Abstract
We have investigated the effects of DNA damage by (±)-anti-benzo[a]pyrene diol epoxide (BPDE) and UV light on the formation of a positioned nucleosome in the Xenopus borealis 5S rRNA gene. Gel-shift analysis of the reconstituted products indicates that BPDE damage facilitates the formation of a nucleosome onto this sequence. Competitive reconstitution experiments show that average levels of 0.5, 0.9, and 2.1 BPDE adducts/146 bp of 5S DNA (i.e., the size of DNA associated with a nucleosome core particle) yield changes of −220, −290, and −540 cal/mol, respectively, in the free energy (ΔG) of nucleosome formation. These values yield increases of core histone binding to 5S DNA (Ka) of 1.4-, 1.6-, and 2.5-fold, compared with undamaged DNA. Conversely, irradiation with UV light decreases nucleosome formation. Irradiation at either 500 or 2500 J/m2 of UV light [0.6 and 0.8 cyclobutane pyrimidine dimer/146 bp (on average), respectively] results in respective changes of +130 and +250 cal/mol. This translates to decreases in core histone binding to irradiated 5S DNA (Ka) of 1.2- and 1.5-fold compared with undamaged DNA. These results indicate that nucleosome stability can be markedly affected by the formation of certain DNA lesions. Such changes could have major effects on the kinetics of DNA processing events.
In eukaryotic cells, DNA processing (e.g., transcription, repair, and replication) occurs in DNA packaged in chromatin. The primary function of chromatin is to organize nuclear DNA in a manner that is compact yet accessible to cellular “machinery.” The fundamental unit of chromatin structure (the nucleosome) is composed of an octamer of four pairs of core histone proteins, a linker histone, and ≈180–200 bp of DNA. This may be subdivided into a “core particle” of 146 bp of DNA that is tightly associated with the histone octamer, and 35–50 bp of “linker DNA” that connects the neighboring cores. In addition to the general packaging of DNA, nucleosomes are involved in both general and specific gene repression and regulation (reviewed in refs. 1 and 2). Positioned nucleosomes are frequently involved in the specific repression of gene activation (3, 4) and, in at least one system, facilitation of gene expression (5). It is believed that nucleosomes are able to repress genes by blocking the access of trans-acting factors to their binding sites in gene promoter regions (reviewed in ref. 2). Agents that alter the binding (or positioning) of these critical nucleosomes could affect the ability of nucleosomes to repress or facilitate gene expression.
In addition to packaging DNA, nucleosomes modulate or protect DNA from damage (refs. 6 and 7). In the case of bulky chemical carcinogens, such as benzo[a]pyrene diol epoxide (BPDE) (Fig. 1A), it has been shown that the presence of a nucleosome suppresses the damage levels within the central area of the nucleosome by up to 60% (8–10). Much of this suppression is the result of differential reaction kinetics and can be overcome at long incubation times (8). The binding or damage by small alkylating agents, however, such as dimethyl sulfate, may not be inhibited by the presence of a nucleosome (10, 11).
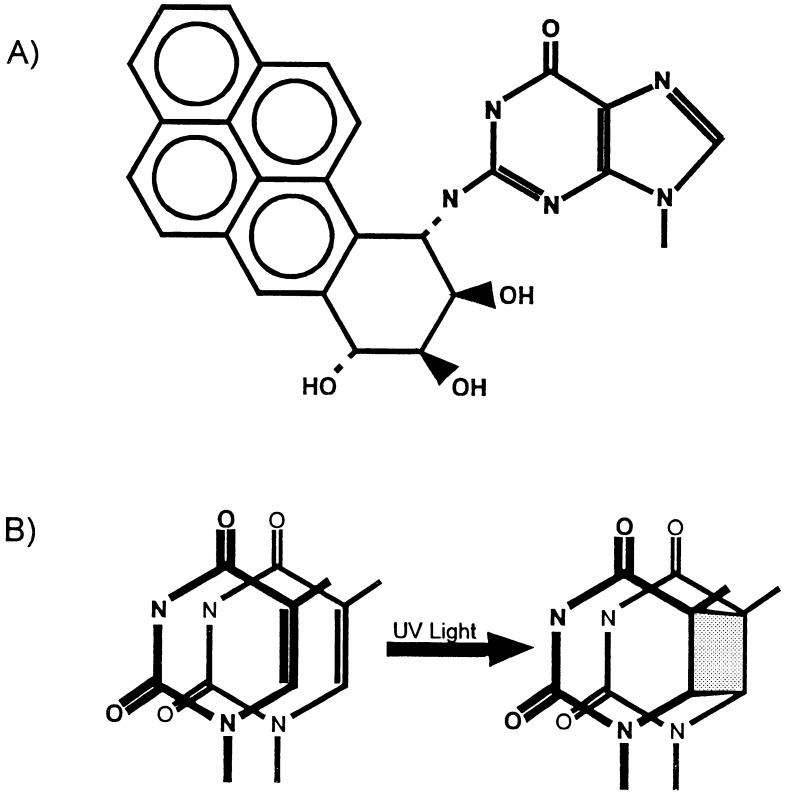
Figure 1.
Major products of benzo[a]pyrene diol epoxide and UV damage. (A) (+)-7R,8S-dihydroxy-9S,10R-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene bound to the N-2 position of guanine. (B) Cyclobutane pyrimidine dimer.
UV damage is also modulated by the presence of nucleosomes. Our laboratory has demonstrated that the most common UV lesions, cyclobutane pyrimidine dimers (CPDs; Fig. 1B), form preferentially where the DNA backbone is furthest from the histone surface in mixed sequence nucleosomes irradiated in intact cells, nuclei, or isolated nucleosome core particles (12). The second most common stable UV lesion, the 6–4 pyrimidine–pyrimidone dimer, severely distorts DNA (13–15) and is found preferentially in nuclease-sensitive DNA, such as linker regions between nucleosome core particles (16–18).
