: 10.1021/bi00094a009. [DOI] [PubMed] [Google Scholar]
- 20.Di Primo C, Hoa G H-B, Douzou P, Sligar S G. J Biol Chem. 1990;265:5361–5363. [PubMed] [Google Scholar]
- 21.Poulos T L, Finzel B C, Howard A J. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 22.Ravichandran K G, Boddupalli S S, Hasemann C A, Peterson J A, Deisenhofer J. Science. 1993;261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 23.Hasemann C A, Ravichandran K G, Peterson J A, Deisenhofer J. J Mol Biol. 1994;236:1169–1185. doi: 10.1016/0022-2836(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 24.Di Primo C, Deprez E, Hoa G H-B, Douzou P. Biophys J. 1995;68:2056–2061. doi: 10.1016/S0006-3495(95)80384-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frauenfelder H, Sligar S G, Wolynes P G. Science. 1991;254:1508–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Poulos T L. Acta Crystallogr D. 1995;51:21–32. doi: 10.1107/S0907444994009194. [DOI] [PubMed] [Google Scholar]
- 27.Andersen J F, Tatsuta K, Gunji H, Ishiyama T, Hutchinson C R. Biochemistry. 1993;32:1905–1913. doi: 10.1021/bi00059a004. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein F C, Koetzle T F, Williams G J B, Meyer E F, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 29.Abola E E, Bernstein F C, Bryant S H, Koetzle T F, Weng J. In: Data Commission of the International Union of Crystallography. Allen F H, Bergerhoff G, Sievers R, editors. Cambridge, U.K.: International Union of Crystallographers; 1987. pp. 107–132. [Google Scholar]
- 30.Ferrin T E, Huang C C, Jarvis L E, Langridge R. J Mol Graphics. 1988;6:13–37. [Google Scholar]
- 31.Couch G S, Pettersen E F, Huang C C, Ferrin T E. J Mol Graphics. 1995;13:153–158. doi: 10.1016/0263-7855(95)00003-o. [DOI] [PubMed] [Google Scholar]
- 32.Hummer G, García A E, Soumpasis D M. Biophys J. 1995;68:1639–1652. doi: 10.1016/S0006-3495(95)80381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bairoch A, Boeckmann B. Nucleic Acids Res. 1994;22:3578–3580. [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson D R, Koymans L, Kamataki T, Feyereisen R, Waxman D J, Waterman M R, Gotoh O, Coon M J, Estabrook R W, Gunsalus I C, Nebert D W. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Fersht A R. J Mol Biol. 1972;64:497–509. doi: 10.1016/0022-2836(72)90513-x. [DOI] [PubMed] [Google Scholar]
- 37.Anderson D E, Becktel W J, Dahlquist F W. Biochemistry. 1990;29:2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 38.Hendsch Z S, Tidor B. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pratt L R, Hummer G, García A E. Biophys Chem. 1994;51:147–165. [Google Scholar]
- 40.Atkins W M, Sligar S G. J Biol Chem. 1988;263:18842–18849. [PubMed] [Google Scholar]
- 41.Wade R. J Comput Aided Mol Des. 1990;4:199–204. doi: 10.1007/BF00125318. [DOI] [PubMed] [Google Scholar]
- 42.Goodford P J. J Am Chem Soc. 1985;28:849–56. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 43.Helms V, Wade R C. Biophys J. 1995;69:810–824. doi: 10.1016/S0006-3495(95)79955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geourjon C, Deleage G. Comput Appl Biosci. 1993;9:87–91. doi: 10.1093/bioinformatics/9.1.87. [DOI] [PubMed] [Google Scholar]
- 45.Loida P J, Sligar S G, Paulsen M D, Arnold G E, Ornstein R L. J Biol Chem. 1995;270:5326–5330. doi: 10.1074/jbc.270.10.5326. [DOI] [PubMed] [Google Scholar]