Abstract
Ribosomal protein S8 of Escherichia coli plays a key role in 30S ribosomal subunit assembly through its interaction with 16S rRNA. S8 also participates in the translational regulation of ribosomal protein expression through its interaction with spc operon mRNA. The binding site for protein S8 within the 16S rRNA encompasses nucleotides G588 to G604 and C634 to C651 and is composed of two base paired helical regions that flank a phylogenetically conserved core element containing nine residues. We have investigated the structure of the rRNA binding site for S8 both in the free state and in the presence of protein using NMR spectroscopy. The integrity of the two helical segments has been verified, and the presence of G597·C643 and A596·U644 base pairs within the conserved core, predicted from comparative analysis, have been confirmed. In addition, we have identified a base triple within the core that is composed of residues A595·(A596· U644). The NMR data suggest that S8–RNA interaction is accomplished without significant changes in the RNA. Nonetheless, S8 binding promotes formation of the U598·A640 base pair and appears to stabilize the G597·C643 and A596·U644 base pairs.
Keywords: protein–RNA interaction, RNA structure/heteronuclear NMR
The structural features of RNA that are important for RNA-specific protein recognition have only recently come under investigation using solution state methods (1–3). The assembly and maturation of ribosomes are critically dependent upon a large network of protein–RNA interactions, and a detailed description of the underlying structures is essential for understanding the biological activity of these particles in protein synthesis (4, 5). The binding of ribosomal protein S8 within the central domain of the 16S rRNA constitutes one of the first steps in 30S subunit assembly in Escherichia coli (6). In addition, the specific interaction of protein S8 with spc operon mRNA mediates translational regulation of the expression of S8 and a number of other ribosomal proteins (7). The association of S8 with its rRNA and mRNA binding sites has been extensively characterized by nuclease protection (8, 9), comparative sequence analysis (9), chemical modification (10–13), cross-linking (14), and site-directed mutagenesis (9, 11, 15). While these investigations have provided important information about the elements of RNA primary and secondary structure that are involved in S8–RNA interaction, we present here the first three-dimensional (3D) structure of the binding site for protein S8 within the 16S rRNA determined using NMR techniques.
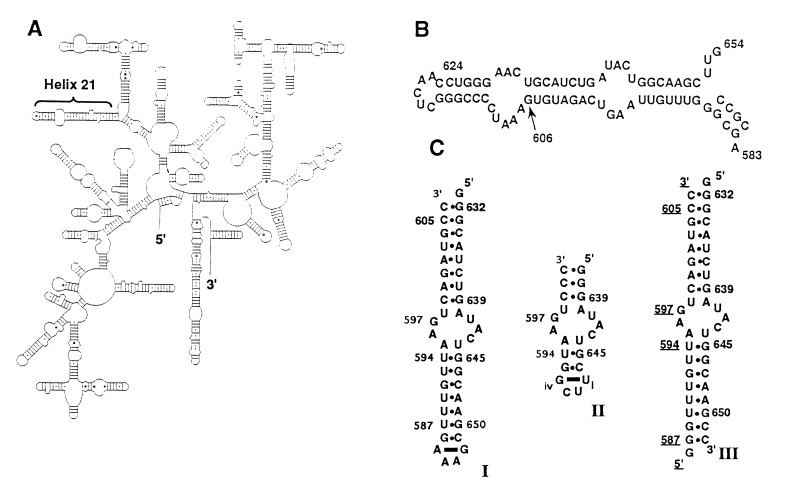
The binding site for protein S8 is located within helix 21 of the 16S rRNA (Fig. 1A) and comprises two helical segments interrupted by a core element of irregular structure that spans residues 595–598 and 640–644 (Fig. 1B). The nucleotides of the core element are very highly conserved among prokaryotic 16S rRNAs (16), and nearly all of them have been shown to be crucial for recognition by protein S8 through specific base substitutions (9, 11, 13). Significantly, the regulatory binding site for protein S8 in spc mRNA can adopt a secondary structure analogous to helix 21 and, although the primary structure of the helical regions differs from that of the rRNA binding site, eight of the nine core nucleotides are the same (9). A model of the S8 binding site derived from phylogenetic analysis indicates that base pairs within the core are exclusively of the Watson–Crick type (16). An alternative model, based primarily on chemical modification studies, suggests the presence of a noncanonical U·U base pair in this region (11, 13, 17). Given the intimate involvement of the RNA core element in the interaction with protein S8, precise structural definition of this feature is of fundamental importance for understanding S8–RNA interaction.
Figure 1.
The binding site for ribosomal protein S8 in E. coli 16S rRNA. (A) Secondary structure of E. coli 16S rRNA (16). (B) Helix 21 of the 16S rRNA. Conserved bases are in boldface type. (C) The three RNA molecules used in this study. RNAs I and III contain all structural elements necessary for protein S8 binding. RNA II permitted a more detailed analysis of the core element in the free RNA. The residues have been numbered to correspond to the 16S rRNA numbering system. The bases of the loop in RNAs I and II, which do not occur in E. coli 16S rRNA, are numbered i–iv.
