Abstract
Lactoferrin (LF), traditionally known as an iron-binding protein present in high concentrations in milk and various secretions, has emerged as a multifunctional protein involved in many aspects of the host defense against infection. Recently, LF has been shown to inhibit the growth of solid tumors and reduce experimental metastasis in mice, suggesting that LF also may play a role in the defense against tumorigenesis. Here we provide the sequence of the cDNA and promoter region, the chromosome assignment, and tissue expression pattern of a novel form of LF mRNA (ΔLF). The sequence of ΔLF mRNA is nearly identical to that of LF mRNA; however, at the 5′ end, we find a novel sequence that replaces the N-terminal signal peptide sequence of LF mRNA. We map the ΔLF mRNA to human chromosome 3 and find that both ΔLF and LF sequences colocalize to the same cloned 90- to 150-kb genomic DNA fragment. We further show that the ΔLF mRNA is the product of alternative splicing of the LF gene and likely is specified by use of an alternative promoter. Although we find ΔLF mRNA at various levels in 20 of 20 adult and fetal human tissues, we do not find ΔLF mRNA in any of 14 diverse tumor-derived cell lines.
Keywords: signal peptide, alternative splicing
Lactoferrin (LF) is an 80-kDa glycoprotein that can bind two molecules of iron very strongly (1, 2). It is present in high concentrations in human milk and in lower concentrations in a variety of secretions derived from glandular epithelium cells such as the prostate and salivary glands (2). LF also is found in high concentrations in the secondary granules of neutrophils (3, 4). The primary functions of LF appear to be iron transport, storage, and chelation. This latter function is thought to be responsible for the bacteriostatic activity of LF. Free iron required for bacterial growth is depleted when neutrophilic granulocytes release LF (3).
Many other effects of LF also have been described, several of which are thought to be related to host defense mechanisms. For example, LF is associated with activation of natural killer cells (5, 6), induction of colony-stimulating activity (7), activation of polymorphonuclear leukocytes and monocytes (8), regulation of granulopoiesis (9), enhancement of antibody-dependent cell cytotoxicity activity (10), stimulation of lymophokine-activated killer cell activity (5), potentiation of macrophage cytotoxicity (8), and maturation of splenic B cells (11).
LF is frequently found in the malignant counterparts of tissues that normally secrete it, such as the salivary gland (12), stomach (13), bone marrow (14), breast (15), and endometrium (16). However, in some malignancies, the levels of LF are lower (17, 18). In addition, methylation patterns in and around the LF gene are altered in malignant compared with normal breast cells (19). These studies suggest that there may be a deregulation of LF expression in some forms of cancer.
LF has been shown to affect the growth of mammalian cells in vitro and in vivo. The effects of LF in vitro are varied. In most cases, LF is stimulatory (20–23); however, in some cases, LF is inhibitory (24). The effect of LF in vivo also has been studied. Bezault and coworkers (25) found that injections of LF reduced tumor growth in mice containing transplanted solid tumors induced by v-ras-transformed fibroblasts and methylcholanthrene-induced fibrosarcomas. They also found substantially reduced lung colonization (experimental metastasis).
A relationship between LF expression and breast cancer susceptibility has been suggested by Furmanski and coworkers (26). They found a lower level of LF-associated RNase activity in the milk obtained from a group of consanguineous women in India who have a high incidence of breast cancer.
Here we identify and clone a novel form of LF mRNA (ΔLF) and its promoter. We also show that ΔLF mRNA is the product of alternative splicing of the LF gene. Further, we compare the tissue expression pattern of ΔLF and LF mRNA in normal tissues and tumor-derived cell lines. We discuss these results in the context of the myriad of effects and functions attributed to LF.
MATERIALS AND METHODS
PCR System and Primer Sequences.
All PCR amplifications were conducted with long and accurate PCR polymerase mixtures (27, 28), using a commercially available kit (CLONTECH). Primers used in this study had the following sequences: Primer LF 1, antisense, 5′-GGC TGT CTT TCG GTC CCG TAG ACT TCC-3′; primer LF 2, sense, 5′-AGG CCA CAA AAT GCT TCC AAT GGC AAA-3′; primer LF 3, antisense, 5′-ACA CCC ACG GGG AGC AGG GCA GGA AT-3′; primer LF 4, antisense, 5′ATT CCA CAG GGC AAC TGC CAC CCT TT-3′; primer LF 5, sense, 5′-GTA CCC CAA AGT GCC ATT GCA ACC CTT-3′; primer LF 6, sense, 5′-TGT CTT CCT CGT CCT GCT GTT CCT CG-3′; primer LF 7, antisense, 5′-CTG CCT CGT ATA TGA AAC CAC CAT CAA-3′; primer LF 8, sense, 5′-AGA GCC TTC GTT CGC CAA GTC GCC TCC-3′; glyceraldehyde-3-phosphate dehydrogenase, sense, 5′-ACC ACA GTC CAT GCC ATC AC-3′, antisense, 5′-TCC ACC ACC CTG TTG CTG TA-3′.
Reverse Transcription-PCR (RT-PCR) and Rapid Amplification of cDNA Ends (RACE).
RT-PCR and 5′ and 3′ RACE (29) were performed with cDNA (Marathon-Ready cDNA, CLONTECH) at a concentration of approximately 1 ng/50 μl of PCR mixture. Cycle parameters were denaturation, 94°C for 25 sec; primer annealing and extension, 68°C for 2 min for various numbers of cycles with a first denaturation time of 1 min, and a final extension time of 5 min. 5′ and 3′ RACE of the ΔLF mRNAs were conducted with primers LF 1 and LF 2, respectively. 3′ RACE of the LF mRNA was conducted with primer LF 8.