The effect of DNA damage on nucleosome formation and structure is important, both in terms of how damage is recognized and repaired, as well as how damage may alter protein–DNA interactions prior to repair. As chromatin structure of the control regions of many genes is altered upon gene activation (2, 19), damage that disrupts nucleosomes or enhances nucleosome stability may affect gene regulation (as proposed in ref. 20). Studies with specific antibiotics (21) or ethidium bromide (22) have shown that at damage levels of several molecules per nucleosome these agents strongly disrupt nucleosome structure. UV damage also alters the interactions of DNA and histones upon nucleosome formation. When nucleosomes are reconstituted onto UV-damaged mixed sequence DNA, they favor a rotational setting that places the lesions away from the histone surface (23). These nucleosomes also tend to adopt a translational setting that excludes lesions from the central three helical turns of DNA at the nucleosome dyad axis (23). Recently, in a defined-sequence nucleosome containing several long T-tracts, it was found that the modulation by nucleosome structure of CPD formation in some of the T-tracts differs from that predicted by mixed sequence nucleosomes (24). Furthermore, there was no effect of CPDs on the rotational setting of DNA irradiated after nucleosome formation (24). Thus, at least some nucleosomes may be able to accommodate the structural alterations in DNA at CPD sites.
Several laboratories have reported an enhancement of protein binding to DNA following DNA damage. Essigmann and coworkers (25) found that cisplatin-adducted random sequence DNA binds human upstream binding factor with nearly the same affinity as its native binding site. More recently, MacLeod et al. (26) observed a similar effect by the carcinogen BPDE on transcription factor Sp1. These authors reported that when a fragment containing the Sp1 binding site was modified by BPDE, the affinity of the protein for the binding site increased 5- to 10-fold (26). Modified DNA fragments, which did not contain the Sp1 recognition sequence, bound Sp1 with about the same affinity as the unmodified Sp1 site. However, in a more recent study where the Sp1 GC box was specifically modified with BPDE, Sp1 binding was reduced or eliminated (27). This indicates that the protein recognizes an altered DNA structure rather than the adduct itself. In both of these cases, it appears that these agents either alter the DNA structure in a manner that facilitates binding to the protein’s binding site, or they introduce a “hinge” (or point of flexibility), which allows these proteins to distort the DNA helix more easily (28).
We have investigated the effects of DNA damage by BPDE and UV light on the formation of a positioned nucleosome in the Xenopus borealis 5S rRNA gene. Analysis of nucleosome formation by competitive reconstitution showed that this formation is dramatically enhanced by BPDE damage in a dose-dependent manner (i.e., 1.4-, 1.6-, and 2.5-fold enhancement in histone binding at BPDE adduct levels of 0.5, 0.9, and 2.1/146 bp, respectively). On the other hand, UV irradiation was found to decrease nucleosome formation, with damage levels of 0.6 and 0.8 CPDs/146 bp resulting in 1.2- and 1.5-fold decreases in histone binding, respectively.
MATERIALS AND METHODS
Plasmid Construction.
Plasmid pBS-5S was constructed by PCR amplification of the segment of pXP-14 (29) containing the X. borealis 5S rRNA gene (i.e., from position −159 to +131 with respect to the start site of the 5S gene; Fig. 2A). Primers used were TGT TCT CGA GTC GTT AGA ACG CGG CTA CAA on the 5′ end and TTC GAA GAA TTC CAA AAG TGC AAA AGC CTA CG on the 3′ end of the amplified area. The underlined sections of each primer are sequences that are not complementary to the pXP-14 DNA and contain the new restriction sites, XhoI and EcoRI, respectively, denoted in bold type. This fragment was cut with both enzymes and ligated into the 2695-bp HindIII/EcoRI fragment of plasmid pBS (Stratagene) along with a 15-bp HindIII/XhoI linker in a three-way forced cloning. The products of this reaction were transfected into Escherichia coli strain JM109 and isolated using a standard alkaline lysis protocol (33). Plasmid pKS-5S was constructed by ligating the 296-bp XhoI/EcoRI fragment of pBS-5S into the 2928-bp XhoI/EcoRI fragment of pBluescript II KS(+) phagemid (Stratagene).
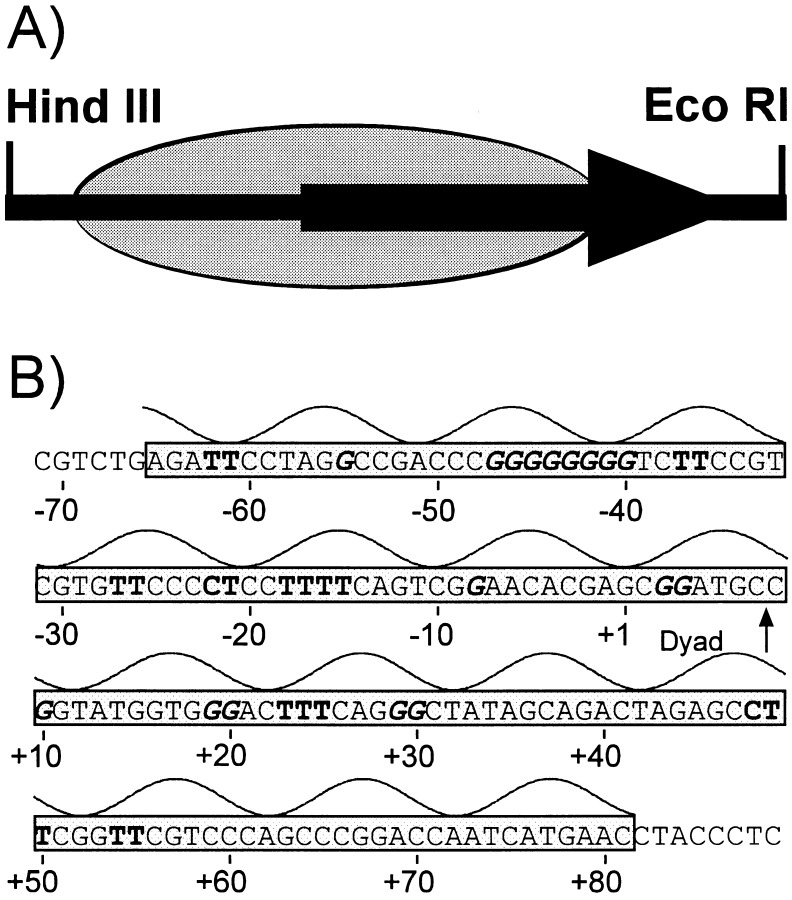
Figure 2.