In this report, we describe the use of two-dimensional (2D) and 3D heteronuclear NMR spectroscopy to probe the structure of the binding site for ribosomal protein S8 both free in solution and in the S8–RNA complex. Our results confirm the secondary structure of the S8 binding site proposed on the basis of comparative analysis (16) and, in addition, demonstrate the presence of a base triple in the core element. The results also indicate that much of the structure present in the free RNA is retained upon association with protein S8. Finally, while Mg2+ is required for protein–RNA complex formation, the divalent cation was also found to stabilize the structure of the core nucleotides by binding to each of three RNA molecules investigated.
MATERIALS AND METHODS
Materials.
All enzymes were purchased from Sigma with the exception of T7 RNA polymerase, which was prepared as described (18). Deoxyribonuclease I type II, pyruvate kinase, adenylate kinase, and the nucleotide monophosphate kinase were obtained as powders; dissolved in 15% glycerol, 1 mM DTT, and 10 mM Tris·HCl (pH 7.4); and stored at −20°C. The guanylate kinase and nuclease P1 were obtained as solutions and stored at −20°C. Phosphoenolpyruvate (potassium salt) was obtained from Bachem.
Preparation of the 15N- and 13C-Labeled RNAs.
RNAs I, II, and III (Fig. 1C) were transcribed in vitro with T7 RNA polymerase using synthetic DNA templates (19, 20) and 13C- and/or 15N-labeled 5′-nucleoside triphosphates. The labeled triphosphates were prepared as described (20). Gel-purified RNA was dialyzed extensively against a solution containing 10 mM NaCl, 10 mM potassium phosphate (pH 6.8), and 0.05 mM EDTA using a Centricon-3 concentrator (Amicon). The RNA was diluted to a volume of 500 μl using the dialysis buffer and lyophilized to a powder.
Preparation of Protein S8.
A mutant of E. coli S8, in which Cys-126 was replaced by Ala to prevent aggregation (21), was used in these experiments. The RNA-binding properties of S8 C126A are indistinguishable from those of the wild-type protein. S8 C126A was overexpressed and purified by chromatography on CM Sephadex (Pharmacia) as described (21).
Formation of the S8–RNA Complex.
RNAs I and III were tested for protein S8 specificity by using filter binding assays as detailed elsewhere (15). Protein S8 was titrated into a solution of 15N-labeled RNA I at RNA/S8 ratios of 1:0.25, 1:0.5, 1:0.8, 1:1.2, and 1:1.4 and 15N–1H heteronuclear multiple quantum coherence (HMQC) spectra were recorded at each ratio to monitor S8–RNA complex formation. No spectral changes were observed beyond the ratio 1:1.2. For additional NMR samples, RNA I and protein S8 were diluted separately to concentrations of approximately 40 μM in 90% 1H2O/10% 2H2O/1 mM DTT/25 mM MgCl2. The pH of the solutions was 6.6. Equal volumes of the solutions were combined at 4°C and concentrated 14-fold. Residual free RNA, as determined by 15N–1H NMR spectroscopy, was titrated with additional protein S8 and was monitored using 2D 15N–1H HMQC spectra.
NMR Spectroscopy.
Spectra were acquired on an NMR spectrometer (Bruker, Billerica, MA; model AMX-500) equipped with a 1H-{13C/15N} triple resonance probe. HMQC and nuclear Overhauser effect spectroscopy (NOESY) experiments in 90% 1H2O were performed at 10°C (RNA II), 15°C (RNAs I and III), and 21°C (protein–RNA I complex) using binomial 11̄ or 13̄31̄ solvent suppression schemes (22) with maximum excitation at 12.5 ppm. NOESY and 3D {15N}-NOESY-HMQC spectra were collected in 90% 1H2O at mixing times of 260 ms for RNA I and 180 ms and 380 ms for RNA II. NOESY and 3D {13C}-NOESY-HMQC spectra were collected at 28°C in 99.9% 2H2O at mixing times of 80 and 280 ms for RNA I and 60, 180, and 400 ms for RNA II. The 13C-1H double half-filtered NOESY spectrum (23) was collected for RNA III at 25°C at a mixing time of 280 ms.
Structure Determination.
2H2O-derived NOE cross peak intensities for RNA II were ranked as strong, medium, and weak and were converted into distance constraints of 1.8–2.5, 2.5–4.0, and 3.0–5.5 Å. Distance constraints for NOEs involving exchangeable protons were set between 1.6 and 6.0 Å. A total of 539 NOE-derived distance constraints (including 105 inter-residue and 111 intraresidue constraints for the 9 core nucleotides) were used in the structure calculations. Base pair constraints were imposed for core residues A596·U644 and G597·C643 and for the 5 bp of the stems flanking the core for which experimental data had been obtained. Torsion angle constraints derived from double quantum filtered correlated spectroscopy data were used for the ribose moieties and centered about either ideal 2′-endo or 3′-endo ring puckers (±30°). RNA II was modeled with A-form geometry using insight ii (Biosym Technologies, San Diego) and was subjected to 20 cycles of simulated annealing that included 15 ps of restrained molecular dynamics at 1200 K followed by 5 ps at 300 K and minimization. Each cycle began with a different random number seed, and no dielectric force field was employed. Eight of the final 20 structures with the lowest energies were averaged, and the average structure was minimized.
