Abstract
The inactivation of the von Hippel-Lindau (VHL) gene predisposes affected individuals to VHL syndrome and is an early genetic event associated with sporadic renal cell carcinoma and CNS hemangioblastomas. The VHL protein (pVHL) has been shown to form a stable complex with elongin B and elongin C, two factors that stabilize and activate the transcription elongation factor elongin A. Here, Hs-CUL-2, a member of the recently identified multigene family, the cullins, is shown to specifically associate with the trimeric pVHL-elongin B-C (VBC) complex in vitro and in vivo. Nearly 70% of naturally occurring cancer-predisposing mutations of VHL disrupt this interaction. The pVHL-Hs-CUL-2 association is strictly dependent on the integrity of the trimeric VBC complex. Immunofluorescence studies show Hs-CUL-2 to be a cytosolic protein that can be translocated to the nucleus by pVHL. Recently it has been shown that a yeast Hs-CUL-2 homolog, Cdc53, is part of a ubiquitin protein ligase complex that targets cell cycle proteins for degradation by the ubiquitin proteolytic pathway. In Caenorhabditis elegans, a null mutation of another Hs-cul-2 homolog, Ce-cul-1, results in hyperplasia in all tissues and is required for cell cycle exit. Hence, Hs-cul-2 may be required for VHL function and, therefore, may be a candidate human tumor-suppressor gene.
Inactivation of the von Hippel-Lindau (VHL) gene is responsible for VHL syndrome, a rare hereditary cancer syndrome (incidence 1 in 36,000) that predisposes affected individuals to develop a variety of tumors including retinal angiomas, CNS hemangioblastomas, renal cell carcinomas (RCCs), pheochromocytomas, and pancreatic cysts (1, 2). Mutation or transcriptional silencing of the VHL gene and subsequent loss of the remaining VHL allele are associated with sporadic, clear cell renal carcinoma, CNS hemangioblastomas, and other VHL-associated tumors, which make it a bona fide tumor-suppressor gene (3, 4).
Most germline VHL mutations are missense alterations (5). Mutations that have been detected in VHL patients or sporadic RCCs are found throughout the coding region of the VHL gene pointing to the functional importance of almost the entire open reading frame (2, 6, 7). However, in the germ line, although there are mutations in each exon, there is a marked preponderance of mutations in the 5′ end of exon 3 (6). The human VHL gene encodes a 213-aa protein expressed in all tissues (8). pVHL has no homology to other known proteins [i.e., the sequence gave no information as to its function(s)]. For this reason, we have searched for clues to the function of VHL, in part, by identifying and characterizing the interaction of pVHL with other proteins. We first showed that pVHL stably associates with the two regulatory subunits, B and C, of the trimeric transcription elongation factor, elongin (9, 10). We demonstrated that pVHL can act as a transcription elongation inhibitor in vitro by virtue of its ability to compete with elongin A for the association with the two regulatory elongin subunits (10). However, there is no evidence that pVHL competes in vivo with elongin A for the B-C subunits; we find most of pVHL in a stable complex with elongin B and C, and exchange of pVHL with elongin A has not been observed. The fact that pVHL is stably associated with elongin B-C led us to speculate that this trimeric complex has a cellular function distinct from transcription elongation. The elongin B-C binding site, which spans 13 aa, is conserved between pVHL and elongin A (10–12). Notably, this region, found at the beginning of exon 3, is a mutational hotspot in VHL families (6). The finding that up to 70% of VHL families have mutations predicted to disrupt VHL binding to elongin B-C (13) suggests the functional importance of this interaction for the tumor-suppressor activity of VHL.
Recent studies have provided insights into the functional consequences of VHL expression in cells. Several groups have shown that the absence of wild-type (wt) VHL in a variety of cells in culture results in the deregulated expression of a set of genes, including the genes encoding vascular endothelial growth factor (VEGF), platelet-derived growth factor-β, and GLUT1 (14–16). Each of these genes is normally controlled by a number of factors, including hypoxia, which induces the accumulation of the respective mRNAs via stabilization of the transcripts (17, 18). Wt pVHL is involved in the negative regulation of these hypoxia-inducible genes via destabilization of their mRNAs under normoxic conditions (16). In addition, we have recently observed that VHL is required for the normal regulation of VEGF mRNA levels in response to changes in serum levels and cell confluency (ref. 15; S.L., unpublished observations). These observations implicate pVHL in a pathway that is involved in sensing changes in oxygen tension and other factors in the local environment of cells. Indeed, tumors associated with VHL mutations are highly vascular and are characterized by VEGF overexpression (ref. 15 and references therein). To gain further insight into the function of VHL, we have continued our search for proteins with which it interacts. In this study we have searched for proteins that interact with the trimeric pVHL-elongin B-C (VBC) complex. This approach led to the identification of Hs-CUL-2, a member of a recently described, highly conserved gene family involved in cell-cycle control in yeast and in the control of cell growth in Caenorhabditis elegans.
