Abstract
Bacteriophage N4 virion RNA polymerase (N4 vRNAP) promoters contain inverted repeats, which form a 5- to 7-base-pair stem, 3-base loop hairpin that is required for vRNAP recognition. We show that, contrary to certain theoretical predictions, hairpin extrusion can occur at physiological superhelical densities in a Mg2+-dependent manner. Specific sequences on the template strand are required for hairpin extrusion. These sequences define stable DNA hairpins that are relatively unreactive to single strand-specific probes. In addition, a specific stable hairpin-inducing sequence can regulate transcription in vivo. Thus, a DNA structure, in its natural environment, is involved in transcriptional regulation.
DNA supercoiling affects transcription by facilitating or inhibiting RNA polymerase (RNAP) binding and/or formation of the open complex (1–3). In addition, by stabilizing DNA bending and/or looping (4–7), supercoiling enhances or inhibits the interaction of activators or repressors with the transcriptional machinery. Supercoiling also promotes the formation of noncanonical DNA structures such as cruciforms, Z-DNA, or triple helices (8) which can affect transcription in vitro (9–11). In no instance, however, is there convincing evidence that such structures play a role in vivo in regulating transcription (reviewed in ref. 12).
The three bacteriophage N4 early promoters utilized by the virion (v)RNAP share sequence from −18 to +1 containing small inverted repeats centered at −12 (ref. 13). On single-stranded DNA templates, these promoters are utilized efficiently by N4 vRNAP (13, 14). Mutational analyses of the promoter template strands indicated that specific sequences and a 5- to 7-base-pair stem, 3-base loop hairpin are important for transcriptional activity (14). Transcription on double-stranded DNA requires supercoiling and Escherichia coli single-stranded DNA-binding protein (EcoSSB) (15). On the basis of these results, we proposed a model in which negative supercoiling extrudes a hairpin to yield an active promoter conformation that is recognized by N4 vRNAP (14). On the contrary, the current theory of the energetics of hairpin extrusion predicts that such small hairpins will extrude only at high, unphysiological superhelical densities (16, 17). Here, we show that extrusion of N4 early promoter hairpins occurs at physiological superhelical densities, that this event requires Mg2+ and specific sequence, and that extrusion occurs in vivo.
MATERIALS AND METHODS
Preparation of Circles Containing N4 Early Promoters.
Cloning of the 2.2-kb SpeI–PstI fragment, containing the N4 early promoters P1 and P2 followed by their respective terminators t1 and t2, into pKB652 to yield pXD102, was as described (18). To generate mutant P1 promoters, pXD102 was digested with SmaI and partially digested with NsiI; the 4844-bp fragment lacking the P1 promoter was ligated to the M13 mp11 SmaI–PstI fragment carrying the wild-type or mutant P1 promoters (14). In vivo generation of circles, isolation, preparation of topoisomers, and determination of the average superhelical density were as described (18).
Probing Structural Changes at the N4 Early Promoters As a Function of Superhelical Density.
Chloroacetaldehyde (CAA), mung bean nuclease (MBN), and T7 endonuclease I (T7 endo I) reactions were carried out as described (18). Primers that hybridize specifically to the template and nontemplate strands, 40–60 bp away, of each promoter [5′-GTAATCCCAGACAAAAGG (nontemplate) and 5′-CAAATGGAGGCTCCTCG (template) of P2 and 5′-GTCATAGTATTAGTCTCC (nontemplate) and 5′-GGCATGCAAGCTTTGTA (template) of P1] were used to detect modifications or cleavages by primer extension analysis. NEB Vent (exo−) DNA polymerase (New England Biolabs) was used to carry out multiple rounds of primer extension, with end-labeled primer, in a thermal cycler as described (18). Sequencing dye mix was added directly to the PCR tubes and samples were loaded onto 8% polyacrylamide/7 M urea gels, alongside double-stranded sequencing reactions of the unmodified DNA using the same primer. Gels were dried and exposed to x-ray film. Whenever mutant promoters were probed, the wild-type P2 promoter present in the same circle was used as a control. The site of modification or cleavage by a specific reagent was confirmed by Maxam–Gilbert DNA sequencing reactions (19).
Preparation and Two-Dimensional Gel Electrophoretic Analysis of Topoisomers.
N4 promoter-containing circular DNA was digested with HindIII, dephosphorylated, isolated on a 1% agarose gel, and end-labeled with T4 polynucleotide kinase. 32P-end-labeled DNA in 50 mM Tris·HCl, pH 7.6/10 mM MgCl2/1 mM ATP/1 mM DTT/5% (vol/vol) polyethylene glycol 8000 was incubated with T4 ligase and various concentrations (0–10 μM) of ethidium bromide at 16°C for 12 hr, purified by extraction with phenol, precipitated with ethanol, and resuspended in TE (10 mM Tris·HCl, pH 8/1 mM EDTA). The different topoisomers were combined and treated with T7 endo I in 50 mM Tris·HCl, pH 8.0/10 mM MgSO4/1 mM DTT containing 50 μg/ml BSA for 10 min at 37°C. The topoisomers were loaded onto a 2-mm circular well in a 0.5× TBE/10 mM MgCl2/1.5% Metaphor agarose (FMC BioProducts) gel (10 × 14 × 0.5 cm; 1× TBE = 90 mM Tris borate, pH 8.3/2 mM EDTA, pH 8.0). Electrophoresis was performed as described (20). Electrophoresis in the first dimension was at 12.8 V/cm for 3 hr at 4°C. Lanes were cut from the gel and turned 90°, and a 2% Metaphor gel (0.5× TBE, 0.25 μg/ml chloroquine) was poured around them (20 × 24 × 0.5 cm). The gel was soaked in 0.5× TBE, 25 μg/ml chloroquine for 1 hr prior to electrophoresis at 8.3 V/cm for 24 hr at 4°C. Gels were dried and exposed to film.