RESULTS
We have used three oligoribonucleotides (Fig. 1C) to characterize the binding site for protein S8 in the free and protein-bound states. RNA I includes all of the features required for specific interaction of S8 with the 16S rRNA (15). In RNA II, the helical segments were truncated to facilitate a detailed structural investigation of the core element of the binding site. RNA III permitted strand-specific isotopic enrichment with 15N or 13C. RNAs I and III exhibit Ka values of 1–2 × 107 M−1 for S8 association and are consistent with previous measurements of S8–rRNA affinity (9, 11, 15).
Exchangeable Proton Spectra of the Free RNA.
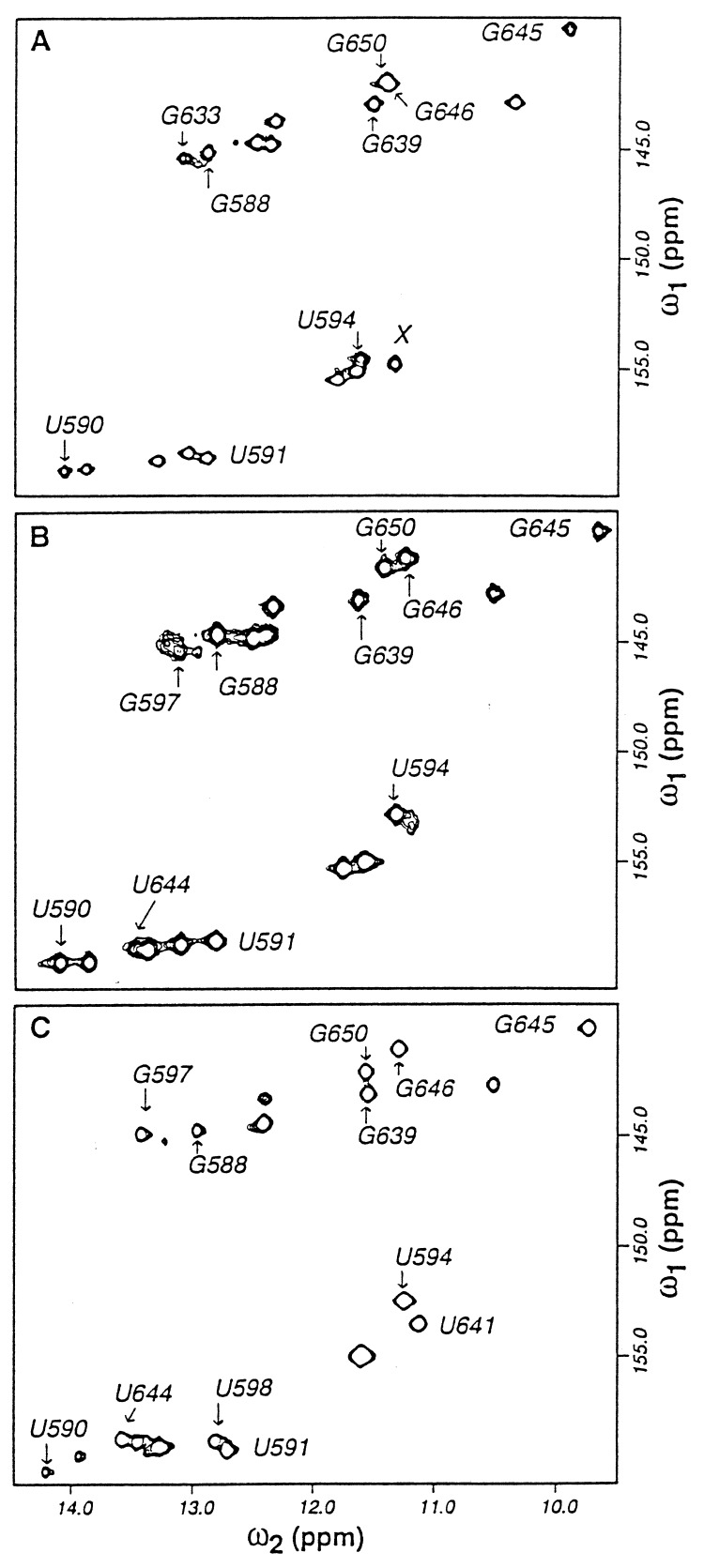
The imino protons of the three RNAs were assigned using 2D and 15N-separated 3D NOESY experiments. The 2D {15N–1H} HMQC spectra of RNA I in the absence and presence of Mg2+ are compared in Fig. 2. In the absence of Mg2+, only imino resonances of stem nucleotides were observed (Fig. 2A). Upon addition of Mg2+, resonances corresponding to nucleotides in the core region appeared, and several of the resonances associated with the stem nucleotides underwent chemical shift changes. The resonances corresponding to nucleotides G639 and G645 exhibit unusual chemical shift properties. G639 participates in a G·C pair, but its imino proton chemical shift resonates in a region of the spectrum typical of guanine imino protons of G·U base pairs. The imino proton of G645, which participates in a G·U base pair, is shifted 2 ppm upfield from other G·U guanine imino protons in the molecule. Of particular interest are the imino resonances of G588, G646, and U594, whose chemical shifts are significantly perturbed when Mg2+ binds to the RNA molecule (Fig. 2B). The imino resonances of G597, U641, and U644 are very weak or absent when Mg2+ is not present. However, the imino resonance of G597 becomes more intense and that of U644 becomes observable in the HMQC spectrum when Mg2+ is added (Fig. 2B). U641 gives rise to a weak resonance at 11.28 ppm in the presence of Mg2+ and was identified definitively using assignments from the spectrum of the protein–RNA complex. The corresponding imino resonances and their chemical shift perturbations were also observed in RNA II.