MATERIALS AND METHODS
VBC Complex.
pVHL and elongin B and C were expressed in Escherichia coli and purified as described (19). After purification, the three proteins were mixed in 6 M guanidine·HCl in equimolar ratios and renatured by dialysis against decreasing concentrations of guanidine·HCl, starting with 4 M guanidine·HCl/2 mM EDTA/0.1 M Tris·HCl, pH8, for 3 hr at 4°C, followed by a 12-hr dialysis against the same buffer containing 0.1% 2-mercaptoethanol. The protein complex was then dialyzed against a buffer containing 3 M guanidine·HCl, 2 mM EDTA, 0.1 M KCl, 50 mM Tris·HCl (pH8), 10% glycerol, 0.1% 2-mercaptoethanol for 3 hr at 4°C, followed by sequential 3-hr dialyses in the same buffer containing 2 M guanidine·HCl, 1 M guanidine·HCl, followed by a 12-hr dialysis in the same buffer containing no guanidine·HCl. The protein concentration was adjusted to 1 mg/ml by concentration in Centricon devices (Amicon) and stored at −80°C.
Plasmids.
For production of recombinant pVHL, a FLAG epitope was added to the C terminus (VHL-F) or N terminus (F-VHL) of the human VHL wild type (9) or (F-VHL)157Δ cDNA (20) and subcloned into the bacterial expression vector pQE 30 (Qiagen, Chatsworth, CA) as described (19). The naturally occurring 157Δ mutant contains the first 157 aa of pVHL followed by a stop codon. Human elongin B and C cDNAs were subcloned into the bacterial expression vector, pET-15b (Novagen). cDNAs for Hs-cul-1 and Hs-cul-2 were cloned by reverse transcription–PCR from human kidney poly(A)+ RNA with primers that introduced a hemagglutinin (HA) epitope at the C terminus of the proteins. The reported Hs-CUL-2 sequence was incomplete at the N terminus. To complete the sequence of the N terminus, a human HeLa cell cDNA UNI-Zap library (Stratagene) was amplified by PCR using a T3 primer and an Hs-cul-2 specific primer. A 550-bp fragment contained the missing 98 N-terminal aa as well as the 5′ noncoding region of human cul-2. This fragment was fused to the C-terminal fragment of Hs-cul-2 at an NheI site. Both cDNAs were subcloned into a pcDNA3 (Invitrogen) vector for in vitro translation and transient transfection into COS-7 cells and were sequenced on both strands (accession no. U83410U83410 for full-length Hs-cul-2). The plasmid pSX-VHL-GFP used for immunofluorescence analysis was derived from the pSX-Fg7 plasmid described earlier (9). The cDNA for the green fluorescent protein (GFP) was subcloned at the C terminus of VHL to produce the VHL-GFP protein.
Cells, Immunoprecipitation Analysis, and Microsequencing.
Cultures of 786-0 RCC cells; HeLa cells stably transfected with either rat VHL-F, human VHL-F, or a human VHL-F (R167W) missense mutation (9, 10); or COS-7 cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS). Cells were labeled with [35S]methionine and lysed in Triton X-100 lysis buffer as described (9, 10). For affinity purification of pVHL binding proteins from 786-0 cells, VBC complex (20 μg) and M2 beads (25 μl; IBI) were added to 1 ml lysate of 1 × 107 [35S]methionine-labeled cells. This mixture was processed as described for immunoprecipitations of transfected pVHL from HeLa or COS-7 cells (9, 10). Subcellular fractionation of HeLa cells was performed as described (9). Preparation of Hs-CUL-2 samples for microsequencing was performed as described (10). Four peptides that matched the published human CUL-2 sequence were obtained: (i) KFYQEIFESPFLTETGEYYK (amino acids 201–220), (ii) ALTSVVNYREPK (amino acids 370–382), (iii) FNNIFIK (amino acids 489–495), and (iv) YLHPSSYTK (amino acids 252–261).
In vitro Translation Analysis.
pcDNA3-Hs-cul-1 and pcDNA3-Hs-cul-2 (1 μg) were translated in the presence of [35S]methionine in vitro using a coupled transcription/translation system (50 μl; Promega). cDNAs for VHL and elongin B and C were translated simultaneously in the presence of [35S]methionine as described (50 μl; ref. 10), mixed with the Hs-CUL-1 or 2 lysates and incubated for 3 min at 30°C, followed by the addition of 25 μl protein G beads (Pharmacia) and 1 μl of HA11 (Babco) or M2 antibody (IBI), and incubated for 2 hr at 4°C with tumbling. The reactions were then processed for autoradiography as described (10).
Immunofluorescence Analysis.
Transient transfections were carried out as described (20). Briefly, COS-7 cells were electroporated with varying amounts of pcDNA3-Hs-cul-2 and pSX-VHL-GFP (5–20 μg), and plated overnight in 10% FCS/DMEM. The cells were washed with PBS, fixed in 1% formaldehyde for 15 min at room temperature, washed in PBS, incubated 15 min in 10% FCS, and incubated 60 min in the presence of HA11 antibody diluted 1/3000 in PBS/10% FCS/0.2% saponin. The cells were washed in PBS and incubated for 60 min in the presence of an anti-mouse secondary antibody linked to Texas Red dye (The Jackson Laboratory). The cells were washed in PBS and mounted on slides. Charge-coupled device camera images were obtained as described (21).
RESULTS
VBC Interacting Proteins.
We and others observed that the majority of pVHL is associated with elongin B-C in the case of both endogenous pVHL as well as in stable lines expressing pVHL (10, 12). Therefore, we reasoned that the trimeric VBC complex might be a functional unit since it is very stable once it is formed and pVHL is not displaced by the addition of elongin A (10).