Schematic diagram and sequence of 5S rRNA gene fragment used in this study. (A) Schematic of 214-bp HindIII/EcoRI fragment of pKS-5S. Large arrow indicates the location of the 120-bp 5S gene. The shaded oval is the predominant binding site of the positioned nucleosome as determined by Hayes and Wolffe (30). (B) Sequence of the transcribed strand of the nucleosomal region of the 5S fragment and its predicted orientation with respect to the histone surface. The curve above the sequence gives the rotational orientation of DNA with respect to the histone surface, with the top of the curve denoting bases positioned away from the histone surface as determined by Hayes and Wolffe (31). The gray area is the predominant binding site of the positioned nucleosome (30). Numbers refer to the transcription start site of the 5S rRNA gene. Boldfaced Cs and Ts are pyrimidines damaged at >3% of the total UV damage; boldfaced, italic Gs are guanines containing >2% of the total BPDE damage for the transcribed strand as determined by T4 DNA polymerase–exonuclease mapping (12, 32).
BPDE Adduction.
Plasmid DNA or restriction fragments at concentrations of 0.5 mg/ml were incubated with 0–250 μM of 3H-(±)-anti-7R,8S-dihydroxy-9S,10R-epoxy-7,8,9,10-tetrahydrobenzopyrene (BPDE; obtained from the National Cancer Institute Chemical Carcinogen Repository operated by Chemsyn Science Laboratories, Lenexa, KS) for 2 hr in 10 mM Tris·HCl (pH 7.1) at 4°C. Unreacted BPDE and its byproducts were removed from the DNA by 10 extractions with an equal volume of water-saturated diethyl ether, 2 extractions with phenol:chloroform:isoamyl alcohol (25:24:1), and 1 extraction with chloroform:isoamyl alcohol (24:1). The samples were precipitated twice with ethanol (0.1 volume 3 M sodium acetate, 2 volumes ethanol) to complete the cleanup. The concentration of recovered DNA was determined by UV spectroscopy (33). Adduction levels were determined by liquid scintillation counting in biodegradable counting scintillant (Amersham) and measured in units of base pair per adduct or adducts per nucleosome core size DNA (146 bp). Locations and relative levels of BPDE adducts were determined by T4 DNA polymerase–exonuclease digestion (Fig. 2B; ref. 32).
Ultraviolet Damage.
Restriction fragments or plasmid DNA were irradiated with 500 J/m2 or 2500 J/m2 of predominately 254-nm UV light at a flux of ≈11 W/m2 [measured with a Spectronic model DM 254N UV meter (Spectronic, Westbury, NY)]. Samples were exposed at a DNA concentration of 50 μg/ml in a volume of 100 μl. CPD levels were determined by the method of Bohr et al. (34). The intensities and locations of UV photoproducts were determined by T4 DNA polymerase–exonuclease digestion (Fig. 2B; ref. 12).
Chicken Erythrocyte (CE) Core Particle Isolation.
Core particles used in the reconstitutions were prepared from CEs by the method of Libertini et al. (35).
Nucleosome Reconstitution.
Reconstitutions were carried out by a modification of the method of Moyer et al. (10). Briefly, 80–100 ng of end-labeled 5S DNA (214-bp HindIII/EcoRI fragment; Fig. 2A) was mixed with CE core particles at molar ratios of 1:200, 1:10, or 1:5 (free DNA:core particles) in 1 M NaCl, 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride (PMSF) for 30 min at 4°C. The samples were dialyzed against 600 mM NaCl, 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, and 0.2 mM PMSF at 4°C for 4–6 hr. A second overnight dialysis step against 50 mM NaCl, 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, and 0.2 mM PMSF completed the reconstitution. Samples were run on nondenaturing polyacrylamide (nucleoprotein) gels, as described below, to check the fidelity of the reconstitutions.
Nucleoprotein Gels.
For characterization of reconstitutes, samples were run on a 5.5% polyacrylamide gel in 0.5× or 1× TBE (1 × TBE: 89 mM Tris/89 mM borate/2 mM EDTA, pH 8.3). Preelectrophoresis at 10 mA until the voltage no longer changed with time (≈30 min) was run before loading the samples on the gel. Electrophoresis was carried out for 4.5–7 hr at 150–200 V for a 20-cm gel length.
Competitive Reconstitution.