Figure 2.
2D 15N–1H HMQC spectra of RNA I. (A) The free RNA in the absence of Mg2+. (B) The free RNA in the presence of 15 mM Mg2+. (C) The RNA complexed with protein S8. The peak marked X in A could not be definitively assigned as no NOE interactions were observed for this resonance, but it most likely corresponds to the imino resonance of U641.
Nonexchangeable Proton Spectra of the RNA.
All base and ribose protons of RNA II and most of the base and 1′ protons of RNAs I and III were assigned using standard methods (24–26). As with the imino protons, the chemical shifts of some nonexchangeable protons of nucleotides in the core are perturbed in the presence of Mg2+. The most dramatic changes were recorded for the 1′ protons of A595 and U641, which resonate at 7.11 and 6.78 ppm, respectively, in the presence of Mg2+. In the absence of Mg2+, these protons resonate at about 5.8 ppm. The H2 resonances of the four adenine residues in the core provide structurally important information, including H2 to imino proton NOEs and H2 to H1′ cross strand NOEs (Fig. 3). Intra- and interstrand NOEs involving these H2 resonances were confirmed by performing 13C ω1/ω2 double half-filtered NOESY experiments (23) on RNA III that was unlabeled from residues 588 to 606 and uniformly 13C-labeled from residues 631 to 651 (Fig. 1C).
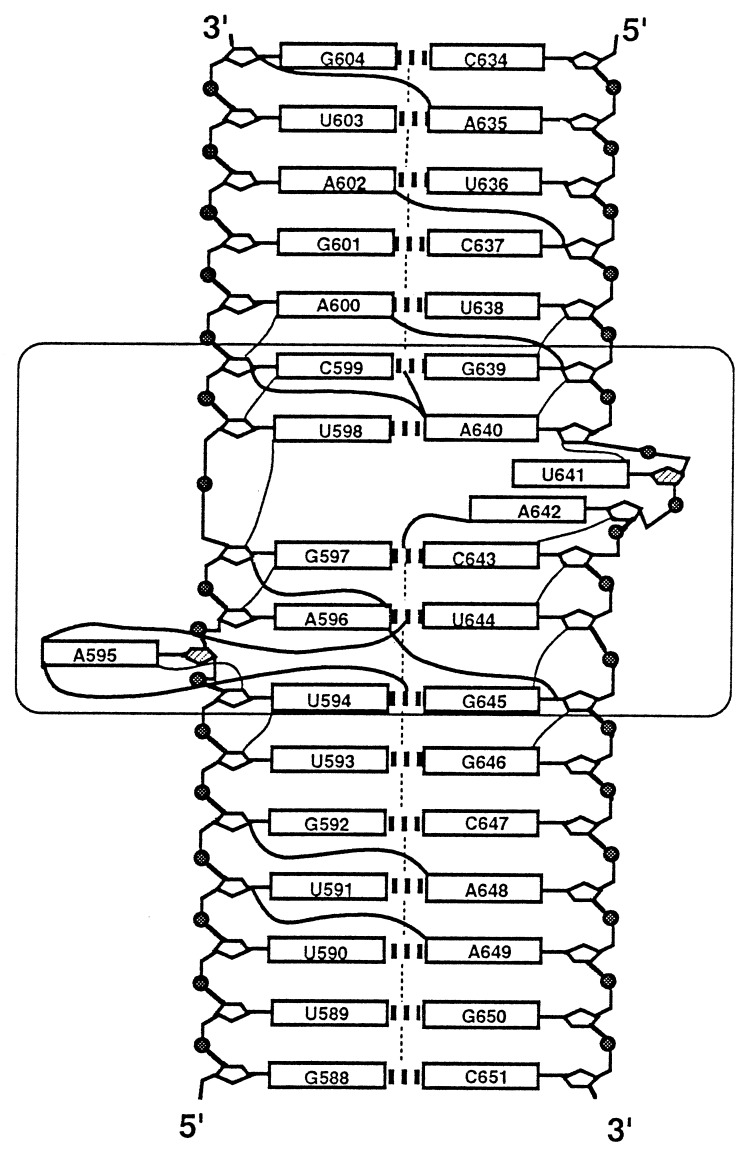
Figure 3.