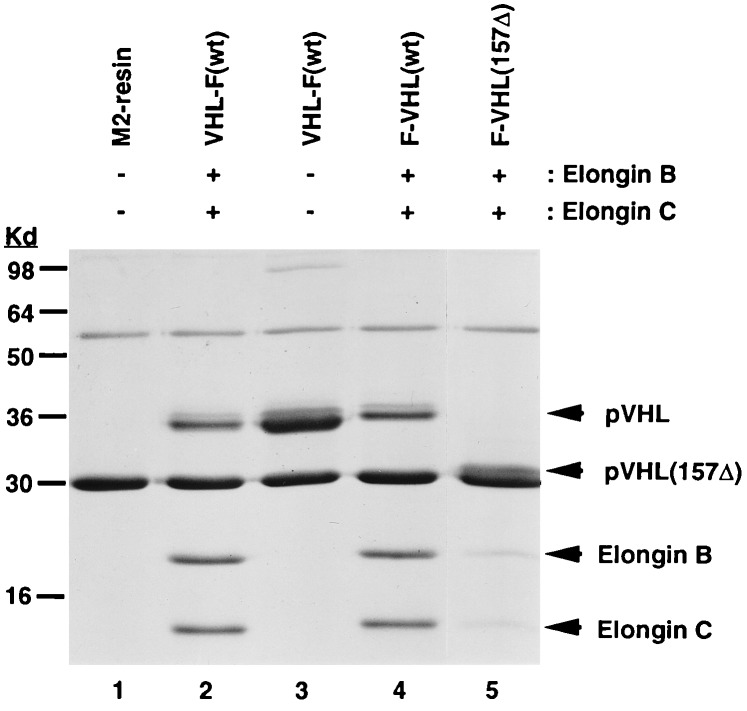
Sucrose gradient centrifugation showed that VBC in cells is part of a larger complex (9), suggesting that this complex associates with other proteins. To test this hypothesis, we generated a VBC affinity column using recombinant proteins to purify interacting proteins from cell lysates. We also utilized columns containing either pVHL alone or a naturally occurring pVHL deletion mutant (157Δ) that only weakly associates with elongin B-C. We introduced a FLAG epitope tag to either the N or C terminus of pVHL that is recognized by the monoclonal antibody, M2. The different complexes were incubated with beads coupled to M2 antibody and then immunoprecipitated. The integrity of the VBC complex and the significant reduction of binding of elongin B-C to the 157Δ mutant is shown in Fig. 1 (compare lane 4 with lane 5).
Figure 1.
Immunoprecipitation analysis of different recombinant pVHL complexes. pVHL complexes VHL-F, F-VHL, and F-VHL-157Δ were renatured in the presence or absence of recombinant elongin B-C followed by immunoprecipitation with M2 antibody and analysis on a SDS/14% polyacrylamide gel stained with Coomassie Blue (F, FLAG epitope). The bands at 55 and 30 kDa, present in all lanes, represent the heavy and light IgG chains, respectively.
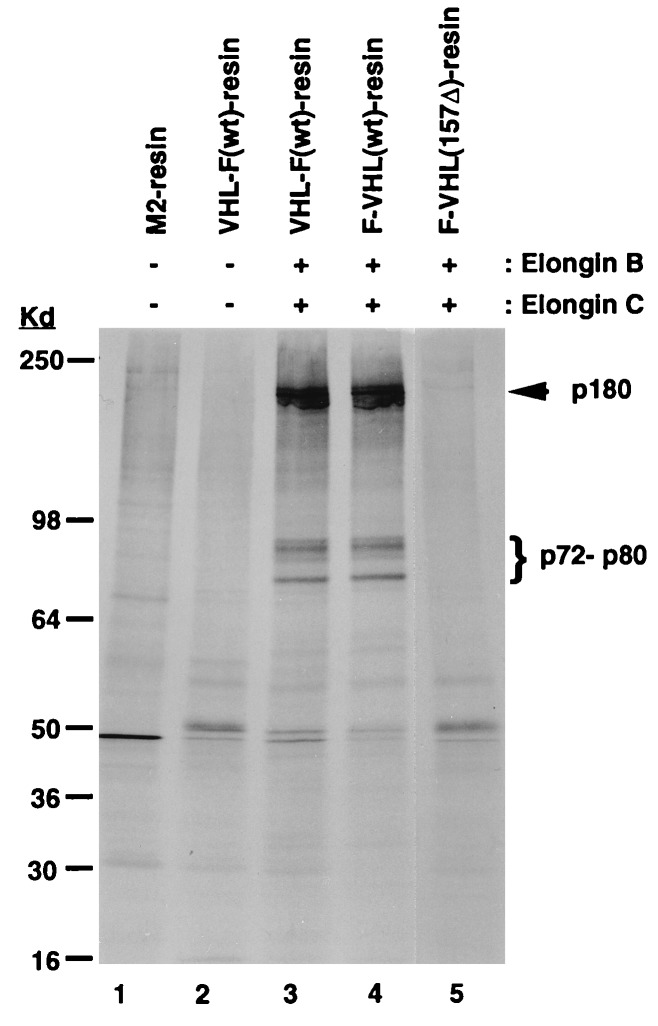
We utilized these columns to affinity-purify possible VBC binding proteins from RCC cells (786-0) that express a nonfunctional pVHL. pVHL alone, as well as the 157Δ mutant, interact with a protein of 50 Kd (Fig. 2; lanes 2 and 5). pVHL alone also interacts with elongin B-C (data not shown). The recombinant VBC complex, however, interacted specifically with a cluster of proteins between 70 and 80 kDa and a 180-kDa protein (lanes 3 and 4). These proteins interacted neither with VHL alone nor with the 157Δ mutant (lanes 2 and 5).