Determination of Oligonucleotide Melting Temperature (Tm).
Data are the average of three independent measurements made in 25 mM sodium cacodylate, pH 7.4. Oligonucleotide concentrations ranged from 1 to 10 μM. UV absorbance melting profiles were monitored at 260 nm in a Hewlett–Packard 8452A diode array spectrophotometer connected to a Hewlett–Packard 89090A Peltier thermal controller, using a heating rate of 0.5°C/min. The Tm, estimated as the midpoint of the transition, was concentration independent.
Determination of the in Vivo Utilization of N4 vRNAP and rrnB P1 Promoters.
E. coli W3350, carrying plasmids containing wild-type P1 or P1 G-13 promoter and the control promoter P2, was grown at 37°C to OD600 = 0.3 in M9 medium (0.2% casamino acids, 5 μg/ml thiamin, 50 μg/ml ampicillin) (21). Cells were collected by centrifugation, resuspended in fresh medium, incubated at 37°C for 10 min, and infected with N4 phage at a multiplicity of infection = 10. Cells were collected 20 min after infection and RNAs were purified as described (22). RNAs were analyzed by primer extension using 5′-CCTACAGTCATACGAACGTTAGCCTGAGTAG (for P1 or P1 G-13) or 5′-GAGACTACTTTTGTCGGTAAGTAATCCCAGAC as primer (22). Extension products were analyzed on 8% polyacrylamide/7 M urea gels in TBE buffer next to double-stranded sequencing reaction products of the same DNA and visualized by autoradiography. Double-stranded oligonucleotide-directed mutagenesis was performed on pRLG1478 (23) to replace the sequences between the −10 and −35 hexamers of the rrnB P1 promoter. E. coli W3350 was transformed with pRLG1478 or the mutated plasmids. Transcription from the plasmid-borne wild-type or mutant rrnB P1 promoters was measured by growing cells at 37°C in Luria–Bertani medium with ampicillin (50 μg/ml) to OD600 = 0.4, isolating RNAs, and performing primer extension with the primer 5′-CTTCCATCAGCGTTTATAGT. When present, novobiocin (1 mg/ml) was added 30 min before RNA isolation.
RESULTS
To study hairpin extrusion at the N4 early promoters, we used the site-specific recombination system of phage λ (24) to produce 2.2-kb DNA circles of the N4 early region containing the vRNAP P1 and P2 promoters. This approach, which isolates N4 sequences from vector sequences, avoids competition with other DNA structural transitions that might occur in vector sequences. Circles were incubated with different concentrations of ethidium bromide and topoisomerase I to generate topoisomers with a range of superhelical densities, and were allowed to react with DNA conformation-specific chemical and enzymatic probes in the absence of proteins. Probes included CAA, which reacts with unpaired cytosine, adenine, and to a lesser extent guanine (25); MBN, which cleaves phosphodiester bonds in single-stranded DNA (26); and T7 endo I, which recognizes DNA four-way junctions (27). Sites of modification or cleavage on both DNA strands were determined by primer extension analysis of the treated circles using primers hybridizing at positions adjacent to the promoters.
Supercoiling-Induced, Mg2+-Dependent Hairpin Extrusion.
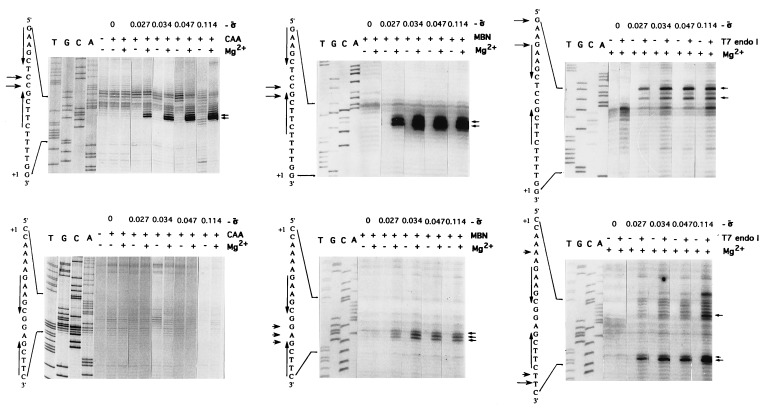
Relaxed DNA did not react with any of the probes tested (Fig. 1, σ̄ = 0) and, in the absence of Mg2+, no reactivity to CAA or MBN was detected even at the highest superhelical density tested (σ̄ = −0.114) (Fig. 1 Left and Center). This is expected because hairpin extrusion at these inverted repeats requires a superhelical density of approximately −0.110 (28). However, when Mg2+ (10 mM) was present, susceptibility to CAA and MBN at specific bases between the inverted repeats was noted in the nontemplate strand at a superhelical density as low as −0.027, which is lower than the superhelical density of these circles when isolated from E. coli cells (−0.034). Surprisingly, the template strand was not reactive to CAA and only weakly reactive to MBN at high superhelical densities, even in the presence of Mg2+, suggesting that the hairpins on the two strands adopt different conformations (see below). Removal of Mg2+, by chelation with EDTA prior to probe treatment, abolished hairpin extrusion (data not shown). Similarly, linearization of the DNA after incubation of the supercoiled circles with Mg2+ abolished the transition (data not shown). These results indicate that Mg2+ must be present in the final structure, and that template supercoiling is required for maintenance of the structure. The lack of extensive reactivity to single-strand-specific probes in sequences immediately flanking the inverted repeats indicated that these regions remained double-stranded, consistent with the formation of a cruciform structure. The reactivity to T7 endo I on both strands suggests the presence of a DNA four-way junction (Fig. 1 Right).