Illustration of the secondary structure of the S8 RNA binding site and some of the NOE constraints used to calculate the 3D structure. The core element and flanking base pairs common to RNAs I, II, and III are boxed. Vertical dashed lines between base pairs indicate inter-base-pair imino NOEs observed in the free and protein-bound forms of the RNA and were extracted from NOESY and 3D NOESY-HMQC spectra of RNA I. The H6/H8-H2′ NOEs (thin solid lines) were extracted from the 60-ms NOESY spectrum of RNA II. NOEs involving adenine H2s (thick solid lines) were extracted from NOESY and double half-filtered NOESY spectra of RNA II and RNA III, respectively. The hatched ribose moeities adopt a C2′-endo ring pucker.
Assignment of Exchangeable RNA Protons in the S8–RNA Complex.
Inter-base-pair NOEs were used to assign the imino proton resonances of the protein–RNA complex. Several imino resonances exhibited discrete chemical shift changes upon titration of the RNA with protein S8, including those corresponding to nucleotides U590, U591, U594, and G650 (Fig. 2C). An additional uridine imino resonance was observed in the HMQC spectrum and has been assigned to residue U598. U598-H3, which resonates in a region of the imino spectrum typical of U·A base pairs, displays NOEs in the base and ribose regions of the NOESY spectrum consistent with those observed for other U·A base pairs. The U641 imino resonance becomes much stronger in the presence of the protein and resonates in a region of the 1H spectrum typical of U·G, U·U, and non-base-paired uridine imino protons. U·G and U·U base pairs give rise to strong intra-base-pair NOEs because of the proximity of U-H3 and G-H1 nuclei. U641 does not give rise to any cross peaks in the imino region of the NOESY spectrum, suggesting that U641 is not involved in a G·U or U·U base pair.
Structure of the Binding Site for Protein S8.
NOE data for RNA II were used to obtain distance constraints to refine the model of the core element of the S8 binding site. Fig. 3 illustrates a few of the NMR-derived distance constraints used in the calculations. The starting structure for molecular dynamics was constructed using insight ii. All residues were modeled in an anti configuration about the glycosidic bond except Giv of the tetraloop, for which NOE data clearly indicate a syn configuration. G-H1 to C-N4H2 NOEs and A-H2 to U-H3 NOEs were used to elucidate the base pairs corresponding to G597·C643 and A596·U644. The base moieties of U598, A640, U641, and A642 were oriented into the helix, but no base pair constraints were imposed upon these residues. Double quantum filtered correlated spectroscopy data for RNA II were used to determine ribose ring puckers. Nucleotides A595 and U641 as well as Uii, and Ciii of the tetraloop were found to be largely in the C2′-endo conformation, and all others were determined to be in the C3′-endo conformation. Breaks in the sequential H6/H8-H2′ NOEs at U641 and A595 are consistent with the C2′-endo conformations of these ribose rings. Although A595 was initially modeled between base pairs U594·G645 and A596·U644, our NMR data suggested that A595 forms a base triple with the A596·U644 base pair. The chemical shifts of the A596 amino protons (7.84 and 8.15 ppm) indicate that both protons are hydrogen-bonded and support the presence of the base triple. The NOE cross peaks A595-H2/A596-N6H2, A595-H2/U644-H3, and G645-H1/U644-H3 provide further evidence for the presence of the base triple and help define its orientation.
DISCUSSION
The protein binding domain of the RNA, which encompasses nucleotides G588 to G604 and C634 to C651, is shown schematically in Fig. 3, and a superposition of eight structures calculated using NMR data coordinates is shown in Fig. 4. The free RNA consists of two regular A-form helical segments extending from base pairs G588·C651 to G597·C643 and from base pairs C599·G639 to G604·C634. The helix axis remains largely unperturbed through the core element. All residues in the binding site adopt an anti configuration about their glycosidic bonds and all G·C and A·U base pairs were found to be of the Watson–Crick type.