Genomic DNA Walking.
To clone genomic sequences immediately upstream from the 5′ end of the ΔLF mRNA, we used an improved PCR-based method for DNA walking in uncloned genomic DNA (30). For this purpose, we used two antisense primers (primers LF 3 and LF 4), which were designed from the 5′ end of the ΔLF mRNA. The resulting PCR products were cloned and partially sequenced.
Northern Blot Analysis.
Multiple tissue Northern blots and hybridization solutions were obtained from CLONTECH. Hybridization was conducted with DNA probe at 1× 106 cpm/ml for 4 h at 68°C. Filters were washed twice at 65°C in 0.5× standard saline citrate (SSC)/0.5% SDS for 30 min, followed by a high-stringency wash in 0.1× SSC/0.5% SDS at 65°C for 20 min.
Chromosome Mapping.
Mapping was performed by PCR analysis of a panel of DNAs obtained from human–mouse somatic cell hybrids (Bios, New Haven, CT). The concentration of template was 250 ng/50 μl of PCR mixture. Cycle parameters were denaturation, 94°C for 25 sec, primer annealing and extension, 68°C for 4 min for 32 cycles with a first denaturation time of 1 min and a final extension time of 10 min.
Isolation of Human P1 Artificial Chromosome (PAC) Clones.
A PCR-derived cDNA hybridization probe from the 5′ end of the ΔLF mRNA was used to isolate two PAC clones (31) each containing about 90–150 kb of human genomic DNA (Genome Systems, St. Louis).
Cloning and Sequencing of PCR Products.
PCR products were cloned into the plasmid vector PCR II (Invitrogen). Plasmid DNA was obtained from bacterial cultures with a commercially available kit (Qiagen, Chatsworth, CA). DNA sequencing was performed on purified double-stranded plasmid DNA with a commercially available kit (Sequenase Version 2.0 DNA Sequencing Kit, United States Biochemical). Because PCR was used in this study, cDNA sequences were confirmed by sequencing both strands of at least two cloned cDNA fragments.
Analysis of DNA Sequence Data.
cDNA sequences were compared against the GenBank database using blast (32). Promoter sequences were evaluated for transcription factor binding sites through the Transfac database.
RESULTS
Identification and Cloning of a Novel Form of Lactoferrin mRNA.
During an attempt to identify the second breast cancer susceptibility gene (BRCA2) by a new positional cloning method (33), we obtained several candidate cDNAs from normal breast tissue. Sequencing of the cDNAs revealed that they all were derived from different regions of the LF mRNA. A review of the literature indicated that the human LF gene is located on chromosome 3q21. The BRCA2 gene, however, maps to chromosome 13q12–13 (34) and recently has been cloned (35). It shares no homology to LF.
The previously cited reports indicate that LF has numerous functions and that there are biochemically distinct isoforms. This suggests that an alternative form of LF may exist on chromosome 13q12–13. Therefore, we performed several experiments directed toward discovering novel LF genes.
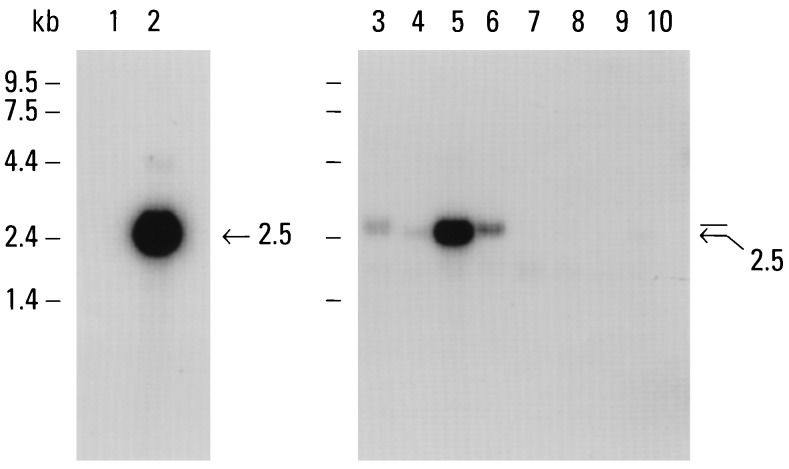
Careful examination of Northern blots using several partial LF cDNA probes revealed the consistent presence of a slightly larger mRNA in human spleen. An example of one of the Northern blots is shown in Fig. 1. We cloned the 5′ end of the larger LF-like mRNA by 5′ RACE using spleen cDNA as a template and primers designed from the LF mRNA. 5′ RACE revealed the presence of two different 5′-end lengths: a minor band of about 400 bp (the size predicted for LF mRNA) and a major band at about 100 bp larger. The sequence of the larger spleen 5′-RACE fragment was found to be nearly identical to LF throughout 90 amino acids of the N-terminal region of the mature protein. However, the 5′ end of the cDNA lacked the sequence encoding the 5′-untranslated region and signal peptide of LF (exon 1). Instead, we found an alternative 160-nt sequence shown in Fig. 2 (nt +1 to nt +160). The divergence in nucleotide sequence occurs exactly at the junction of exon 1 and exon 2 with respect to the mouse LF gene (36). A search of the available DNA sequence database failed to reveal any homologies to this alternative 160-nt sequence.
Figure 1.
Multiple-tissue Northern blot analysis. A partial LF mRNA cDNA probe was used on a Northern blot containing 2 μg of poly(A)+ RNA obtained from the following tissues: liver (lane 1), breast (lane 2), spleen (lane 3), thymus (lane 4), prostate (lane 5), testis (lane 6), ovary (lane 7), intestine (lane 8), colon (lane 9), and peripheral blood leukocytes (lane 10). The positions of RNA size markers are indicated.