Figure 1.
Dependence on superhelical density of the reactivity of promoter P2 nontemplate (Upper) and template (Lower) strands to different probes: CAA (Left), MBN (Center), and T7 endo I (Right). The sequences of the two strands (arrows indicate inverted repeats) are shown next to the gels. Short arrows, modification or cleavage sites; σ̄ is superhelical density.
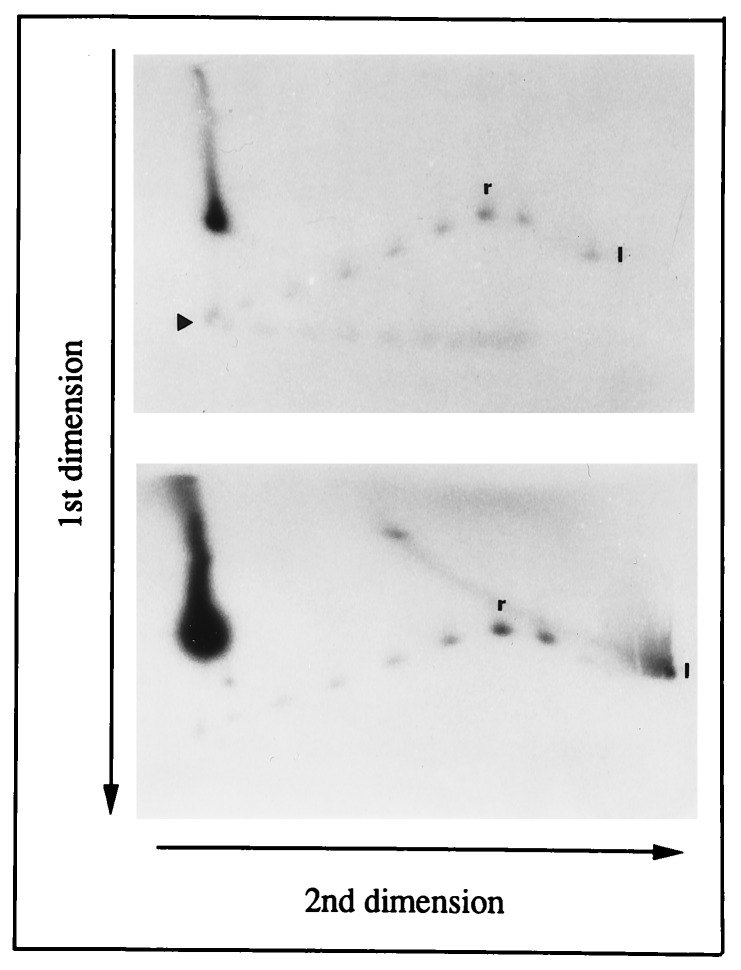
To identify the superhelical density at which extrusion occurred, a uniform distribution of topoisomers of a 2.2-kb circle containing a single promoter was subjected to two-dimensional agarose gel electrophoresis in 10 mM Mg2+ in the first dimension. Since the hairpin at the promoter encompasses 15 bp, hairpin extrusion should lead to the relaxation of 1.44 turns. Due to the difficulty in detecting such a change, and since it was expected that topoisomers which had supported hairpin extrusion would be cleaved by T7 endo I, a sample was treated with T7 endo I prior to analysis. The results presented in Fig. 2 show that topoisomers with more than 7 superhelical turns were cleaved with the concomitant appearance of linear DNA, and indicate that cruciform formation, as detected by cleavage with T7 endo I, occurs at a threshold superhelical density of −0.035.
Figure 2.
Two-dimensional gel electrophoresis of N4 promoter-containing circles. Topoisomers of a 2.2-kb circle containing a single N4 promoter were subjected to two-dimensional gel electrophoresis before (Upper) or after (Lower) treatment with T7 endo I. The arrowhead marks the topoisomer with a linking number of 7, r indicates the relaxed topoisomer, and l indicates linear DNA.
Effect of Cations on Hairpin Extrusion.