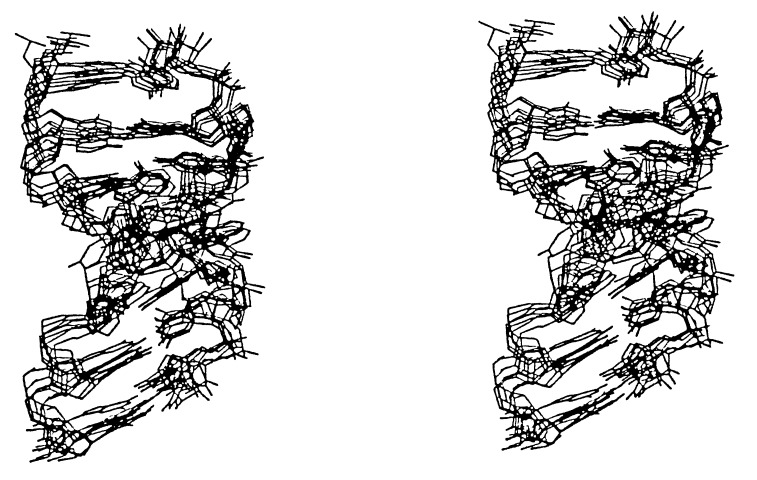
Figure 4.
Superposition of nucleotides 638–646 and 593–600 from eight lowest energy structures generated using simulated annealing. The structures shown contain no NOE violations. The all atom r.m.s. deviations between the eight structures shown and the average of the eight structures range from 1.5 to 1.8 Å.
Structure of the Core Element.
All of the core nucleotides, residues 595–598 and 640–644, have been implicated in protein S8 binding by site-directed mutagenesis (refs. 9, 11, and 13; unpublished work) and/or chemical modification experiments (13, 17). Moreover, all of these nucleotides except U641 are highly conserved in prokaryotic 16S rRNAs (ref. 16; R. R. Gutell, personal communication; see also Fig. 1B). The A596·U644 and G597·C643 base pairs, predicted from comparative sequence analysis (16), have been confirmed by the NMR data presented here. These base pairs are observed both in the free RNA and in the protein–RNA complex. Furthermore, disruption or exchange of base pairing partners at these positions leads to a sharp decrease in S8 binding (9, 11, 13).
The data reported in this study indicate that the base of residue A595 forms a base triple with the A596·U644 base pair. As depicted in Fig. 5, the purine ring of A595 is located in the major groove of the lower helix and is coplanar with that of A596. A hydrogen bond is oriented along an axis between the N3 of A595 and the second amino proton of A596 (Fig. 5). Interestingly, the 595·(596·644) base triple is also predicted by comparative phylogenetic evidence. In the more than 2500 16S rRNAs sequenced, these positions are occupied by A·(A·U) approximately 80% of the time and by G·(C·G) 10% of the time (R. R. Gutell, personal communication). The deletion of A595 severely impairs the binding of E. coli S8 (11), indicating that this nucleotide is essential for formation of the RNA structure that interacts with the protein.
Figure 5.
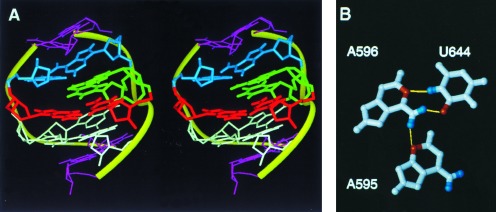
Stereoview of the core element of the protein S8 RNA binding site (A) and illustration of the A·(A·U) base triple (B). In A, the following colors are used: white, A595, A596, and U644; red, G597 and C643; blue, U598 and A640; green, U641 and A642; pink, U594, C599, G639, and G645. The r.m.s. deviation for all atoms between the structure shown in A and the average of the eight superimposed structures shown in Fig. 4 was 1.6 Å.
The conformations of nucleotides U598, A640, and A642 are poorly defined. The NOE cross peaks from A640-H2 to C599-H1′ and G639-H1 are consistent with those NOEs expected from A-form regions of duplex RNA helices. The continuity of sequential NOE cross peaks between H6/H8 and H2′ resonances from A596 through C599 and from G639 through A640 in the 60-ms NOESY spectrum is also indicative of an A-form helix and suggests that inter-base-pair stacking is maintained in this region of the core element. Thus, U598 stacks on C599 and A640 stacks on G639, with A640 oriented so as to form a base pair with U598. The absence of a resonance corresponding to the imino proton of U598 in the spectra of the free RNA limits our ability to determine the base pairing status of U598 from the NMR data alone. However, the existence of the U598·A640 pair is supported by covariance analysis (16) as well as by the NMR analysis of the S8–RNA complex. Presumably, the rapid exchange of U598-H3 with solvent protons prevents the observation of this imino resonance. Although there are relatively few constraints for A642, an NOE between A642 H2 and G597 H1 suggests that the base of A642 occupies the major groove of the helix.
Position 641 exhibits fewer restrictions on nucleotide identity than other positions in the core region and may be occupied by U or A without a significant change in protein-binding activity (11). However, when C is introduced at position 641, a sharp decrease in affinity for S8 is observed (11). This substitution results in the appearance of multiple conformations within the core region of the RNA (data not shown) which probably interfere with the recognition or binding of S8. Although a base pair between U598 and U641 was proposed earlier on the basis of chemical modification experiments (17), the present results do not support this interaction in either the free RNA or the S8–RNA complex. However, the NMR data suggest that U641 may be involved in a base triple. In approximately 50% of the structures calculated, U641 stacks on A640 with its imino proton within hydrogen bonding distance (2.2 Å) of the C6 carbonyl oxygen of G597. The C2′-endo sugar pucker of U641 may facilitate this interaction. Although the chemical shift of the U641 imino proton lies in a spectral region typical of G·U base pairs, no bond constraints between G597 and U641 were used for the structure calculation. The possible occurrence of a U641·(G597·C643) triple in the core is also suggested by a recent mutational analysis of the S8 binding site (27) but receives little support from comparative structural data (R. R. Gutell, personal communication).