Figure 2.
Immunoaffinity purification of pVHL-associated proteins using recombinant pVHL complexes. Different pVHL complexes were incubated with a lysate of [35S]methionine-labeled 786-0 RCC cells and precipitated with M2 resin, followed by analysis on a SDS/14% polyacrylamide gel and autoradiography.
To test whether these interactions also occur in vivo, we immunoprecipitated pVHL from HeLa cells that stably express either rat pVHL, human pVHL, or the naturally occurring hotspot mutation, R167W, to compare possible associated proteins with the VBC column precipitates. First, we observed that the pVHL-associating proteins between 70 and 80 kDa are composed of three proteins termed p72, p76, and p80, reflecting their Mrs on SDS/PAGE. These proteins coimmunoprecipitated with the VBC column in the RCC cells (Fig. 3; lane 2) as well as in the rat and human pVHL expressing HeLa lines (lanes 5 and 6). To test the stability of the VBC-p72–76-80–180 complexes, the immunoprecipitates were washed with either 0.5% sarcosyl (lane 3) or 1 M NaCl (lane 4). No significant loss of the complexes was detected, suggesting a very stable interaction (compare lane 2 with lanes 3 and 4). pVHL in the HeLa immunoprecipitates associates with elongin B-C in a nearly equimolar ratio, whereas elongin binding is strongly reduced in the R167W hotspot mutation (compare lanes 5 and 6 to lane 7). The R167W mutation in pVHL resulted in a significant reduction of the association of p72, 76, and 80, as well as p180 (compare lanes 5 and 6 with lane 7).
Figure 3.
pVHL associates with p76 in the cytosol of cells. Lanes 1–4 and 8, VBC complexes were incubated with lysates of [35S]methionine-labeled 786-0 RCC cells and precipitated with M2 resin. The precipitates in lanes 3 and 4 were washed in the presence of 0.5% Sarkosyl or 1 M NaCl, respectively. Lanes 5–7, HeLa cells expressing either rat VHL-F, human VHL-F, or a R167W mutant VHL-F were labeled with [35S]methionine and lysed, and pVHL was immunoprecipitated with M2 antibody. Lanes 9–12, HeLa cells expressing the rat VHL protein were labeled with [35S]methionine and fractionated into a nuclear fraction (600 × g pellet), cytosolic fraction (100,000 × g supernatant), and a membrane fraction (100,000 × g pellet). Total lysate (lane 12) and subcellular fractions were solubilized in Triton X-100 lysis buffer and immunoprecipitated with M2 antibody. All reactions were analyzed on a split SDS/polyacrylamide gel (7% on the top/14% on the bottom) followed by autoradiography.
We have shown previously that pVHL can localize to the cytosol as well as the nucleus of cells, depending on cell density (20). We then wanted to determine where the pVHL protein complexes localize in the cell. Subcellular fractionation of HeLa cells that stably express rat pVHL showed that the bulk of VBC complex is in the cytosolic fraction, where it is predominantly associated with p76 (Fig. 3, lane 10). Note that in the cytosol there is relatively little p180, p80, or p72 seen in association with VBC. A small fraction of VBC is found in the Triton X-100 soluble nuclear fraction that contained some attached plasma membrane (lane 11) and in membrane fractions (lane 9) where it was associated with p72, 76, 80, and p180. When we analyzed the different fractions from HeLa cells that did not express exogenous pVHL with the VBC affinity column, we found the bulk of p72, p80, and p180 in the Triton X-100 soluble nuclear and membrane fraction (data not shown). The nuclear fraction contained some attached plasma membrane, and it is therefore possible that p72, p80, and p180 are embedded in the plasma or nuclear membrane, whereas p76, together with the vast majority of VBC, is soluble either from the cytosol or released from the nucleus upon fractionation. We also tested whether the association of these proteins with VBC occurs after lysis of cells. When labeled and unlabeled cells were mixed before lysis, all of the detected p76 was associated with VBC in the cell before lysis; whereas, p72 and p80 bound specifically, but after the cells were lysed (data not shown). These data suggest that the VBC complex in the cell is associated with p76, which prompted us to identify p76.
The Identification of Hs-CUL-2 as a VHL-BC Interacting Protein.
p76 was immunoaffinity-purified in sufficient quantity to determine its identity. The sequence of four different peptides were obtained after microsequencing, all of which were present in the predicted protein, termed Hs-CUL-2, in GenBank. Hs-cul-2 is a member of a recently identified multigene family that encodes proteins of 70–80 kDa, termed cullins, that are present in yeast (3 members), in C. elegans (5 members) and in humans (6 members; ref. 22). Phylogenetic analysis suggested that successive duplications of an ancestral cullin gave rise to three main branches containing cul-1/cul-2, cul-3/cul-4A,B, and cul-5, respectively, that predate the separation of chordates and nematodes ≈800 million years ago (22).