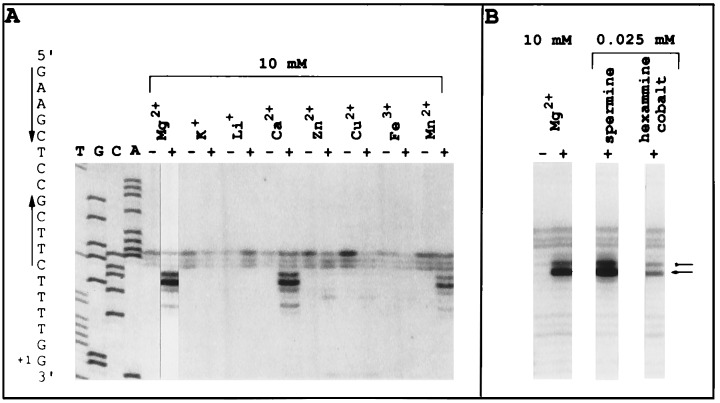
To determine the role of Mg2+ in hairpin extrusion, other cations were tested. Only Mn2+ and Ca2+ were able to replace Mg2+, as determined by CAA modification (Fig. 3A). Previous work with four-way junctions has established that Mg2+, Mn2+, and Ca2+ stabilize the junction to yield a well-stacked, folded ×-like structure (29, 30). Complex ions and polyamines, specifically hexammine cobalt(III) and spermine, also stabilize the four-way junction, albeit at much lower concentrations (31, 32). Indeed, hexammine cobalt(III) and spermine were able to induce the same pattern of MBN reactivity as that induced by Mg2+ at low concentrations (25 μM) (Fig. 3B). Maximal CAA and MBN reactivities were achieved at 1–2 mM Mg2+. These results indicate that Mg2+, within the physiological free concentration range (T. Record, personal communication), associates with and stabilizes the four-way junction (33).
Figure 3.
Effect of cations on hairpin extrusion at promoter P2. (A) Effect of cations on CAA reactivity of promoter P2 nontemplate strand. Reactions with CAA were performed in the absence and presence of cations as indicated at the top of the lanes. (B) Effect of complex cations on MBN reactivity of promoter P2 nontemplate strand. The short arrows indicate the sites at which Vent DNA polymerase terminates as a result of modification or cleavage.
Strand Asymmetry.
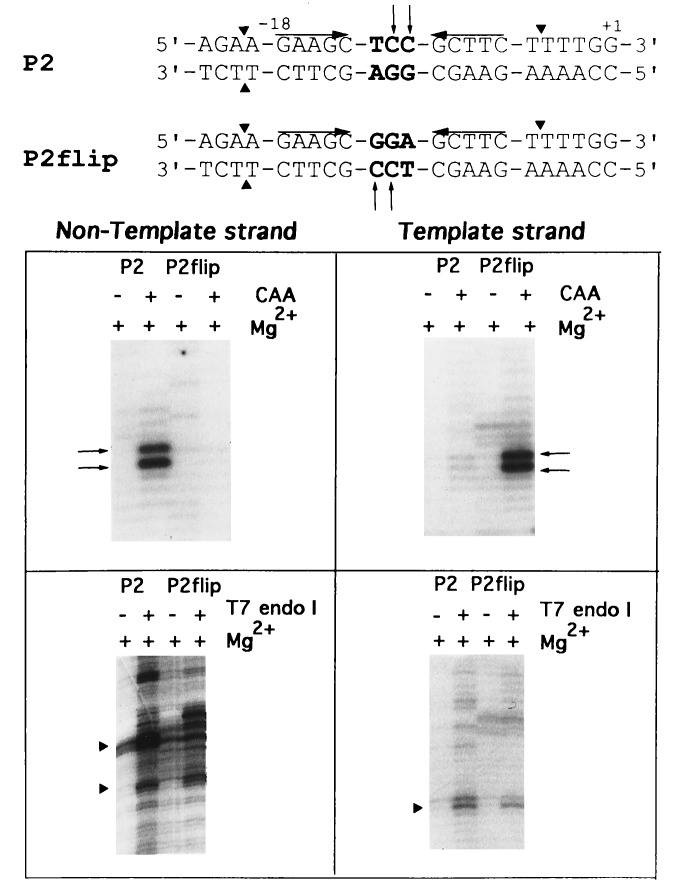
The template- and non-template-strand hairpins exhibit different reactivities toward single-strand-specific chemical and nuclease probes, presumably reflecting a difference in their conformations. Since the hairpins on the two strands differ only in their loop sequences, the bases within the loop must contribute to the observed strand asymmetry. A mutant promoter, P2flip, was generated in which the bases within the loops of the template and non-template strand hairpins were switched (Fig. 4). In contrast to the wild-type promoter, where CAA reactivity occurred only on the nontemplate strand, the mutant promoter showed sensitivity to CAA only on the template strand (see top of Fig. 4). T7 endo I cleaved both strands of P2flip outside the inverted repeats, indicating cruciform formation at the mutant promoter (Fig. 4 gels). These results indicate that bases within the loop cause the observed strand asymmetry.
Figure 4.