Effects of Mg2+ on the RNA Binding Site.
Mg2+ appears to facilitate the S8–RNA interaction by stabilizing the conformation of the RNA. In the absence of Mg2+, H2 and C2 nuclei of A642 exhibit multiple resonances, indicative of multiple conformations, but resolve to single resonances in the presence of Mg2+. The addition of Mg2+ also results in the appearance of the imino resonance of U644 and causes the H1′ resonances of A595 and U641 to shift downfield to 7.11 and 6.78 ppm, respectively. These spectral changes suggest that Mg2+ stabilizes a unique conformation of the RNA core. While addition of Mg2+ perturbs chemical shifts throughout the helical regions of the RNAs used in this study, the most significant effects involve residues G588, G646, and U594, which either compose or are adjacent to G·U base pairs. The tendency of Mg2+ to associate with G·U wobble pairs has previously been noted (28). Although G·U base pairs occur frequently in helix 21 of the prokaryotic 16S rRNAs, they are not absolutely conserved (16) and can be replaced by Watson–Crick base pairs without significantly disrupting S8–RNA association. Indeed, the regulatory binding site for S8 in the spc operon contains no G·U base pairs proximal to the core yet exhibits an apparent Ka of approximately 1 × 106 M−1 for protein S8 (15).
Effects of Protein S8 on the RNA Binding Site.
The inter-base-pair NOEs between imino protons of the S8–RNA complex indicate that the secondary structure characteristic of the free RNA is maintained in the protein–RNA complex. The conformation of the core element is of particular interest owing to its role in S8 recognition (9, 11, 13, 17). The 15N–1H HMQC and NOESY spectra of the complex lack any indication of a U·U base pair but demonstrate the presence of a U·A base pair between U598 and A640. It is possible that no stable base pair involving U598 exists in the free RNA but instead is formed or stabilized in the presence of the protein. The chemical shift of the imino resonance of G639, which is proximal to U598, exhibits a difference of 0.13 ppm between the free and protein-bound forms of RNA I (Fig. 2), suggesting that the RNA core element of the binding site undergoes only minor conformational changes upon complex formation. Moreover, the imino proton resonance of U641 becomes stronger in the protein-bound state but only exhibits an exchange peak with the solvent and a single NOE to a resonance in the ribose region of the NOESY spectrum of the complex. In addition, the imino resonances of U644, G645, and U594 exhibit modest intensity changes upon interaction with S8. We note that no intermolecular NOEs that involve the RNA imino protons were identified. This is not unexpected, though, as the imino protons are sequestered into the RNA helix. Importantly, each imino group gives rise to only one resonance in the HMQC spectrum, indicating that the S8-bound RNA is in a unique conformation. Extensive mutational analysis (9, 11, 15), a Ka of 1–2 × 107 M−1 between RNA I and protein S8, and the presence of only two discrete states throughout the titration of RNA with S8 combine to support a specific interaction between the two. Together, these data suggest that protein binding is accomplished without significant alterations in the structure of the RNA core.
Mutational analysis has established that stems several base pairs in length above and below the core element are required for efficient association with S8 (15). Although no NOEs were observed between RNA imino resonances and protein S8, changes in the chemical shift of several resonances were noted. In particular, U591 H3 is shifted ≈0.2 ppm upfield and G650 N3 is shifted ≈0.8 ppm upfield in the protein–RNA complex relative to the free RNA. Thus, it is possible that protein S8 forms a nonspecific contact(s) with the RNA near the base of the lower helix, perhaps involving the phosphate backbone of the RNA. Moreover, a protein–RNA contact in this region would account for the inability of S8 to associate with RNA II. Although RNA II contains the structural features of the core that are necessary for protein binding, the lower helix is truncated three base pairs below the core element. We have recently determined that the U589·G650 base pair, as well as the invariant G588·C651 base pair adjacent to it, can be replaced by any Watson–Crick base pair combination without affecting S8–RNA interaction. In contrast, deletion of either of these base pairs significantly reduces the affinity of the RNA for the protein (unpublished work). Thus, while the nucleotide identity and the tertiary structure of the core element are highly constrained, the distal regions of the S8 binding site appear to only require a double helical segment for S8–RNA complex formation. The results of this investigation, together with the recently reported crystallographic structure of S8 from Bacillus stearothermophilus (29), should provide new insights into the S8–RNA interaction.