Cul-1 recently has been identified in a C. elegans genetic screen as a negative regulator of the cell cycle (22). A null mutation in Ce-cul-1 is associated with hyperplasia of all tissues. Ce-cul-1 is required for developmentally programmed transitions from the G1 phase of the cell cycle to the G0 phase or the apoptotic pathway. The mutant phenotype also reveals an acceleration of the G1-S phase transition, overriding mechanisms for mitotic arrest and producing abnormally small cells (22).
In yeast there are three cullins, with Sc-CUL-A being most similar to Hs-CUL-1 (36% identity) and less similar to Hs-CUL-2 (30% identity). CUL-B is most similar to CUL-3, and CUL-C, an outlying member of the family, has only 15–18% identity to other known cullins (22). Mutants of Sc-cul-A, originally identified as cdc53, are arrested in the late G1 phase of the cell cycle with unreplicated DNA and multiple elongated buds (23, 24). Cdc53 has more recently been shown to be part of a protein complex that targets phosphorylated G1 cyclins, as well as the phosphorylated cdk inhibitor, p40SIC1, for degradation by the ubiquitin-proteasome pathway (24–26). Cdc53 is thought to be an E3 ubiquitin protein ligase, as it is auto-ubiquitinated and binds to its targets as well as to an E2 ubiquitin conjugating enzyme, Cdc34.
To verify the VBC-Hs-CUL-2 interaction, both Hs-cul-1 and Hs-cul-2 were cloned by RT-PCR. The reported Hs-cul-2 sequence was incomplete and lacked ≈100 aa at the N terminus, as judged from the C. elegans homolog (22). The missing piece, as well as the 5′ untranslated region of Hs-cul-2, were cloned by PCR from a human cDNA library and assembled to generate a full-length Hs-cul-2 cDNA (Fig. 4). Comparison of Hs-CUL-2 with Ce-CUL-2 revealed a 45% identity, similar to the identity score of the CUL-1 orthologs (47% identity). Hs-CUL-1 compared with Hs-CUL-2 revealed only 39% identity, suggesting that the Hs-cul-2 gene is the ortholog of the Ce-cul-2 gene.
Figure 4.
Amino acid sequence comparison between predicted full-length human CUL-2 and C. elegans CUL-2 proteins. Predicted sequences of the human Hs-CUL-2 (745 aa) and C. elegans Ce-CUL-2 (743 aa) are shown. Amino acids identical between human and C. elegans CUL-2 proteins are boxed.
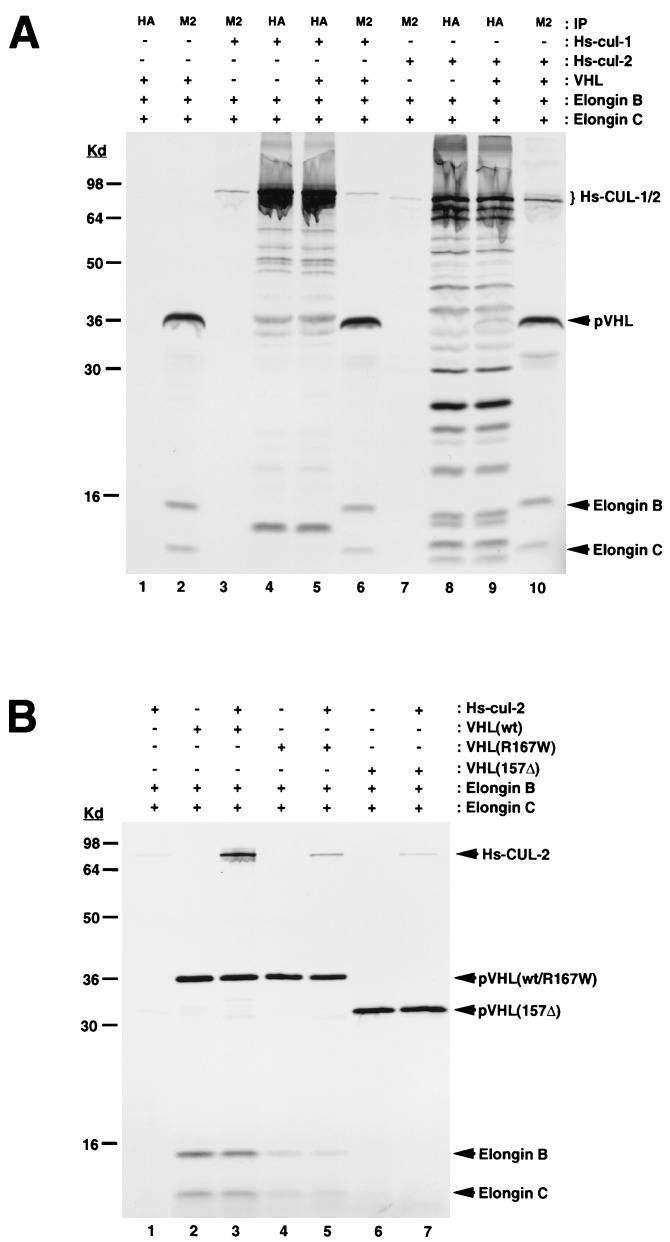
We then tested whether the Hs-CUL-2 homolog, Hs-CUL-1, is also capable of binding to VBC complexes. For in vitro binding experiments, we added epitope tags to VHL and Hs-CUL-1 or 2 (FLAG and HA, respectively) and translated the various combinations of VHL, elongin B, elongin C, Hs-cul-1, and Hs-cul-2 cDNAs in rabbit reticulocyte lysates followed by immunoprecipitations with anti-Flag or anti-HA antibodies (Fig. 5A). We observed no coprecipitation of Hs-CUL-1 with VBC (lanes 5 and 6). However, Hs-CUL-2 bound specifically to the VBC complex and could be coimmunoprecipitated with the FLAG and HA antibodies (compare lanes 7 and 8 with lanes 9 and 10). The VBC-Hs-CUL-2 interaction was reduced or strongly reduced by using the cancer-predisposing VHL missense mutation, R167W (Fig. 5B, lane 5), and the frameshift mutation, 157Δ, respectively (lane 7). These VHL mutations also resulted in a weak or almost undetectable pVHL-elongin BC association, since they affect the elongin binding site in pVHL (10). We also tested whether Hs-CUL-2 could interact with elongin B and C alone without pVHL. Cotranslation of Hs-cul-2 and elongin B-C followed by HA immunoprecipitation did not result in detectable Hs-CUL-2-elongin B-C binding, suggesting that Hs-CUL-2 only associates with the trimeric VBC complex (Fig. 5A, lane 8) and that pVHL is a necessary component of this complex.