Reactivity of wild-type promoter P2 and mutant promoter P2flip to different probes. At the top, promoter sequences; mutated bases in P2flip are shown in boldface. Triangles and small arrows indicate sites of T7 endo I and CAA modification, respectively. Upper pair of gels, CAA; lower pair, T7 endo I.
Sequence Requirements for Hairpin Extrusion.
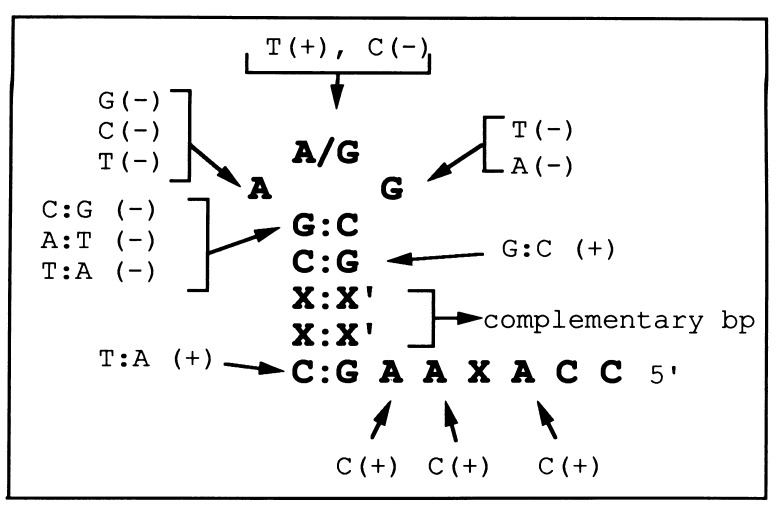
To determine if specific sequences are required for extrusion, we performed extensive mutational analyses of promoters present on circles and probed for extrusion using various DNA conformation-specific probes (X.D., M.B.G., and L.B.R.-D., unpublished work). A minimum stem length of 4 bp, provided that the stem is composed exclusively of G·C base pairs, and specific sequences at the stem-loop junction and in the loop (5′-C-GDA-G-3′, where D = G, A, or T) are required (Fig. 5). Surprisingly, C at the center of the loop blocks extrusion. In general, the sequences required for extrusion are those that yield the most stable DNA hairpins (Table 1). Mg2+ (10 mM) had no effect on the relative Tm of the hairpins (data not shown). A decrease in hairpin stability was observed for mutations at the base pair closing the loop and bases at positions −11 and −13 (Table 1). In addition, the template-strand hairpins were more stable than the non-template-strand hairpins by 4–8°C. Elucidation of the structure of template-strand hairpins by NMR spectroscopy indicates that the hairpin loops exist as well-ordered, stacked structures (ref. 34 and M.B.G., unpublished results). This conformation explains the unusual stability and the lack of reactivity of template-strand hairpin loops to single-stranded DNA-specific reagents (this paper and ref. 35).
Figure 5.
Promoter sequences required for hairpin extrusion. The sequence of the wild-type promoter template strand is shown. X and X′ can be any nucleotide as long as they can base pair. Mutations tested for hairpin extrusion are shown indicating the presence (+) or absence (−) of extrusion from circular templates of physiological superhelical density.
Table 1.
Tm of template- and non-template-strand hairpins and the effect of mutations
| Promoter | Sequence | Tm, °C |
|---|---|---|
| P1 | 5′-AGTTGC-G−11 A A−13-GCAACG-3′ | 76.7 ± 0.5 |
| G- -C | 69.8 ± 0.2 | |
| C | 69.4 ± 0.6 | |
| T | 66.0 ± 0.6 | |
| C G | 75.6 ± 0.3 | |
| A G | 68.5 ± 0.3 | |
| T- -A | 65.5 ± 0.4 | |
| A- -T | 63.6 ± 0.2 | |
| G | 74.9 ± 1.9 | |
| T | 75.7 ± 0.3 | |
| C | 76.7 ± 0.3 | |
| 5′-CGTTGC-T T C-GCAACT-3′ | 67.5 ± 0.5 | |
| P2 | 5′-AGAAGC-G G A-GCTTCT-3′ | 73.6 ± 1.0 |
| G- -C | 61.2 ± 0.7 | |
| C | 67.5 ± 1.0 | |
| T | 63.6 ± 1.1 | |
| C G | 73.9 ± 0.5 | |
| T | 72.0 ± 0.4 | |
| C | 74.9 ± 0.2 | |
| A | 75.0 ± 0.4 | |
| 5′-AGAAGC-T C C-GCTTCT-3′ | 69.4 ± 0.6 | |
| P3 | 5′-AGCTGC-G G A-GCAGCT-3′ | 84.0 ± 2.0 |
| 5′-AGCTGC-T C C-GCAGCT-3′ | 76.3 ± 0.6 |
Numbering of sequence on template strand corresponds to numbering of promoter template strand.
Hairpin Extrusion in Vivo.