Figure 2.
(A) Sequence of the ΔLF promoter region and unique 5′ end region of the ΔLF mRNA (exon 1β). In the promoter region, the putative TATA box is boxed. Consensus sequences for transcription factors and regulatory elements are underlined and noted. In exon 1β, the ATG, a stop codon, and the beginning of exon 2 of the LF gene are indicated. (B) Sequence of the exon/intron boundary sequences of the LF gene. (Upper sequence) Last 18 nts of exon 1β followed by the immediate genomic sequence. (Lower sequence) Genomic sequence immediately upstream from exon 2 of the LF gene followed by the beginning sequence of exon 2. The invariant dinucleotide splicing junctions are underlined.
Interestingly, an ATG codon was not found in the novel sequence that was in frame with the coding region of LF exon 2. The sequence did contain a ATG codon, which was out of frame at nucleotide 42 and was followed by stop codon (TGA) 15 codons later. This suggests that if this LF-like mRNA is translated, it will occur internally with respect to the LF mRNA. The use of the first available ATG codon in frame with the coding region of the LF mRNA predicts translation in exon 2 at amino acid 25 of the mature protein.
To compare the remaining nucleotide sequences of the ΔLF and LF mRNAs, we performed 3′ RACE with human lung cDNA. Lung cDNA was chosen instead of spleen, because it was obtained from a single human being. 3′ RACE of the LF mRNA was performed with an upstream primer designed from the beginning of the established LF mRNA sequence. 3′ RACE of the ΔLF mRNA was performed with an upstream primer representing the 5′ end of the novel ΔLF mRNA sequence. Except for the 5′ ends, the sequences of the ΔLF and LF mRNAs from lung were found to be identical. When compared with one of the published LF mRNAs obtained from breast (37), we found three differences. At amino acid 130 of the mature LF protein, an ATA (isoleucine) replaces ACA (threonine). At amino acid 404, a GGC (glycine) replaces TGC (cysteine). In the 3′ untranslated region (182 nt), we found an insertion of one nucleotide.
We designate this alternative LF-like mRNA and its putative translation product delta lactoferrin (ΔLF).
Sequence of the Promoter Region of the ΔLF mRNA.
We cloned genomic sequences immediately upstream of the 5′ end of the ΔLF mRNA by using a PCR-based method for genomic DNA walking (30). This method is analogous to RACE, but uses endonuclease-restricted human genomic DNA as a template. Three distinct PCR products were obtained from the same adaptor-ligated DNA and were cloned and partially sequenced. All three were found to have identical 3′-end sequences. We selected the largest DNA fragment (about 1 kb) for more detailed examination. A partial 363-nt sequence upstream from the 5′ end of the ΔLF mRNA is provided in Fig. 2A (nucleotide −1 to nucleotide −363).
Although the exact transcriptional start was not identified, a putative TATA box was found approximately 27 nt upstream from the 5′ end of the ΔLF mRNA. Several potential mammalian regulatory elements also were identified by comparison to a transcription factor binding sequence database. These included a consensus sequence for SP1 and the glucocorticoid receptor, an enhancer protein binding sequence found upstream from the NF-κ gene, a GA box found in the c-myc gene, as well as a NF-E2, CCAAT-binding protein (hsp 70) and IRF binding site. Other consensus sequences that were nearly perfect included c-ets-1, NF-IL6, NRF-2, LYF-1, and RVF.
Genomic Relationship Between the ΔLF and LF mRNAs.
We determined the chromosomal origin of the ΔLF mRNA by PCR analysis of genomic DNA obtained from human–mouse somatic cell hybrids. For this experiment, we used a primer designed from the 5′ end of the ΔLF mRNA and paired it with the same primer used for the earlier 5′ RACE experiment. The results are shown in Fig. 3A. Positive PCR signals (with a size of ≈ 7 kb) were obtained from the human control DNA and DNA obtained from the chromosome 3 hybrid DNA. Signals were absent from the mouse control DNA and human chromosome 1, 11, and 13 hybrid DNAs. This indicates that the ΔLF mRNA is encoded by a gene present on human chromosome 3.
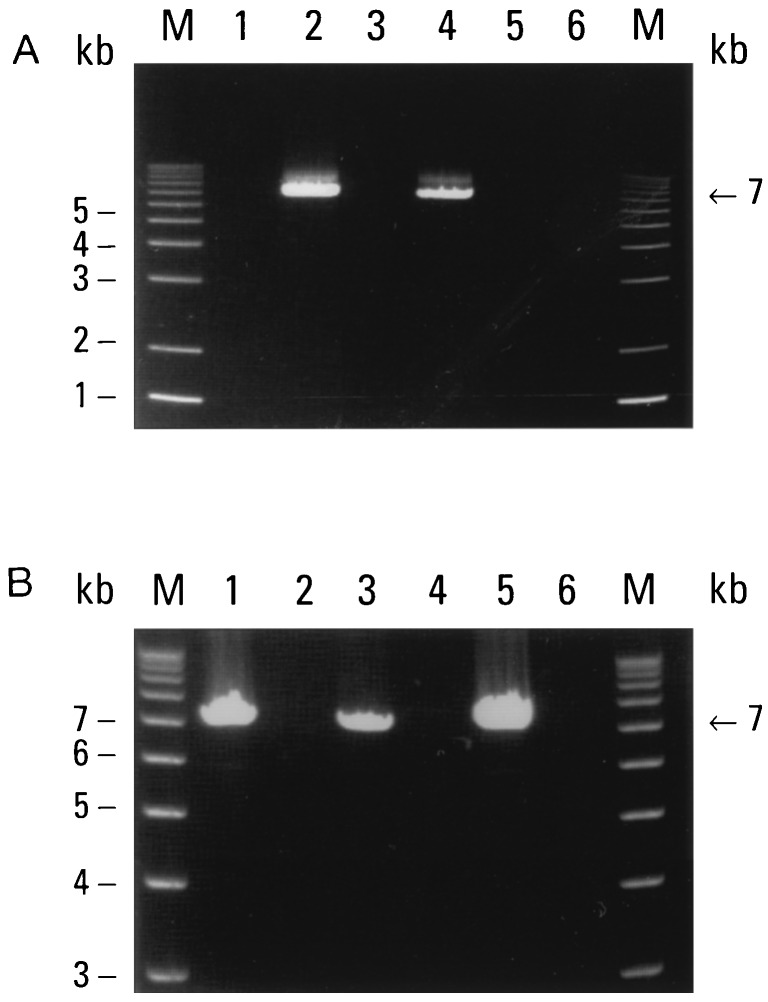
Figure 3.