Competitive reconstitutions were carried out by two methods. In the method of Schild et al. (ref. 5; see also refs. 36 and 37), radiolabeled 5S DNA (≈10 ng) was mixed with 1 μg of CE core particles (based on DNA concentration) and various amounts of nonspecific competitor DNA (deproteinized CE DNA, ranging from 0.5 to 8 μg) in 1 M NaCl, 10 mM Tris·HCl (pH 8.0), 0.1% Nonidet P-40, 0.1% BSA, and 0.2 mM PMSF in a 10-μl reaction volume. The samples were incubated for 30 min at 37°C to allow the core histones to exchange. Salt concentrations were decreased to 100 mM by three additions of 30 μl of TE buffer (10 mM Tris·HCl, pH 8.0/1 mM EDTA) containing 0.2 mM PMSF, 30 min apart at room temperature. Aliquots of the reconstitutes were assayed for radioactivity and equal numbers of disintegrations per minute loaded onto nucleoprotein gels as described above.
In the second method, equal amounts (≈10 ng) of radiolabeled, damaged 224-bp HindIII/SmaI fragment were combined with radiolabeled, undamaged 214-bp HindIII/EcoRI fragment. The samples were reconstituted as described above at a 1:1 ratio of CE core particles:CE DNA (1 μg each) and separated on a 1% agarose, 1× TBE gel. The bands containing the free DNA and reconstituted fractions of each sample were recovered and run on denaturing and nondenaturing acrylamide gels as described above.
Calculation of ΔΔG.
Autoradiographs were scanned by either a LKB Ultrascan XL densitometer (Pharmacia LKB) or a Molecular Dynamics model PDSI-P90 personal densitometer. Alternatively, images were developed using a Molecular Dynamics model 445-P90 PhosphorImager. The data generated by the LKB densitometer were analyzed using Microsoft’s excel4.0 combined with Jandel’s (San Rafael, CA) peakfit3.1. Data generated by the Molecular Dynamics instruments were analyzed with imagequant Version 4.1 (Molecular Dynamics). For each lane, the area under each peak (or the volume of a defined area) was determined and the ratio of reconstituted to free DNA calculated (Figs. 3 and 4). For the reconstituted samples, the area indicated as such was selected to give the best resolved bands. Very slow migrating bands or smears, considered to be due to aggregates of DNA and histones, were not included in the calculations. Due to the damage-dependent smearing in both UV and BPDE samples, the area used for the DNA bands was selected by determining the minimum value between the DNA and nucleosomal peaks with scanning densitometry, and this location was used as the dividing point. The difference in the free energy (ΔG) of formation between undamaged and damaged samples can be calculated from the equation: ΔΔG = −RT ln(KD/KU), where KD is the ratio of nucleosomal to free damaged DNA and KU is the ratio of nucleosomal to free undamaged DNA.
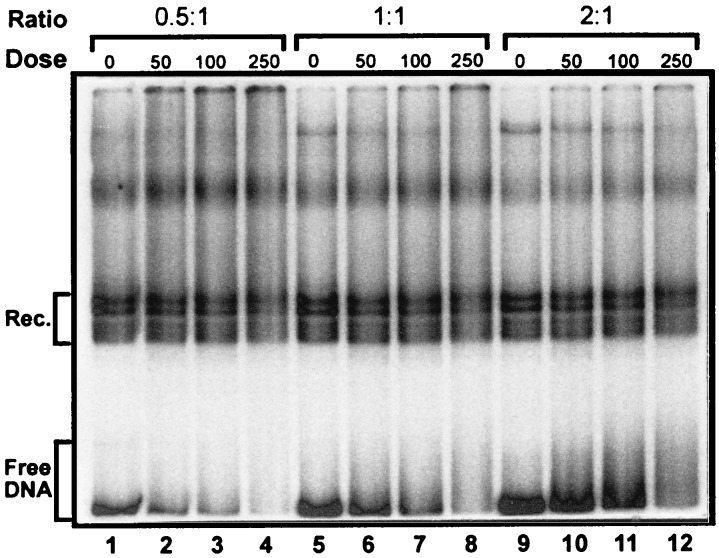
Figure 3.
Competitive reconstitution of UV-damaged DNA. Comparison of the effect of different levels of UV damage on 214-bp 5S DNA reconstitution. Lanes: 1–3, free DNA:core particle ratio of 0.5:1; 4–6, free DNA:core particle ratio of 1:1; 7–9, free DNA:core particle ratio of 2:1. UV dose levels are indicated above each lane. Rec., approximate area used to calculate reconstituted fraction of 5S DNA; Free DNA, approximate area used to calculate DNA fraction of 5S DNA. Multiple bands in the reconstituted region on these gels are due (at least in part) to multiple translational positions of the nucleosome on the 5S DNA.
Figure 4.
Competitive reconstitution of BPDE-damaged DNA. Comparison of effect of different levels of BPDE damage on 214-bp 5S rRNA gene fragment reconstitution. Lanes: 1–4, free DNA:core particle ratio of 0.5:1; 5–8, free DNA:core particle ratio of 1:1; 9–12, free DNA:core particle ratio of 2:1. BPDE dose levels are indicated above each lane. Rec., approximate area used to calculate reconstituted fraction of 5S DNA; Free DNA, approximate area used to calculate DNA fraction of 5S DNA. Multiple bands in reconstituted region are discussed in legend to Fig. 3.
For the second competitive reconstitution method, gels were scanned as above and the relative amounts of damaged 224-bp vs. undamaged 214-bp DNA in both the free DNA and reconstitute calculated for each damage level as: (224Reconstitute/214Reconstitute) ÷ (224Free DNA/214Free DNA) to yield KD or KU for damaged or undamaged DNA, respectively.
RESULTS
BPDE and UV Damage.
Incubation of linear or supercoiled plasmid DNA (either pBS-5S or pKS-5S) with BPDE resulted in concentration-dependent levels of adduction, as determined by tritium incorporation of [3H]BPDE, primarily (≈90%) to the N-2 position of guanine (ref. 38; Fig. 1A). Doses used in these experiments varied from 10 to 250 μM. At the highest dose (250 μM), an average adduction level of 1 adduct per 71 bp was achieved (Table 1), corresponding to an average of 2.1 adducts per nucleosome core-size DNA (146 bp), assuming a random distribution between fragments this size. We note that the GC content of the plasmid DNA and the 214-bp fragment are 50 and 60%, respectively. Therefore, somewhat more damage is expected in the 214-bp fragment.