Figure 5.
(A) Coimmunoprecipitation of VBC with Hs-CUL-2 but not with Hs-CUL-1 in vitro. Various combinations of the cDNAs encoding human F-VHL, elongin B, elongin C, Hs-CUL-1-HA, and HS-CUL-2-HA were expressed in a coupled transcription–translation system in the presence of [35S]methionine. The translation products were immunoprecipitated with either M2 or HA antibodies and analyzed on a SDS/14% polyacrylamide gel followed by autoradiography. (B) Coimmunoprecipitation of Hs-CUL-2 with wild-type (wt) or mutant VHL (R167W, 157Δ) in vitro. cDNAs encoding human wt F-VHL, R167W, and 157Δ mutant VHL, elongin B, elongin C, and Hs-CUL-2-HA were expressed in a coupled transcription–translation system in the presence of [35S]methionine. The translation products were immunoprecipitated with M2 antibody and analyzed on a SDS/14% polyacrylamide gel, followed by autoradiography.
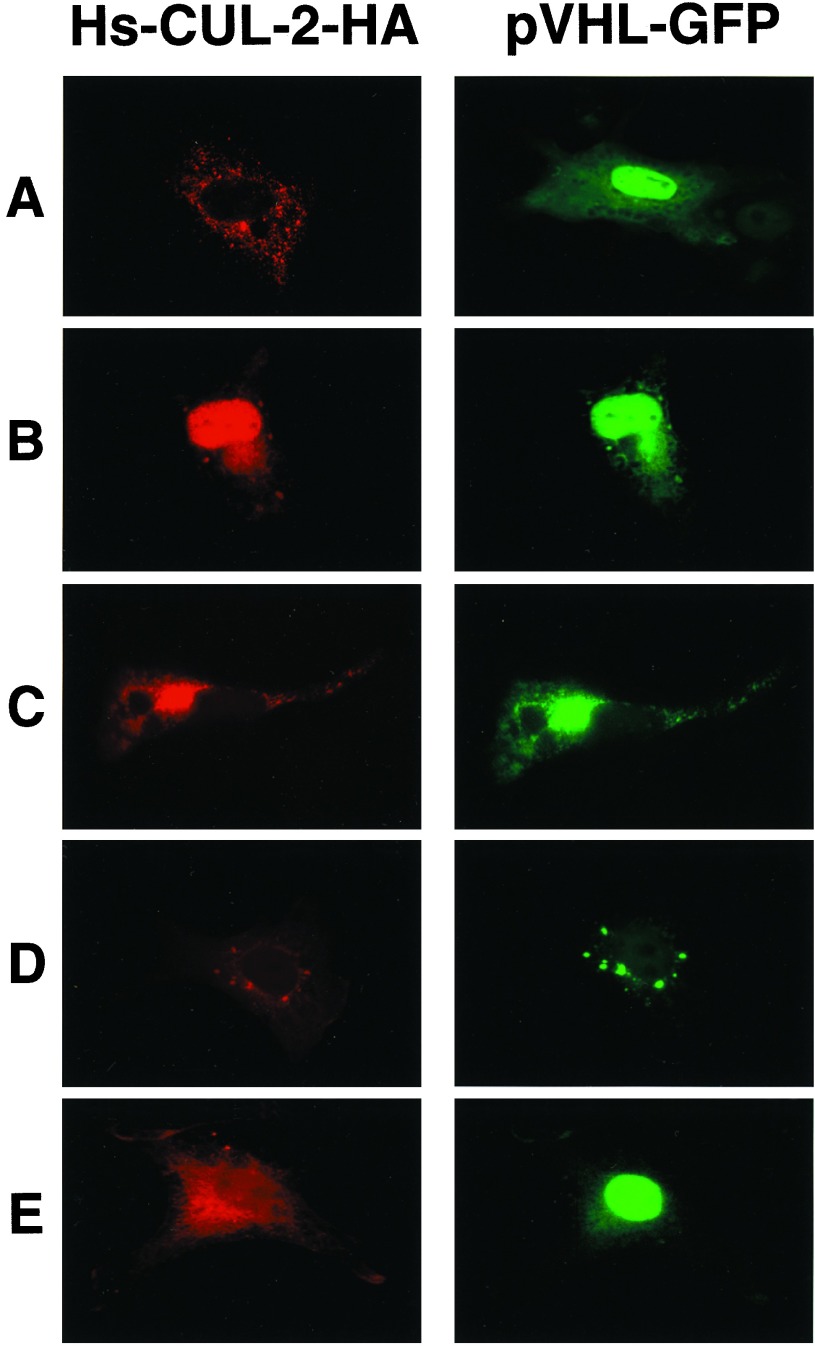
In cultures of sparse cells, wt VHL is found in the nucleus, whereas in confluent cells in culture, it is largely excluded from the nucleus (20). Mutational analysis has defined a possible nuclear localization signal in exon 1 that is required for nuclear entry (20). We verified the VBC-Hs-CUL-2 interaction in vivo by assessing whether the two proteins colocalized within cells by using a pVHL-GFP expression vector as well as an HA epitope-tagged HS-CUL-2 expression vector. Overexpression of Hs-CUL-2 alone resulted in the exclusive localization to the cytosol, regardless of the growth conditions (Fig. 6A Left). In contrast, pVHL alone localized to the cytosol and predominantly to the nucleus (Fig. 6A Right). Hs-CUL-2 was localized to the nucleus when it was coexpressed with wt pVHL-GFP (Fig. 6B). Hs-CUL-2 was observed only in the nucleus of those cells expressing nuclear VHL. In contrast, pVHL alone localized variably to either the cytosol or the nucleus. Coexpression of Hs-CUL-2 and a pVHL mutant, Δ60, which lacks the putative pVHL nuclear localization signal and therefore cannot enter the nucleus, resulted in colocalization of both proteins (Fig. 6C) or partial colocalization of both proteins in characteristic punctate cytosolic structures that we have previously observed for this VHL mutant (Fig. 6D; ref. 20). Coexpression of Hs-CUL-2 with the 157Δ mutant resulted in the localization of significantly less Hs-CUL-2 to the nucleus (compare Fig. 6 B with E), presumably because 157Δ interacts with Hs-CUL-2 only weakly (see Fig. 5B). These data suggest that the VBC-Hs-CUL-2 association is specific and occurs in vivo, the complex remains intact in both the nucleus and the cytosol, and that VBC is capable of relocalizing Hs-CUL-2 to the nucleus.