(A) Chromosome mapping of the ΔLF mRNA. DNAs prepared for mouse, human, and human-mouse somatic cell hybrids were used as templates for PCR with primers designed to specifically amplify ΔLF mRNA sequences (primers 5 and 7). Mouse DNA (lane 1), human DNA (lane 2), human chromosome 1 hybrid (lane 3), human chromosome 3 hybrid (lane 4), human chromosome 11 hybrid (lane 5), and human chromosome 13 hybrid (lane 6). Lane M contained 1-kb DNA size markers. Thirty-two cycles of PCR were performed. PCR products were resolved on a 0.8% agarose/ethidium bromide gel. (B) Genomic relationship between ΔLF and LF mRNA sequences. Human DNA and DNA prepared from PAC clones were used as templates for PCR with two different primer combinations. Human DNA (lanes 1 and 2), PAC clone 1 DNA (lanes 3 and 4), and PAC clone 2 (lanes 5 and 6). Primer combinations were ΔLF 5′ end, sense (primer LF 5) and exon 3, antisense (primer LF 1) (lanes 1, 3, and 5); ΔLF 5′ end, antisense (primer LF 3) and exon 3, sense (primer LF 6) (lanes 2, 4, and 6). For human DNA, 32 cycles were conducted. For PAC clone DNA, 18 cycles were performed. PCR products were resolved on a 0.8% agarose/ethidium bromide gel.
To further study the genomic relationship between ΔLF and LF mRNAs, we designed two PCR primers from the 5′ end of the ΔLF mRNA, one sense and one antisense, and paired them with antisense and sense primers, respectively, which were designed from exon 3 of the LF gene. As a template, we used total human DNA. The results are shown in Fig. 3B, lanes 1 and 2. PCR products (about 7 kb) were obtained only when the sense ΔLF mRNA primer was paired with the antisense primer designed from exon 3. This places the 5′ end of the ΔLF mRNA about 7 kb upstream from exon 3 of the LF gene.
To further define the genomic relationship between the ΔLF and LF mRNAs, we isolated two human PAC clones, each containing approximately 90–150 kb of DNA, using a hybridization probe prepared from the unique 5′ end of the ΔLF mRNA. We then used DNA prepared from these PAC clones as templates for PCR with the same sets of primers used in Fig. 3B, lanes 1 and 2. The results are shown in Fig. 3B, lanes 3–6. The pattern of PCR products obtained from both PAC clones was the same as that obtained from whole human DNA.
We also determined the nucleotide sequence of a portion of the 7-Kb PAC clone PCR product corresponding to the region immediately downstream from the novel ΔLF mRNA sequence. This revealed the invariant dinucleotide donor splice sequence, gt. We also sequenced the region immediately upstream from exon 2 of the same PAC clone, which revealed the invariant dinucleotide splice acceptor sequence, ag. These results are presented in Fig. 2B.
Together these genomic studies indicate that sequences specific to the ΔLF and LF mRNAs are very closely linked and that these mRNAs could be derived from alternative splicing. We designate the novel ΔLF mRNA 5′ end sequence exon 1β.
Expression Pattern of ΔLF and LF mRNA in Normal Human Tissues.
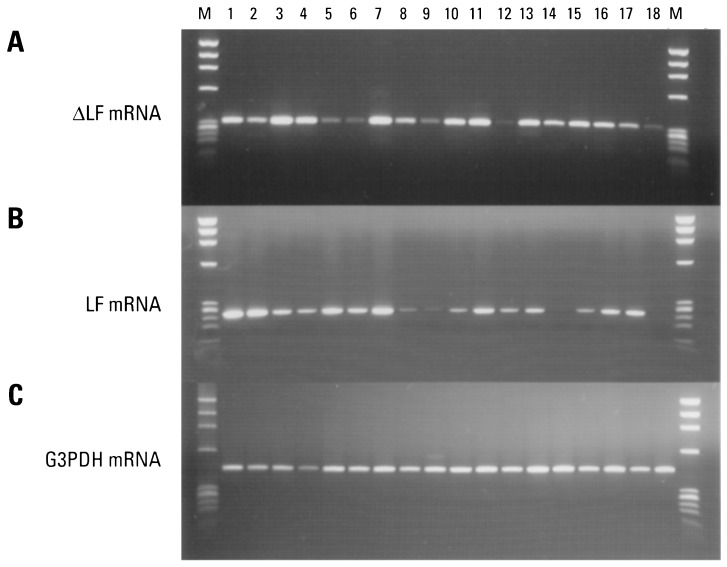
Using RT-PCR, we examined the expression pattern of LF and ΔLF mRNAs in 20 normal human tissues. The results are shown in Fig. 4. To examine ΔLF mRNA, we used an upstream primer designed from exon 1β of the ΔLF mRNA and a downstream primer in common with the LF and ΔLF mRNAs. The results are shown in Fig. 4A. ΔLF mRNA was highest in spleen, pancreas, kidney, and lung. ΔLF mRNA was only barely detectable in brain, testis, liver, leukocytes, and fetal brain. As a control, we amplified the housekeeping mRNA glyceraldehyde-3-phosphate dehydrogenase (Fig. 4C). Subsequently, we found relatively high levels of ΔLF mRNA in colon and pituitary gland (data not shown).