Table 1.
UV and BPDE damage levels in X. borealis 5S rDNA
| Mutagen | Base pairs/lesion* | Lesions/146-bp DNA |
|---|---|---|
| BPDE | ||
| 10 μM | 1840 ± 24 | 0.08 ± 0.001 |
| 50 μM | 306 ± 53 | 0.49 ± 0.08 |
| 100 μM | 166 ± 5 | 0.88 ± 0.03 |
| 250 μM | 71 ± 10 | 2.09 ± 0.32 |
| UV | ||
| 500 J/m2 | 255 ± 40 | 0.57 ± 0.09 |
| 2500 J/m2 | 193 ± 18 | 0.76 ± 0.07 |
DNA was damaged by UV or BPDE at indicated dose (see Materials and Methods). Error represents SD for three (UV) or four (BPDE) different preparations. UV-induced CPD levels were measured by the method of Bohr et al. (34). BPDE adduct levels were measured based on [3H]BPDE levels (see Materials and Methods).
Irradiation of plasmid DNA with UV doses of 500 J/m2 and 2500 J/m2 resulted in CPD levels of ≈1 per 255 bp and 193 bp, respectively, within the 214-bp fragment (Table 1), as determined by digestion with T4 endonuclease (endo) V (34). These damage levels correspond to an average of 0.6 and 0.8 CPDs per 146 bp of DNA. In addition, 6–4 pyrimidine-pyrimidone photoproducts, which are not detected by the T4 endo V assay, are expected to be present at levels of about 1 per 3000 bp and 600 bp at UV doses of 500 and 2500 J/m2, respectively (39).
Gel Shift Analysis.
Based on previous studies with antibiotics (21) or intercalating agents (22), we anticipated that BPDE and UV damage might interfere with reconstitution of a nucleosome onto the 5S rDNA fragment. Low resolution gel shift experiments with BPDE and UV-damaged 5S fragments (to determine the extent of reconstitution) suggested that no marked changes occur in the level of reconstitution of UV-damaged samples; however, BPDE damage appeared to enhance formation of 5S nucleosomes. Therefore, competitive reconstitutions were carried out with CE DNA, undamaged 5S DNA, and either UV- (Fig. 3) or BPDE- (Fig. 4) damaged DNA, to quantify any changes. In all cases, ratios of free DNA (CE core particle DNA) to CE core particles ranging from 0.5:1 up to 8:1 were used. In both the UV- and BPDE-damaged samples, we observed a damage-dependent retardation and smearing of the naked DNA bands (e.g., compare lanes 7–9 in Fig. 3 and 9–12 in Fig. 4), which arises from bends (or hinges) induced in DNA by these two agents (15, 28, 40, 41). Multiple bands that are observed in the reconstituted nucleosome region on these gels (see Figs. 3 and 4, region designated Rec.) were not observed on lower resolution gels (6% glycerol/PAGE, 1% agarose, or 4% acrylamide/20% glycerol) and are at least partly due to multiple translational positions of the histone octamer on the 5S DNA (ref. 42; unpublished observations).
The intensities of reconstituted and free DNA bands for each sample were quantified and the ratio of these values used to calculate a ΔΔG value between undamaged (Fig. 3, lanes 1, 4, and 7; Fig. 4, lanes 1, 5, and 9) and damaged (Fig. 3, lanes 2 and 3, 5 and 6, and 8 and 9; Fig. 4, lanes 2–4, 6–8, and 10–12) 5S DNA. The ΔΔG is the difference between the free energies of formation (ΔG) of two complexes with different substrates. In this case, it is the difference between the ΔG of formation of the undamaged 5S reconstitutes and that of the damaged 5S reconstitutes, or the difference with mixed sequence CE DNA. The 5S samples damaged at 0.5–2.1 BPDE adducts per 146 bp were found to have average ΔΔGs of −220 to −540 cal/mol with respect to the undamaged 5S DNA (Fig. 5). As KD/KU = exp(−ΔΔG/RT), this yields a 45–150% increase in relative binding affinity (KD/KU) of histones to the BPDE damaged DNA compared with undamaged DNA.
Figure 5.
Changes in free energy (ΔΔG) for BPDE- and UV-damaged 5S DNA. Averages of competitive reconstitution experiments for UV- (•) or BPDE- (▪) damaged 5S DNA. Error bars represent SEM of three or more experiments. □, Values obtained from one experiment with BPDE-damaged and undamaged 5S DNA using the second competition protocol.
Similar changes in ΔG were obtained in experiments in which equal amounts of damaged and undamaged 5S DNA were combined in the same reconstitution reaction. Following recovery of the free DNA and reconstituted bands from an agarose gel (Fig. 6A), the DNA fragments were electrophoresed on denaturing (Fig. 6B) or nondenaturing acrylamide gels to resolve the damaged (224 bp) and undamaged (214 bp) fragments. The longer BPDE-damaged DNA predominated in the reconstitute fraction at each damage level and the undamaged DNA predominated in the free DNA fraction (Fig. 6B). The resulting free energy changes were very similar to those resulting from the competitive reconstitutions (Fig. 5, □). Adduction with 10 μM BPDE, which resulted in damage levels of 1 adduct per 1800 bp (<0.1 adduct/146 bp on average), did not have a measurable effect on ΔΔG compared with undamaged samples (data not shown). Furthermore, DNA lengths of 296, 214, and 160 bp have been used with comparable results, although the longer (296 bp) fragment forms dinucleosomes to some degree (data not shown). However, when these bands are included in the calculation, the total enhancement of the reconstitution is the same as for the shorter fragments (within the accuracy of this assay). A 160-bp 5S DNA fragment damaged with 50 or 100 μM of the (+)-anti-BPDE isomer, which has a higher adduction level than the (±)-anti-BPDE used in the experiments here, resulted in ΔΔG values of about −240 and −500 cal/mol, respectively (D.L.S., unpublished results).