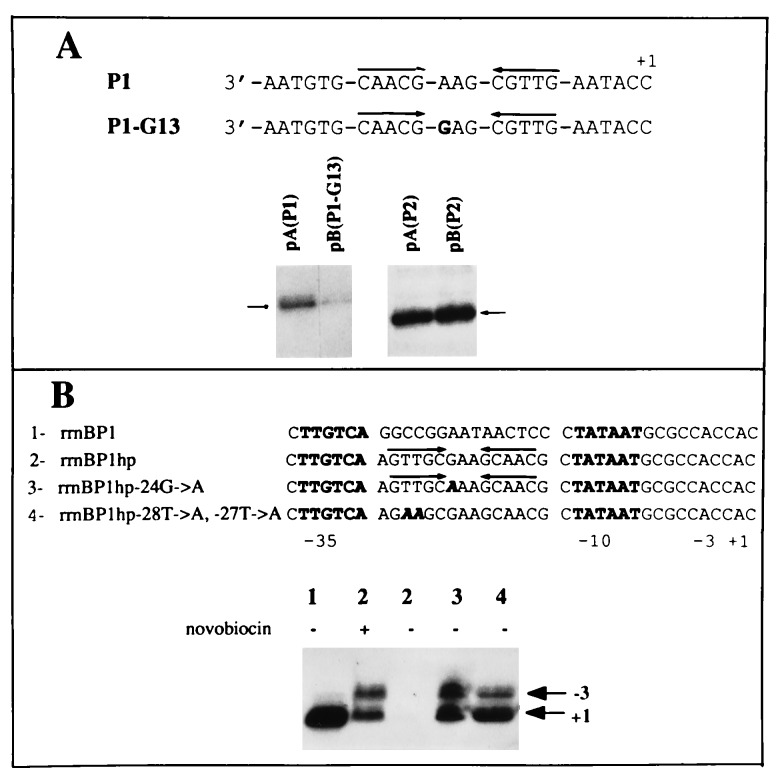
To determine whether hairpin extrusion occurs in vivo, we used two approaches. First, we assessed the transcriptional activity of extruding and nonextruding promoters when present on supercoiled plasmids inside E. coli cells by RNA primer extension analysis (Fig. 6A). Promoter P1 G-13 supports N4 vRNAP transcription at wild-type levels when present on single-stranded templates (14) but does not extrude a cruciform in vitro (Fig. 5). In vivo, transcriptional activity of the nonextruding promoter P1 G-13 was greatly reduced compared with the activity of the extruding promoter (Fig. 6A), indicating that hairpin formation occurs.
Figure 6.
In vivo hairpin extrusion. (A) Primer extension analysis of RNAs synthesized in E. coli cells from promoters P1 or P1 G-13 and the respective P2 control promoters. P1 and P1 G-13 promoter template-strand sequences are shown. Short arrow, primer extension product. (B) Primer extension analysis of RNAs synthesized in E. coli cells from rrnB P1 wild-type or mutant promoters containing extruding and nonextruding hairpin sequences between the −10 and −35 hexamers.
In addition, we assessed the effect of replacing the sequences between the −35 and −10 hexamers of the rrnB P1 promoter with either extruding or nonextruding hairpin sequences (Fig. 6B), with the expectation that cruciform extrusion would inhibit binding of E. coli RNAP at the modified rrnB P1 promoters. Replacement of spacer sequences with extruding hairpin sequences abolished promoter activity, while pretreatment of cells with novobiocin, an inhibitor of DNA gyrase, restored activity. In contrast, replacement with nonextruding hairpin sequences—i.e., sequences with a single base change in the loop (−24G → A) or with disrupted inverted repeats (−28T → A, −27T → A) yielded active promoters. Two sites of transcription initiation (+1 and −3) were detected in constructs where the natural spacer sequences were replaced, due either to the utilization of multiple start sites or to slippage of the nascent RNA during transcription initiation, as has been observed in vitro (36). These results indicate that a supercoil-dependent structure that inhibits the interaction of RNAP with the rrnB P1 promoter can be formed in vivo by the extruding N4 promoter sequences.
DISCUSSION
The extrusion of the small N4 vRNAP promoter hairpins at low superhelical densities is unexpected. The observed reactivity to probes that react with single-stranded DNA (MBN and CAA) and a four-way junction (T7 endo I) indicates that a specific DNA structure forms in a Mg2+- and supercoiling-dependent manner. The pattern of reactivity to T7 endo I suggests cruciform formation. Low or absent template-strand reactivity to single-stranded probes is due to the presence of specific sequences that yield unusually stable DNA hairpins. The fact that mutations which result in a less stable template-strand hairpin also affect extrusion suggests that the unusually stable, template-strand hairpin facilitates the formation and/or stabilizes the structure of the cruciform. While the energy of extrusion is supplied by the level of negative supercoiling, the extent of cruciform formation is influenced by the intrinsic thermostability of the hairpins, Mg2+, and supercoiling. However, the increased thermostability (≈10°C, equivalent to ≈1.4 kcal) of the unusually stable hairpins cannot provide all of the energy required. It is worth pointing out that current calculations of the energetics of supercoiling-induced cruciform formation do not take into account the contribution of localized Mg2+ in these structures. Surprisingly, C at the center of the template-strand hairpin loop did not allow extrusion, although the corresponding hairpin is as stable as the wild type (Table 1). Since the first step in cruciform formation requires melting of base pairs at the center of the inverted repeats followed by intrastrand base pairing, the stability of the duplex at the center of the inverted repeats as well as the rate of intrastrand nucleation will affect hairpin formation (37, 38). Cytidine at the center of the template-strand hairpin loop may inhibit initiation of strand separation, since the GC stacking interaction of the mutant promoter (5′-C-GCA-G-3′) results in a more stable duplex at the nucleation site (39). In this case, the formation of the stacked structure at the loop of the template-strand hairpin might play an essential, yet undefined, role in intrastrand base pairing during extrusion of these small hairpins.