Figure 4.
Tissue expression pattern of ΔLF and LF mRNA in normal human tissues. cDNAs prepared from various normal human tissues were used as templates for PCR.(A) Pattern of ΔLF mRNA: 34 cycles of PCR with primers LF 5 and LF 7 were used, predicted size is 328 bp. (B) Pattern of LF mRNA: 30 cycles of PCR with primers LF 6 and LF 7 were used, predicted size is 254 bp. (C) Housekeeping gene glyceraldehyde-3-phosphate dehydrogenase control: 22 cycles of PCR were used, predicted size is 452 bp. Breast (lane 1), prostate (lane 2), spleen (lane 3), pancreas (lane 4), adult brain (lane 5), testis (lane 6), kidney (lane 7), placenta (lane 8), liver (lane 9), small intestine (lane 10), lung (lane 11), peripheral blood leukocytes (lane 12), skeletal muscle (lane 13), ovary (lane 14), uterus (lane 15), thymus (lane 16), fetal liver (lane 17), and fetal brain (lane 18). DNA size markers (φX174 digested with HaeIII) are shown in lane M.
For comparison, we also examined LF mRNA (Fig. 4B). In this case, we used an upstream primer designed from LF exon 1 and the same downstream primer used in Fig. 4A. LF mRNA was highest in breast, prostate, and kidney. LF mRNA was only barely detectable in placenta, liver, ovary, and fetal brain.
Expression Pattern of ΔLF and LF mRNA in Tumor-Derived Cell Lines.
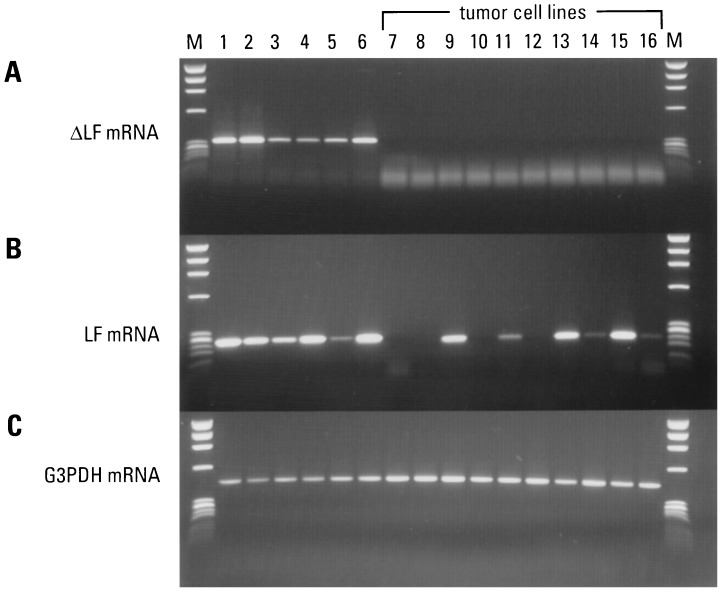
We also examined the expression pattern of ΔLF mRNA in 10 human tumor-derived cell lines by RT-PCR. The results are shown in Fig. 5. Five normal tissues were included as positive controls and to extend the data shown in Fig. 4. In this experiment, we performed PCR on dilutions of the spleen and kidney cDNA, which allowed us to extend the number of PCR cycles without overcycling these samples (36 cycles total). Now, clearly detectable levels of ΔLF mRNA were found in testis and brain. The housekeeping mRNA control is shown in Fig. 5C. We detected no expression of ΔLF mRNA in any of the 10 tumor-derived cell lines we examined, even after continuing to amplify these samples an additional 6 cycles (42 cycles as shown).
Figure 5.
Tissue expression pattern of ΔLF and LF mRNA in normal compared with tumor-derived cell lines. cDNAs prepared from both normal and tumor cells were used as templates for PCR with the same primer combinations used in Fig. 3. (A) Pattern of ΔLF mRNA in 6 normal tissues (36 cycles of PCR). The spleen and kidney cDNAs were diluted 20× and 10×, respectively. The 10 tumor-derived cells are indicated (42 cycles of PCR). (B) Pattern of LF mRNA in 6 normal tissues (33 cycles of PCR) and 10 tumor-derived cells (42 cycles of PCR). (C) Housekeeping gene control (22 cycles of PCR). Normal tissues: breast (lane 1), spleen (lane 2), placenta (lane 3), testis (lane 4), brain (lane 5), and kidney (lane 6). Tumor cell lines: HeLa cell S3 (lane 7), melanoma A2058 (lane 8), promyelocytic leukemia HL-60 (lane 9), T lymphoblastic leukemia MOLT-4 (lane 10), T lymphoblastic leukemia Jurkat (lane 11), lung tumor A549 (lane 12), breast tumor T41D (lane 13), lymphoma Burkitt’s Raji (lane 14), colorectal adenocarcinoma SW490 (lane 15), and erythroleukemia K562 (lane 16). DNA size markers (φX174 digested with HaeIII) are shown in lane M.