Figure 6.
Competition between equal amounts of undamaged and BPDE-damaged reconstitutes. (A) Preparative agarose gel showing one lane of the reconstitution mixture. Free DNA and reconstituted fractions are delineated by DNA and Rec, respectively. (B) Denaturing acrylamide gel of free DNA and reconstituted bands isolated from agarose gel in A. Lanes: 1 and 2, undamaged 224- and 214-bp DNA recovered from free DNA and reconstitute fractions of agarose gel, respectively; 3 and 4, 250-μM BPDE-adducted 224-bp and undamaged 214-bp recovered from free DNA and reconstitute fractions of agarose gel, respectively.
In the experiments with UV-damaged DNA, doses of both 500 and 2500 J/m2 (Fig. 3) were used, corresponding to an average of 0.6 and 0.8 CPDs per 146 bp, respectively (Table 1). These levels of damage resulted in an increase in the ΔΔG values of +130 and +250 cal/mol, respectively, corresponding to 20 and 35% decreases in the level of nucleosome formation compared with undamaged 5S DNA (Figs. 3 and 5).
We also compared the affinity of the 5S rDNA nucleosome formation relative to random sequence DNA (Fig. 3). We found a ΔΔG of −900 ± 230 cal/mol for both the 214- and 296-bp fragments of the 5S rDNA vs. mixed sequence CE DNA (data not shown). This value is somewhat lower than that reported by Schild et al. (ref. 5; −1750 cal/mol), for a 583-bp fragment containing the same 5S sequence. This difference may be due to the difference in fragment length and/or cooperative binding of histone octamers (43).
DISCUSSION
The results presented here clearly indicate that the presence of BPDE adducts in the X. borealis 5S rRNA gene enhances nucleosome formation in a damage-dependent manner, while UV damage partially inhibits nucleosome formation. Based on previous work with other DNA binding agents (21, 22) and work done with UV damage (23, 44, 45), we initially anticipated that both types of damage would have a disruptive effect on nucleosome formation. Recently, however, studies have demonstrated that damaged DNA can enhance the binding of proteins other than repair proteins (see Introduction). In these cases, it is thought that the damaged DNA “mimics” the bent structure of DNA when bound to the proteins, thereby lowering the binding energy to near that of the normal DNA binding site.
The mechanism by which BPDE adducts enhance nucleosome formation is not known. The major product of adduction with racemic BPDE is a (+)-trans adduct to the exocyclic nitrogen (N-2) of guanine (38). This adduct accounts for ≈90% of the total adduction products and appears to be the only adduct of BPDE that significantly bends (or forms a hinge point in) the DNA (28). It is also the most tumorigenic adduct in mice (46). Two dimensional NMR and molecular modeling studies indicate that the major (+)-trans BPDE adduct lies in the minor groove of DNA with the pyrene pointing toward the 5′ end of the damaged strand (47). The effects of an adduct in this position are twofold. First, the BPDE adduct increases the minimum interstrand phosphate-to-phosphate distance of the minor groove (47, 48). Second, the adduct introduces a flexible hinge point in the DNA, as detected by gel migration (28, 40). There is, however, considerable heterogeneity in the structures that this isomer can form with DNA, which appears to be dependent on the local sequence of the adducted base (49).
In DNA sequences that position nucleosomes, such as the 5S rRNA gene, GC-rich sequences tend to be positioned toward the outside of the nucleosome, away from the histone surface (Fig. 2B; ref. 50). The widening of the minor groove width seen with the (±)-trans-BPDE adducts (47, 48) is similar to the widening seen in the DNA helix in which the minor groove faces away from the histone surface (51). Adducts located at these outside sites could potentially “fix” DNA in a conformation favorable for nucleosome formation. Alternatively, the bend (or flexible hinge point) induced by the BPDE adducts may require less energy for the DNA to wrap around the histones (lowering the free energy of nucleosome formation).
Our results demonstrate that a single adduct of BPDE can enhance the level of nucleosome formation by ≈70% (Fig. 5). At physiologic levels of damage in vivo, where this damage is spread throughout the genome with a frequency of about one adduct per 105 bp (52, 53), such a change in nucleosome stability could have significant consequences if damage occurs in a critical location (e.g., a gene promoter). Within a transcribed region of a gene, a more stable nucleosome could affect nucleosome disruption during transcription, or transcription-coupled repair, possibly leading to truncated RNAs or mutation.