Figure 6.
Colocalization of Hs-CUL-2 and pVHL-GFP in transfected cells. COS-7 cells were transiently transfected with cDNAs encoding Hs-CUL-2-HA or pVHL-GFP (wt) separately (A) or concurrently (B–E). (C–E) COS-7 cells were transfected with cDNAs encoding Hs-CUL-2-HA and mutant pVHL-GFP [Δ60 (C and D) and 157Δ (E)]. Cells were stained with the anti-HA antibody to detect Hs-CUL-2-HA or observed under fluorescein isothiocyanate illumination to detect pVHL-GFP.
DISCUSSION
Our data suggest that pVHL, already shown to complex with elongin B and C, can interact with an additional protein, Hs-CUL-2, in the cell. This latter interaction is dependent on the integrity of the trimeric VBC complex, and naturally occurring mutations that interfere with binding of elongins also eliminate binding to this protein. Our experiments suggest that most of the VBC-p72-p80-p180 association occurs post-cell lysis, and it is not clear whether pVHL interacts with these proteins in the cell. We believe that the VBC-Hs-CUL-2 interaction is functionally significant, since it occurs in the cell and in in vitro binding assays, and is abrogated by cancer-predisposing pVHL mutations that are detected in up to 70% of VHL families (13). These results further suggest that the VBC-Hs-CUL-2 interaction may be important for the tumor-suppressor function of pVHL, and we propose that Hs-cul-2 itself might be a candidate tumor-suppressor gene as proposed for Hs-cul-1 (27, 28).
It has been estimated that there are up to 1,100 cysts and 600 microscopical solid tumors in a VHL kidney (29). VHL loss of heterozygosity has been described in solid tumors as well as even the earliest, single-cell-lined VHL renal cyst (30). This scenario is similar to the myriad of polyps observed in the colons of Familial Adenomatous Polyposis patients in which the APC tumor-suppressor gene appears to function as the gatekeeper (31). VHL might have a “gatekeeper” function in certain tissues such as the kidney. Normally, gatekeeper genes are responsible for maintaining a constant cell number in renewing cell populations, ensuring that cells respond appropriately to situations requiring net cell growth (e.g., tissue damage). Loss of function of a gatekeeper would result in a permanent imbalance of cell division over cell death. The Ce-cul-1 phenotype is reminiscent of a gatekeeper function of this gene in the worm, and it is possible that in humans the VBC-Hs-CUL-2 complex is functioning as a gatekeeper in kidney epithelial cells.