For comparison, we examined the expression pattern of LF mRNA in the same 10 tumor cell lines. Low levels of LF mRNA (requiring 42 cycles) were found in the promyelogeneous leukemia cell line HL-60, the breast cancer cell line T410, and the colorectal adenocarcinoma cell line SW490. A faint signal was found in the T lymphoblastic Jurkat cell line.
We also examined ΔLF and LF mRNA in four additional tumor-derived cell lines using whole cDNA libraries as PCR templates. These included the breast cancer cell lines SKBR-3 and ZR-75–1, the fibrosarcoma cell line HT-1080, and the osteosarcoma cell line MG-63. Again, no ΔLF mRNA was detected. LF mRNA, however, was found in the two additional breast cancer cell lines and, to a lesser extent, in the fibrosarcoma cell line (data not shown).
DISCUSSION
In the present study, we identified and cloned a novel form of LF mRNA, designated ΔLF. Except for the 5′ end, the ΔLF mRNA is almost identifical to the LF mRNA obtained from human mammary gland (37). At the 5′ end, however, we find a novel sequence (exon 1β) that replaces the signal peptide of LF (exon 1). The sequence predicts that if the ΔLF mRNA is translated, translation initiation likely would occur at an internal methionine with respect to the LF mRNA. If this is true, the protein encoded by the ΔLF mRNA will lack the LF signal peptide as well as amino acids from the mature protein. The use of the first available internal ATG, which is in frame with the coding sequence of LF, would result in a LF-like protein lacking 25 amino acids from the N terminus of the mature protein. Although the nucleotide sequence around this codon does not strictly adhere to the Kozak consensus sequence for translation initiation (38), the most highly conserved position in the motif is present (a purine at position −3, which is usually A). For comparison, an imperfect Kozak sequence is also present in the established translation initiation site of the LF mRNA. The use of internal methionines further downstream would predict drastically truncated proteins lacking more than half the normal number of amino acids.
Use of alternative translation initiation sites (in the same mRNA) is not uncommon. Examples include the transcription factors c-myc (39), Pit 1 (40), GATA-1 (41), and the prostate protein probasin (42). In the case of probasin, the use of alternative translation initiating sites accounts for the secreted and nuclear forms of the protein with the nuclear form lacking the N-terminal signal peptide (42). Interestingly, the amino acid sequence just following the first methionine of the ΔLF mRNA contains a cluster of basic amino acids, which is similar to nuclear targeting signals (43, 44). This is intriguing in light of the fact that LF can be taken up by erythroleukemic K562 cells, where it localizes to the nucleus (45). It is tempting to speculate, by analogy to probasin, that the use of alternative initiating methionines in the LF and ΔLF mRNA also may determine whether the proteins they encode are secreted from the cell or localized to the nucleus.
Except for the 5′ ends of the LF and ΔLF mRNAs, the two sequences were identical. When our lung LF mRNA sequence is compared with one of the published LF mRNA sequences (from breast), we found three nucleotide differences. Two were found in the coding region that would change amino acids. Threonine replaced isoleucine and glycine replaced cysteine. A single nucleotide change was found in the 3′ untranslated region. It is likely that these few differences are due to polymorphisms, which are common to LF mRNAs (19).
We also cloned and characterized genomic sequences immediately upstream of the 5′ end of the ΔLF mRNA. A TATA box-like sequence was found 27 nt upstream from the 5′ end of the ΔLF mRNA. This distance is typical for TATA boxes (20–30 nt) (46). Besides a TATA box, there were a number of potential regulatory elements, including putative binding sites for the glucocorticoid receptor and the erythroid nuclear factor NF-E2. In addition, we found a GA box element present in the c-myc gene, an IRF binding sequence found in the IFN-β gene, an NF-E2 binding sequence, and an enhancer binding protein sequence near the NF-κ gene.
When the promoter regions of ΔLF (presented here) and LF (36, 47) are compared, both similarities and differences are found. For example, there are a glucocorticoid receptor and SP1 consensus sequence and GA box in the LF promoter. In contrast, putative binding sites for the estrogen receptor, AP-2, and GATA-1 and a retinoic acid response element are found in the LF promoter, but not in the ΔLF promoter.
It remains to be determined by DNA footprinting whether any of these transcription factor binding sites are actually used. Nonetheless, these findings suggest that there may be mechanisms that can both coordinately regulate and differentially regulate transcription from the ΔLF and LF promoters.
The chromosome assignment and genomic relationship of the ΔLF and LF mRNAs also were examined. The ΔLF mRNA was mapped to human chromosome 3; the LF gene was previously assigned to chromosome 3q21 (48). We also found not only that sequences specific to both the ΔLF and LF mRNAs are present on the same cloned 90- to 50-kb PAC genomic DNA fragments and thus are closely linked but also that the ΔLF 5′-end sequence (exon 1β) and LF exon 3 are within 7 kb. These results map the ΔLF mRNA close to or within the LF gene on chromosome 3q21. Further studies (not shown) indicate that at least one additional LF-like gene is also present on both PAC clones.
Our genomic DNA results, together with the findings that the nucleotide sequences of the ΔLF and LF mRNAs are identical (except for their 5′ ends) and that the divergence in sequence occurs exactly at the junction of exon 1 and exon 2 of the LF gene) indicate that the ΔLF mRNA is the product of alternative splicing of the LF gene. Further, our finding of a promoter-like sequence (containing a TATA box) immediately upstream for the 5′ end of the ΔLF mRNA suggests that transcription could initiate from two different promoters within the same LF gene and, as a result, specify alternative first exons. This is the situation that occurs with the human NF-E2 gene (49). In this case, use of alternative promoters specifies the major fetal and adult forms of the NF-E2 mRNA. Another example of the use of alternative promoters in the same gene specifying alternative first exons is the p16 tumor-suppressor gene (50, 51).