The existence of small stable hairpins capable of extrusion at physiological superhelical densities raises three new questions: how prevalent are they, under what conditions do they extrude, and do they play a role in protein recognition? Inspection of the sequence database of bacterial, bacteriophage, and plasmid genomes reveals some sequences that can give rise to small stable hairpins (F. Leclerc and L.B.R.-D., unpublished work). We expect that many of these sequences may extrude in vivo, especially when present upstream of a site of transcription initiation where transcription will transiently increase the level of negative supercoiling behind RNAP (40).
Hairpin extrusion provides a potential supercoiling-dependent recognition site for proteins. In N4 early promoters, it provides a DNA structure that is invaded by E. coli single-stranded DNA-binding protein (EcoSSB). EcoSSB destabilizes the nontemplate hairpin, while the stable template hairpin persists to provide the structure and sequences required for productive transcription initiation by N4 vRNAP (41). This system presents what is to our knowledge the first example of the involvement of a supercoil-driven DNA structural change, in its natural environment, in transcriptional regulation. Why should vRNAP-promoter recognition be regulated by the superhelical density of the N4 genome? vRNAP-promoter recognition is the first event after injection of the genome into the host. We have suggested that the dependence of N4 early RNA synthesis on gyrase activity allows the phage to monitor the ability of the host cell to support phage growth (15), since the level of DNA supercoiling inside the cell is influenced by the [ATP]/[ADP] ratio (42–44). On the other hand, hairpin extrusion could inhibit the binding of sequence-specific DNA binding proteins to their cognate sites if these sites overlap extruding sequences. We show that replacement of sequences between the −10 and −35 hexamers of the rrnB P1 promoter with stable hairpin sequences leads, in vivo, to transcriptional repression of the rrnB P1 promoter in a supercoiling-dependent manner. Finally, the asymmetry of hairpin structures in these cruciforms may be used to mark the two strands of DNA for differential recognition by biochemical machineries and thus provide a directionality for such processes as recombination, replication, and transcription activation. Since only one of the hairpin loops is susceptible to single-strand nuclease, one might envision that after cleavage these structures could provide a specific and asymmetric site for loading of proteins (45).
Acknowledgments
We thank J. Miller for expert technical assistance, Dr. W. Studier for a generous supply of T7 endo I, Dr. W. Ross for pRLG1478 and the corresponding primer, and Drs. S. Mirkin and A. Dayn for instruction in the use of CAA. We thank K. Kazmierczak and Drs. D. Gottschling and J. Roberts for constructive comments on the manuscript, and Drs. N. Cozzarelli, E. P. Geiduschek, D. Levens, S. Mirkin, V. Potaman, R. Sinden, and A. Vologodskii for numerous discussions on cruciform formation energetics and extensive comments on the manuscript. This work was supported by National Institutes of Health grant RO1 AI12575 to L.B.R.-D.; X.D. and K.N.-C. were partially supported by National Institutes of Health Training Grants GM08369 and GM07183, respectively.
ABBREVIATIONS
- RNAP
RNA polymerase
- vRNAP
virion RNAP
- CAA
chloroacetaldehyde
- MBN
mung bean nuclease
- T7 endo I
T7 endonuclease I
References
- 1.Brahms J G, Dargouge O, Brahms S, Ohara Y, Vagner V. J Mol Biol. 1985;181:455–465. doi: 10.1016/0022-2836(85)90419-x. [DOI] [PubMed] [Google Scholar]
- 2.Ohlson K L, Gralla J D. J Biol Chem. 1992;267:19813–19818. [PubMed] [Google Scholar]
- 3.Rojo F, Nuez B, Mencia M, Salas M. Nucleic Acids Res. 1993;21:935–940. doi: 10.1093/nar/21.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec J A, Zhang L, Sasse-Dwight S, Gralla J D. J Mol Biol. 1987;196:101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- 5.Richet E, Raibaud O. J Mol Biol. 1991;218:529–542. doi: 10.1016/0022-2836(91)90699-7. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Martin J, Espinosa M. Science. 1993;260:805–807. doi: 10.1126/science.8387228. [DOI] [PubMed] [Google Scholar]
- 7.Choy H E, Park S-W, Parrack P, Adhya S. Proc Natl Acad Sci USA. 1995;92:7327–7331. doi: 10.1073/pnas.92.16.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank-Kamenetskii M D. In: DNA Topology and Its Biological Effects. Cozzarelli N R, Wang J C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 185–216. [Google Scholar]