Although Hs-cul-2 is a member of the cullin family, as defined by sequence homology, this protein is not yet well characterized, in contrast to Ce-CUL-1 in C. elegans and Cdc53 in Saccharomyces cerevisiae. Loss of function of cul-1 in C. elegans results in increased cell numbers in a variety of lineages, suggesting a tumor-suppressor-like function. Mutants of cdc53 are arrested in the late G1 phase of the cell cycle with unreplicated DNA and multiple elongated buds (23). The cdc53 mutant phenotype implicates that gene product in control of the cell cycle, and recent data point to a specific role for Cdc53 in the degradation of critical cell cycle control proteins (24–26). Sequence analysis suggests that Cdc53 in S. cerevisiae is a homolog of CUL-1 and CUL-2 in multicellular organisms (36% and 30% identity, respectively).
Whether CUL-2 and/or other members of the cullin family are involved in targeted protein degradation remains to be determined. VBC-Hs-CUL-2 is part of a complex that shuttles in and out of the nucleus, perhaps degrading specifically targeted substrates of currently unknown identity. It is possible that such a proposed degradation complex targets certain transcription factors or factors that regulate the stability of hypoxia-inducible mRNAs, such as VEGF, platelet-derived growth factor-β, and GLUT1 (14–16). Furthermore, the presence of a Hs-cul-2 ortholog in C. elegans, should make cul-2 mutant phenotypes in this organism informative. We propose that identification of Hs-CUL-2 as a VBC-interacting protein will prove to be critical in characterizing the function of the VHL tumor-suppressor gene pathway.
Acknowledgments
We thank D. Roxanne Duan and James H. Hurley for advice and the kind gift of the pET-15b-elongin B and C plasmids. We thank William S. Lane and his team at the Harvard Microchemistry Facility (Boston) for microsequencing analysis. A.P. is supported by a fellowship from the Deutsche Forschungsgemeinschaft. D.Y.T.C. was supported by the Howard Hughes Medical Institute–National Institutes of Health Research Scholars Program.
ABBREVIATIONS
- VBC
pVHL-elongin B-C
- RCC
renal cell carcinoma
- HA
hemagglutinin
- FCS
fetal calf serum
- GFP
green fluorescent protein
- RT-PCR
reverse transcription–PCR
- wt
wild type
- VHL
von Hippel-Lindau
- VEGF
vascular endothelial growth factor
References
- 1.Linehan W M, Lerman M I, Zbar B. J Am Med Assoc. 1995;273:564–570. [PubMed] [Google Scholar]
- 2.Gnarra J R, Duan D R, Weng Y, Humphrey J S, Chen D Y, et al. Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 3.Knudson A G. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knudson A G. Cancer. 1995;1:180–181. [PubMed] [Google Scholar]
- 5.Zbar B. Cancer Surv. 1995;25:219–232. [PubMed] [Google Scholar]
- 6.Zbar B, Kishida T, Chen F, Schmidt L, Maher E R, et al. Human Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Shuin T, Kondo K, Torigoe S, Kishida T, Kubota Y, et al. Cancer Res. 1994;54:2852–2855. [PubMed] [Google Scholar]
- 8.Latif F, Tory K, Gnarra J R, Yao M, Duh F, et al. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 9.Duan D R, Humphrey J S, Chen D Y T, Weng Y, Sukegawa J, Lee S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1995;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 11.Aso T, Lane W S, Conaway J W, Conaway R C. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 12.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G., Jr Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 13.Kishida T, Stackhouse T M, Chen F, Lerman M I, Zbar B. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 14.Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 15.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassiliou E, Olfield E H, Klausner R D, Linehan W M. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Jr, Goldberg M A. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein I, Neeman M, Shweiki D, Itin A, Keshet E. Mol Cell Biol. 1995;15:5363–5368. doi: 10.1128/mcb.15.10.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy A P, Levy N S, Goldberg M A. J Biol Chem. 1996;271:2746–2753. doi: 10.1074/jbc.271.5.2746. [DOI] [PubMed] [Google Scholar]
- 19.Pause A, Aso T, Linehan W M, Conaway J W, Conaway R C, Klausner R D. Methods Enzymol. 1996;274:436–441. doi: 10.1016/s0076-6879(96)74035-1. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Chen D Y T, Humphrey J S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole N B, Sciaky N, Marotta A, Song J, Lipincott-Schwartz J. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 23.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 24.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 25.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 26.Mathias N, Johnson S L, Winey M, Adams A E, Goetsch L, Pringle J R, Byers B, Goebl M G. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raff M C. Cell. 1996;86:173–175. doi: 10.1016/s0092-8674(00)80087-2. [DOI] [PubMed] [Google Scholar]
- 28.Jackson P K. Curr Biol. 1996;6:1209–1212. doi: 10.1016/s0960-9822(96)00697-5. [DOI] [PubMed] [Google Scholar]
- 29.Walther M M, Lubensky I A, Venzon D, Zbar B, Linehan W M. J Urol. 1995;154:2010–2014. [PubMed] [Google Scholar]
- 30.Lubensky I A, Gnarra J R, Bertheau P, Walther M M, Linehan W M, Liotta L, Lubensky I A. Am J Pathol. 1996;149:2089–2095. [PMC free article] [PubMed] [Google Scholar]
- 31.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]