Earlier work in this laboratory (23) demonstrated that CPDs are preferentially positioned away from the histone surface in nucleosomes reconstituted with UV-damaged, mixed-sequence DNA. In addition, we observed for mixed-sequence nucleosomes that CPDs are preferentially excluded from the central three turns of the DNA helix about the nucleosome (23). This central section of the nucleosomal DNA is underwound to 10.7 bp per turn of the helix (2) and presumably has difficulty incorporating a CPD within this altered structure. It was also reported that high UV doses interfere with nucleosome formation on supercoiled plasmid DNA molecules (44, 45). A change in rotational positioning of UV-damaged DNA (irradiated prior to nucleosome formation) was also observed by reconstitution of nucleosomes on a specific sequence; however, reduced levels of nucleosome assembly were not observed on that DNA following UV irradiation (24). This may be due to a difference in the DNA substrate, although we also did not observe major differences with an agarose gel system similar to that used by these authors (24). Interestingly, the level of inhibition seen in our studies with polyacrylamide gels (Figs. 3 and 5) is similar to that reported for supercoiled, UV-damaged DNA (44, 45). As with BPDE adducts, CPDs produce structural changes in the DNA molecule. Molecular modeling calculations predict ≈10° unwinding of the helix (54); and gel migration (41) and NMR (15) studies with site-specific CPDs in oligonucleotides indicate they cause a 7–9° bend toward the major groove. Furthermore, based on the rotational setting of 5S rDNA with respect to the histone surface (55, 56), the majority of hot spots for CPD formation in the 5S gene are at sites that would place them near (or at) the histone surface (Fig. 2B; unpublished observations). This would predict that UV lesions in this sequence would destabilize nucleosome formation as our results indicate. Taken together with our BPDE results, this data argues that the type of DNA lesion is very important in determining how damage will affect DNA–protein interactions.
Finally, the relevance of persistent damage in DNA has been demonstrated by studies that have correlated mutation frequency with damage persistence (57, 58). These reports examined the relationship between mutation hot spots due to UV damage in p53 (57) and BPDE damage in hypoxanthine (guanine) phosphoribosyl transferase (58) genes with the relative repair rates in these codons. In both studies it was found that damaged bases in codons with the highest mutation frequencies also had low repair rates, implying that repair efficiency may be a major component in deciding mutation frequency. A more stable nucleosome, such as those found on BPDE-damaged 5S DNA, could interfere with nucleotide excision repair and thereby increase mutation.
Acknowledgments
We thank Dr. Stephen Lloyd for supplying purified T4 endonuclease V, and members of the Smerdon laboratory, Dr. R. Reeves, Dr. F. Thoma, M. Nissen, and U. Schieferstein, for stimulating and critical discussions of this work. This study was supported by National Institutes of Health Grant ES02614 from the National Institute of Environmental Health Sciences (M.J.S.) and Department of Energy contract DE-AC06-76RLO 1830 (D.L.S.). D.B.M. was supported in part by a fellowship through Associated Western Universities, Inc., Northwest Division under Grant DE-FG06-89ER-75522 and DE-FG06-92RL-12451 with the U.S. Department of Energy.
ABBREVIATIONS
- BPDE
benzo[a]pyrene diol epoxide
- CPD
cyclobutane pyrimidine dimer
- PMSF
phenylmethylsulfonyl fluoride
References
- 1.van Holde K E. Chromatin. New York: Springer; 1989. [Google Scholar]
- 2.Wolffe A P. Chromatin: Structure and Function. 2nd Ed. New York: Academic; 1995. [Google Scholar]
- 3.Archer T K, Cordingly M G, Wolford R G, Hagar G L. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montecino M, Lian J, Stein G, Stein J. Biochemistry. 1996;35:5093–5102. doi: 10.1021/bi952489s. [DOI] [PubMed] [Google Scholar]
- 5.Schild C, Claret F-X, Wahli W, Wolffe A P. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smerdon M J. In: DNA Repair Mechanisms and Their Biological Implications in Mammalian Cells. Lambert M, Laval J, editors. New York: Plenum; 1989. pp. 274–291. [Google Scholar]
- 7.Smerdon, M. J. and Thoma, F. in DNA Damage and Repair: Biochemistry, Genetics, and Cell Biology, eds. Hoekstra, M. F. and Nickoloff, J. A. (Humana, Clifton, NJ), in press.
- 8.Thrall B D, Mann D B, Smerdon M J, Springer D L. Biochemistry. 1994;33:2210–2216. doi: 10.1021/bi00174a030. [DOI] [PubMed] [Google Scholar]
- 9.Smith B L, MacLeod M C. J Biol Chem. 1993;268:20620–20629. [PubMed] [Google Scholar]
- 10.Moyer R, Marien K, van Holde K, Bailey G. J Biol Chem. 1989;264:12226–12231. [PubMed] [Google Scholar]
- 11.McGhee J D, Felsenfeld G. Proc Natl Acad Sci USA. 1979;76:2133–2137. doi: 10.1073/pnas.76.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale J M, Nissen K A, Smerdon M J. Proc Natl Acad Sci USA. 1987;84:6644–6648. doi: 10.1073/pnas.84.19.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin W A, Doetsch P W, Haseltine W A. Nucleic Acids Res. 1985;13:5317–5325. doi: 10.1093/nar/13.14.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor J-S, Garrett D S, Cohrs M P. Biochemistry. 1988;27:7206–7215. doi: 10.1021/bi00419a007. [DOI] [PubMed] [Google Scholar]
- 15.Kim J-K, Patel D, Choi B-S. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell D L, Nguyen T D, Cleaver J E. J Biol Chem. 1990;265:5353–5356. [PubMed] [Google Scholar]