We used RT-PCR to examine the expression pattern of both ΔLF and LF mRNA in normal tissues and tumor-derived cell lines. In normal tissues we found both ΔLF and LF mRNA at various levels (albeit very low in some cases) in 20 of 20 adult and fetal tissues. However, the relative levels of the ΔLF and LF mRNA appear to differ in many of the tissues.
Examination of ΔLF mRNA in 12 diverse tumor-derived cell lines revealed the complete absence of this mRNA. This was even true in the breast, lung, and colon tumor-derived cell lines, whose normal tissue counterparts express relatively high levels of ΔLF mRNA. LF mRNA also was examined in the same tumor cell lines. LF mRNA was found to be expressed at low levels in the promyelocytic leukemic cell line and in the breast and colon tumor cell lines. Although our RT-PCR experiments were not quantitative, the absence of a signal for ΔLF in the tumor cell lines, even after excessive cycles of PCR, provides good evidence for an absence of ΔLF mRNA in these cells.
There are a multitude of effects associated with LF, ranging from iron absorption and storage, regulation of cell growth, and the host defense against infection. Further, LF recently has been implicated as playing a role in the host defense against tumorigenesis. It is likely that some of these effects are due to nuclear activities. For example, LF has been shown to alter the transcriptional activity of transfected reporter genes (52) and to down-regulate the activity of the human granulocyte macrophage colony-stimulating factor promoter (53).
It is difficult to imagine how a single LF polypeptide could account for all of these effects. The finding of an alternative form of LF mRNA suggests that some of the effects may be due to protein encoded by this mRNA. The findings that ΔLF mRNA is present in every normal tissue we examined but is undetectable in all tumor-derived cell lines suggests that ΔLF may play an important role in the process or regulation of normal cell growth.
Acknowledgments
We thank Drs. Alex Chenchik and Luda Diatchenko for helpful discussions, Jennifer Fishel for secretarial assistance, Theresa Provost for preparation of the figures, and Stephanie Trelogan for critical review of the manuscript. This work was supported by Small Business Innovation Research Grant MH52941–03 to P.D.S. from the National Institute of Mental Health.
ABBREVIATIONS
- LF
lactoferrin
- ΔLF
delta lactoferrin
- RT-PCR
reverse transcription PCR
- RACE
rapid amplification of cDNA ends
- PAC
P1 artificial chromosome
Footnotes
References
- 1.Aisen P, Leibman A. Biochim Biophys Acta. 1972;257:314–323. doi: 10.1016/0005-2795(72)90283-8. [DOI] [PubMed] [Google Scholar]
- 2.Aisen P, Listowsky I. Annu Rev Biochem. 1980;49:357–393. doi: 10.1146/annurev.bi.49.070180.002041. [DOI] [PubMed] [Google Scholar]
- 3.Arnold R R, Cole M F, McGhee J R. Science. 1977;197:263–265. doi: 10.1126/science.327545. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini M, Duve C D, Masson P L, Heremans J F. J Exp Med. 1970;131:559–570. doi: 10.1084/jem.131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shau H, Kim A, Golub H. J Leukocyte Biol. 1992;15:343–349. [PubMed] [Google Scholar]
- 6.Horwitz D A, Bakke A C, Abo W, Nishiya K. J Immunol. 1984;132:2370–2374. [PubMed] [Google Scholar]
- 7.Sawatzki G, Rich I N. Blood Cells. 1989;15:371–385. [PubMed] [Google Scholar]
- 8.Gahr M, Speer C P, Damerau B, Sawatzki G. J Leukocyte Biol. 1991;49:427–433. doi: 10.1002/jlb.49.5.427. [DOI] [PubMed] [Google Scholar]
- 9.Broxmeyer H E, Gentile P. Blood. 1983;61:982–993. [PubMed] [Google Scholar]
- 10.DeSousa M, Breedvelt F, Dynesius-Trentham R, Trentham D, Lum J. Ann NY Acad Sci. 1988;526:310–322. doi: 10.1111/j.1749-6632.1988.tb55515.x. [DOI] [PubMed] [Google Scholar]
- 11.Zimecki M, Mazurier J, Spik G, Kapp J A. Immunology. 1995;86:122–127. [PMC free article] [PubMed] [Google Scholar]
- 12.Mori M, Ninomiya T, Okada Y, Tsukitami K. Pathol Res Pract. 1989;184:168–178. [PubMed] [Google Scholar]
- 13.Tuccari G, Barresi G, Arena F, Inferrera C. Arch Pathol Lab Med. 1989;113:912–915. [PubMed] [Google Scholar]
- 14.Davey F R, Erber W N, Gatter K C, Mason D Y. Am J Hematol. 1987;26:157–166. doi: 10.1002/ajh.2830260206. [DOI] [PubMed] [Google Scholar]
- 15.Campbell T, Skilton R, Lugmani Y A, Coombes R C. Proc Amer Assoc Cancer Res. 1990;31:209. [Google Scholar]
- 16.Walmer D K, Padin C J, Wrona M A, Healy B E, Bentley R C, Tsao M-S, Kohler M F, McLachlan J A, Gray K D. Cancer Res. 1995;55:1168–1175. [PubMed] [Google Scholar]