- 9.Horwitz M S Z, Loeb L A. Science. 1988;241:703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- 10.Bagga R, Ramesh N, Brahmachari S K. Nucleic Acids Res. 1990;18:3363–3369. doi: 10.1093/nar/18.11.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohwi Y, Kohwi-Shigematsu T. Genes Dev. 1991;5:2547–2554. doi: 10.1101/gad.5.12b.2547. [DOI] [PubMed] [Google Scholar]
- 12.Sinden R R. DNA Structure and Function. New York: Academic; 1994. [Google Scholar]
- 13.Haynes L L, Rothman-Denes L B. Cell. 1985;41:597–605. doi: 10.1016/s0092-8674(85)80032-5. [DOI] [PubMed] [Google Scholar]
- 14.Glucksmann M A, Markiewicz P, Malone C, Rothman-Denes L B. Cell. 1992;70:491–500. doi: 10.1016/0092-8674(92)90173-a. [DOI] [PubMed] [Google Scholar]
- 15.Markiewicz P, Malone C, Chase J W, Rothman-Denes L B. Genes Dev. 1992;6:2010–2019. doi: 10.1101/gad.6.10.2010. [DOI] [PubMed] [Google Scholar]
- 16.McClellan J A, Boublikova P, Palecek E, Lilley D M J. Proc Natl Acad Sci USA. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinden R R, Carlson J O, Pettijohn D E. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 18.Miller A, Dai X, Choi M, Glucksmann-Kuis M A, Rothman-Denes L B. Methods Enzymol. 1996;274:9–19. doi: 10.1016/s0076-6879(96)74004-1. [DOI] [PubMed] [Google Scholar]
- 19.Maxam A M, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 20.Su T T, McClure W R. J Biol Chem. 1994;269:13511–13521. [PubMed] [Google Scholar]
- 21.Bolle A, Epstein R, Salser W, Geiduschek E P. J Mol Biol. 1969;31:325–348. doi: 10.1016/0022-2836(68)90413-0. [DOI] [PubMed] [Google Scholar]
- 22.Cho N-Y, Choi M, Rothman-Denes L B. J Mol Biol. 1995;246:461–471. doi: 10.1006/jmbi.1994.0098. [DOI] [PubMed] [Google Scholar]
- 23.Josaitis C A, Gaal T, Gourse R L. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backman K, O’Connor M J, Maruya A, Erfle M. Bio/Technology. 1984;2:1045–1049. [Google Scholar]
- 25.Singer B, Gruberger D. Molecular Biology of Mutagens and Carcinogens. NY: Plenum; 1983. pp. 45–96. [Google Scholar]
- 26.Kowalski D, Sanford J P. J Biol Chem. 1982;257:7820–7825. [PubMed] [Google Scholar]
- 27.de Massy B, Weisberg R A, Studier F W. J Mol Biol. 1987;193:359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]
- 28.Vologodskii A. Topology and Physics of Circular DNA. Boca Raton, FL: CRC; 1992. pp. 105–109. [Google Scholar]
- 29.Cooper J P, Hagerman P J. J Mol Biol. 1987;198:711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- 30.Cooper J P, Hagerman P J. Proc Natl Acad Sci USA. 1989;86:7336–7340. doi: 10.1073/pnas.86.19.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duckett D R, Murchie A I H, Lilley D M. EMBO J. 1990;9:583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan K M, Lilley D M J. J Mol Biol. 1987;193:397–404. doi: 10.1016/0022-2836(87)90227-0. [DOI] [PubMed] [Google Scholar]
- 33.Olmsted M C, Hagerman P J. J Mol Biol. 1994;243:919–929. doi: 10.1006/jmbi.1994.1692. [DOI] [PubMed] [Google Scholar]
- 34.Hirao I, Kawai G, Yoshizawa S, Y, N, Ishido Y, Watanabe K, Miura K. Nucleic Acids Res. 1994;22:576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshizawa S, Ueda T, Ishido Y, Miura K, Watanabe K, Hirao I. Nucleic Acids Res. 1994;15:2217–2221. doi: 10.1093/nar/22.12.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borukhov S, Sagitov V, Josaitis C A, Gourse R L, Goldfarb A. J Biol Chem. 1993;268:23477–23482. [PubMed] [Google Scholar]
- 37.Murchie A I H, Lilley D M J. Nucleic Acids Res. 1987;15:9641–9654. doi: 10.1093/nar/15.23.9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng G, Sinden R. J Biol Chem. 1988;263:5356–5361. [PubMed] [Google Scholar]
- 39.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1983. [Google Scholar]
- 40.Wang J C. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 1253–1269. [Google Scholar]
- 41.Glucksmann-Kuis M A, Dai X, Markiewicz P, Rothman-Denes L B. Cell. 1996;84:147–154. doi: 10.1016/s0092-8674(00)81001-6. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh L-S, Burger R, Drlica K. J Mol Biol. 1991;219:443–450. doi: 10.1016/0022-2836(91)90185-9. [DOI] [PubMed] [Google Scholar]
- 43.Hsieh L-S, Rouviere-Yaniv J, Drlica K. J Bacteriol. 1991;173:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruhdal P, Loman L, Petra B, van der Weijden C, Westerhoff H V. J Bacteriol. 1995;177:3420–3426. doi: 10.1128/jb.177.12.3420-3426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tinker R L, Wiliams K P, Kassavetis G A, Geiduschek E P. Cell. 1994;77:225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]