- 17.Gale J M, Smerdon M J. Photochem Photobiol. 1990;51:411–417. doi: 10.1111/j.1751-1097.1990.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 18.Suquet C, Mitchell D L, Smerdon M J. J Biol Chem. 1995;270:16507–16509. doi: 10.1074/jbc.270.28.16507. [DOI] [PubMed] [Google Scholar]
- 19.Wallrath L L, Lu Q, Granok H, Elgin S C R. BioEssays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 20.MacLeod M C. Mol Carcinog. 1996;15:241–250. doi: 10.1002/(SICI)1098-2744(199604)15:4<241::AID-MC1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 21.Low C M L, Drew H R, Waring M J. Nucleic Acids Res. 1986;14:6785–6801. doi: 10.1093/nar/14.17.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurray C T, van Holde K E. Proc Natl Acad Sci USA. 1986;83:8472–8476. doi: 10.1073/pnas.83.22.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suquet C, Smerdon M J. J Biol Chem. 1993;268:23755–23757. [PubMed] [Google Scholar]
- 24.Schieferstein U, Thoma F. Biochemistry. 1996;35:7705–7714. doi: 10.1021/bi953011r. [DOI] [PubMed] [Google Scholar]
- 25.Treiber D K, Zhai X, Jantzen H-M, Essigmann J M. Proc Natl Acad Sci USA. 1994;91:5672–5676. doi: 10.1073/pnas.91.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLeod M C, Powell K L, Tran N. Carcinogenesis. 1995;16:975–983. doi: 10.1093/carcin/16.5.975. [DOI] [PubMed] [Google Scholar]
- 27.MacLeod M C, Powell K L, Kuzmin V A, Kilbanovskiy A, Geacintov N E. Mol Carcinog. 1996;16:44–52. doi: 10.1002/(SICI)1098-2744(199605)16:1<44::AID-MC6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 28.Xu R, Mao B, Xu J, Li B, Birke S, Swenberg C E, Geacintov N E. Nucleic Acids Res. 1995;23:2314–2319. doi: 10.1093/nar/23.12.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolffe A P, Jordan E, Brown D D. Cell. 1986;44:381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- 30.Hayes J J, Wolffe A P. Proc Natl Acad Sci USA. 1993;90:6415–6419. doi: 10.1073/pnas.90.14.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes J J, Wolffe A P. Proc Natl Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thrall B D, Mann D B, Smerdon M J, Springer D L. Carcinogenesis. 1992;13:1529–1534. doi: 10.1093/carcin/13.9.1529. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 34.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 35.Libertini L J, Ausio J, van Holde K E, Small E W. Biophys J. 1988;53:477–487. doi: 10.1016/S0006-3495(88)83126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrader T E, Crothers D M. Proc Natl Acad Sci USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayasena S D, Behe M J. J Mol Biol. 1989;208:297–306. doi: 10.1016/0022-2836(89)90390-2. [DOI] [PubMed] [Google Scholar]
- 38.Struab K M, Meehan T, Burlingame A L, Calvin M. Proc Natl Acad Sci USA. 1977;74:5285–5289. doi: 10.1073/pnas.74.12.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Smerdon M J. Anal Biochem. 1995;229:323–328. doi: 10.1006/abio.1995.1420. [DOI] [PubMed] [Google Scholar]
- 40.Hogan M E, Dattagupta N, Whitlock J P., Jr J Biol Chem. 1981;256:4504–4513. [PubMed] [Google Scholar]
- 41.Wang C-I, Taylor J-S. Proc Natl Acad Sci USA. 1991;88:9072–9076. doi: 10.1073/pnas.88.20.9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts M S, Fragoso G, Hager G L. Biochemistry. 1995;34:12470–12480. doi: 10.1021/bi00038a046. [DOI] [PubMed] [Google Scholar]
- 43.Nuebauer B, Hörz W. Methods Enzymol. 1989;170:630–644. doi: 10.1016/0076-6879(89)70069-0. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto H, Takakusu A, Ohnishi T. Photochem Photobiol. 1994;60:134–138. doi: 10.1111/j.1751-1097.1994.tb05079.x. [DOI] [PubMed] [Google Scholar]
- 45.Matsumoto H, Takakusu A, Mori T, Ihara M, Todo T, Ohnishi T. Photochem Photobiol. 1995;61:459–462. doi: 10.1111/j.1751-1097.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 46.Slaga T J, Bracken W J, Gleason G, Levin W, Yagi H, Jerina D M, Conney A H. Cancer Res. 1979;39:67–71. [PubMed] [Google Scholar]
- 47.Cosman M, de los Santos C, Fiala R, Hingerty B E, Singh S B, Ibanez V, Margulis L A, Live D, Geacintov N E, Broyde S, Patel D J. Proc Natl Acad Sci USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de los Santos C, Cosman M, Hingerty B E, Ibanez V, Margulis L A, Geacintov N E, Broyde S, Patel D J. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 49.Mao B, Xu J, Li B, Margulis L A, Smirnov S, Ya N Q, Courtney S H, Geacintov N E. Carcinogenesis. 1995;16:357–365. doi: 10.1093/carcin/16.2.357. [DOI] [PubMed] [Google Scholar]
- 50.Satchwell S C, Drew H R, Travers A A. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 51.Richmond T J, Finch J T, Rushton B, Rhodes D, Klug A. Nature (London) 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 52.Springer D L, Mann D B, Dankovic D A, Thomas B L, Wright C W, Mahlum D D. Carcinogenesis. 1989;10:131–137. doi: 10.1093/carcin/10.1.131. [DOI] [PubMed] [Google Scholar]
- 53.DiGiovanni J, Decina P C, Prichett W P, Fisher E P, Aalfs K K. Carcinogenesis. 1985;6:741–747. doi: 10.1093/carcin/6.5.741. [DOI] [PubMed] [Google Scholar]
- 54.Perlman D A, Holbrook S R, Pirkle D H, Kim S-H. Science. 1985;227:1304–1308. doi: 10.1126/science.3975615. [DOI] [PubMed] [Google Scholar]
- 55.Hayes J J, Clark D J, Wolffe A P. Proc Natl Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pruss D, Hayes J J, Wolffe A P. BioEssays. 1995;17:1–9. doi: 10.1002/bies.950170211. [DOI] [PubMed] [Google Scholar]
- 57.Tornaletti S, Pfeifer G P. Science. 1994;263:1436–1438. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]
- 58.Wei D, Maher V M, McCormick J J. Proc Natl Acad Sci USA. 1995;92:2204–2208. doi: 10.1073/pnas.92.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]