Acknowledgments
We thank M. Michnicka for preparation of the T7 RNA polymerase, M. Michnicka and K. McKinney for synthesis of the labeled oligoribonucleotides, Dr. Robin R. Gutell for phylogenetic analysis of nucleotides in the S8 binding site, and Hervé Moine for providing results prior to publication. This work was supported by Welch Foundation Grant C-1277 and National Institutes of Health Grant GM52115 to E.P.N. and by National Science Foundation Grant MCB-9108104 and National Institutes of Health Grant GM22807 to R.A.Z.
ABBREVIATIONS
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- 2D
two-dimensional
- 3D
three-dimensional
- HMQC
heteronuclear multiple quantum coherence
References
- 1.Hall K B. Methods Enzymol. 1995;261:542–559. doi: 10.1016/s0076-6879(95)61024-3. [DOI] [PubMed] [Google Scholar]
- 2.Aboul-Ela F, Karn J, Varani G. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 3.Puglisi J D, Chen L, Blanchard S, Frankel A D. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann R A. In: Ribosomes: Structure, Function and Genetics. Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M, editors. Baltimore: University Park; 1980. pp. 135–169. [Google Scholar]
- 5.Draper D E. In: In Ribosomal RNA: Structure, Evolution, Processing and Function in Protein Biosynthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1996. pp. 171–197. [Google Scholar]
- 6.Held W A, Ballou B, Mizushima S, Nomura M. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 7.Dean D, Yates J L, Nomura M. Nature (London) 1980;289:89–91. doi: 10.1038/289089a0. [DOI] [PubMed] [Google Scholar]
- 8.Gregory R J, Zeller M L, Thurlow D L, Gourse R L, Stark M J R, Dahlberg A E, Zimmermann R A. J Mol Biol. 1984;178:287–302. doi: 10.1016/0022-2836(84)90145-1. [DOI] [PubMed] [Google Scholar]
- 9.Gregory R J, Cahill P B F, Thurlow D L, Zimmermann R A. J Mol Biol. 1988;204:295–307. doi: 10.1016/0022-2836(88)90577-3. [DOI] [PubMed] [Google Scholar]
- 10.Thurlow D L, Ehresmann C, Ehresmann B. Nucleic Acids Res. 1983;11:6787–6802. doi: 10.1093/nar/11.19.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mougel M, Allmang C, Eyermann F, Cachia C, Ehresmann B, Ehresmann C. Eur J Biochem. 1993;215:787–792. doi: 10.1111/j.1432-1033.1993.tb18093.x. [DOI] [PubMed] [Google Scholar]
- 12.Svensson P, Changchien L-M, Craven G R, Noller H F. J Mol Biol. 1988;200:301–308. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- 13.Allmang C, Mougel M, Westhof E, Ehresmann B, Ehresmann C. Nucleic Acids Res. 1994;22:3708–3714. doi: 10.1093/nar/22.18.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wower I, Brimacombe R. Nucleic Acids Res. 1983;11:1419–1437. doi: 10.1093/nar/11.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, Jiang L, Zimmermann R A. Nucleic Acids Res. 1994;22:1687–1695. doi: 10.1093/nar/22.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutell R R. Nucleic Acids Res. 1993;21:3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mougel M, Eyermann F, Westhof E, Romby P, Expert-Bezançon A, Ebel J, Ehresmann B, Ehresmann C. J Mol Biol. 1987;198:91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- 18.Davanloo P, Rosenburg A H, Dunn J J, Studier F W. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heus H A, Pardi A. J Mol Biol. 1991;217:113–124. doi: 10.1016/0022-2836(91)90615-d. [DOI] [PubMed] [Google Scholar]
- 20.Nikonowicz E P, Sirr A, Legault P, Jucker F M, Baer L M, Pardi A. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Wower I, Zimmermann R A. Biochemistry. 1993;32:4761–4768. doi: 10.1021/bi00069a010. [DOI] [PubMed] [Google Scholar]
- 22.Hore P J. J Magn Reson. 1983;55:283–300. [Google Scholar]
- 23.Otting G, Wuthrich K. Q Rev Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- 24.Scheek R M, Russo N, Boelens R, Kaptein R. J Am Chem Soc. 1983;105:2914–2916. [Google Scholar]
- 25.Nikonowicz E P, Pardi A. J Mol Biol. 1993;232:1141–1156. doi: 10.1006/jmbi.1993.1466. [DOI] [PubMed] [Google Scholar]
- 26.Legault P, Farmer B T, Mueller L, Pardi A. J Am Chem Soc. 1995;116:2203–2204. [Google Scholar]
- 27.Moine, H., Cachia, C., Westhof, E., Ehresmann, B. & Ehresmann, C. (1997) RNA, in press. [PMC free article] [PubMed]
- 28.Allain F H-T, Varani G. Nucleic Acids Res. 1995;23:341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies C, Ramakrishnan V, White S W. Structure. 1996;4:1093–1104. doi: 10.1016/s0969-2126(96)00115-3. [DOI] [PubMed] [Google Scholar]