- 17.Campbell T, Skilton R A, Coombes R C, Shousha S, Graham M D, Luqmani Y A. Br J Cancer. 1992;65:19–26. doi: 10.1038/bjc.1992.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchens T W, Henry J F, Yip T-T, Hachey D L, Schanler R J, Motil K J, Garza C. Pediatr Res. 1991;29:243–250. doi: 10.1203/00006450-199103000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Panella T J, Liu Y, Huang A T, Teng C T. Cancer Res. 1991;51:3037–3043. [PubMed] [Google Scholar]
- 20.Hashizume S, Kuroda K, Murakami H. Biochim Biophys Acta. 1983;763:377–382. doi: 10.1016/0167-4889(83)90099-x. [DOI] [PubMed] [Google Scholar]
- 21.Nichols B L, McKee K S, Henry J F, Putman M. Pediatr Res. 1987;21:563–567. doi: 10.1203/00006450-198706000-00011. [DOI] [PubMed] [Google Scholar]
- 22.Azuma N, Mori H, Kaminogawa S, Yamauchi K. Agric Biol Chem. 1989;53:31–35. [Google Scholar]
- 23.Hagiwara T, Shinoda I, Fukuwatari Y, Shimamura S. Biosci Biotechnol Biochem. 1995;59:1875–1881. doi: 10.1271/bbb.59.1875. [DOI] [PubMed] [Google Scholar]
- 24.Rejman J J, Oliver S P, Muenchen R A, Turner J D. Cell Biol Int Rep. 1992;16:993–1001. doi: 10.1016/s0309-1651(06)80052-4. [DOI] [PubMed] [Google Scholar]
- 25.Bezault J, Bhimani R, Wiprovnick J, Furmanski P. Cancer Res. 1994;54:2310–2312. [PubMed] [Google Scholar]
- 26.Furmanski P, Li Z-P, Fortuna M B, Swamy C V B, Das M R. J Exp Med. 1989;170:415–429. doi: 10.1084/jem.170.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnes W M. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng S, Fockler C, Barnes W M, Higuchi R. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chenchik A, Diatchenko L, Moqadam F, Tarabykin V, Lukyanov S, Siebert P D. BioTechniques. 1996;21:526–535. doi: 10.2144/96213pf02. [DOI] [PubMed] [Google Scholar]
- 30.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannou P A, Amemiya C T, Garnes J, Kroisel P M, Shizuya H, Chen C, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 32.Altschul S F, Gish W, Miller W, Myers W W, Lipman D J. J Mol Biol. 1990;215:404–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Chen-Liu L W, Huang B C, Scalzi J M, Hall B K, Sims K R, Davis L M, Siebert P D, Hozier J C. Genomics. 1995;30:388–392. doi: 10.1006/geno.1995.0038. [DOI] [PubMed] [Google Scholar]
- 34.Wooster R, Neuhausen S L, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 35.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, et al. Nature (London) 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 36.Shirsat N V, Bittenbender S, Krider B L, Rovera G. Gene. 1992;110:299–233. doi: 10.1016/0378-1119(92)90653-7. [DOI] [PubMed] [Google Scholar]
- 37.Rey M W, Woloshuk S L, deBoer H A, Pieper F R. Nucleic Acids Res. 1990;18:5288. doi: 10.1093/nar/18.17.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. Nucleic Acids Res. 1987;15:8125–8132. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hann S T, Dixit M, Sears R C, Sealy L. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 40.Voss J W, Yao T P, Rosenfeld M G. J Biol Chem. 1991;266:12832–12835. [PubMed] [Google Scholar]
- 41.Calligaris R, Bottard S, Cogoi S, Apezteguia I, Santoro C. Proc Natl Acad Sci USA. 1995;92:11598–11602. doi: 10.1073/pnas.92.25.11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spence A M, Sheppard P C, Davie J R, Matuo Y, Nishi N, McKeehan W L, Dodd J G, Matusik R J. Proc Natl Acad Sci USA. 1989;86:7843–7847. doi: 10.1073/pnas.86.20.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 44.Osborne M A, Silver P A. Annu Rev Biochem. 1993;62:219–254. doi: 10.1146/annurev.bi.62.070193.001251. [DOI] [PubMed] [Google Scholar]
- 45.Garre C, Bianchi-Scarra G, Sirito M, Musso M, Ravazzolo R. J Cell Physiol. 1992;153:477–482. doi: 10.1002/jcp.1041530306. [DOI] [PubMed] [Google Scholar]
- 46.Sawadogo M, Sentenac A. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- 47.Lee M-O, Liu Y, Zhang X-K. Mol Cell Biol. 1995;15:4194–4207. doi: 10.1128/mcb.15.8.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng C T, Pentecost B T, Marshall A, Solomon A, Bowman B H, Lalley P A, Naylor S L. Somatic Cell Mol Genet. 1987;13:689–693. doi: 10.1007/BF01534490. [DOI] [PubMed] [Google Scholar]
- 49.Pischedda C, Cocco S, Melis A, Marini M G, Kan Y W, Cao A, Moi P. Proc Natl Acad Sci USA. 1995;92:3511–3515. doi: 10.1073/pnas.92.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stone S, Jiang P, Dayananth P, Tavtuguan S V, Katcher H, Parry D, Peters G, Kamb A. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- 51.Mao L, Merlo A, Bedi G, Shapiro G I, Edwards C D, Rollins B J, Sidransky D. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 52.He J, Furmanski P. Nature (London) 1995;373:721–724. doi: 10.1038/373721a0. [DOI] [PubMed] [Google Scholar]
- 53.Penco S, Pastorino S, Bianchi-Scarra G, Garre C. J Biol Chem. 1995;270:12263–12268. doi: 10.1074/jbc.270.20.12263. [DOI] [PubMed] [Google Scholar]