Abstract
The vitamin K-dependent matrix Gla protein (MGP) is synthesized in a wide variety of tissues such as lung, heart, kidney, cartilage, and bone. Expression of the MGP gene is regulated by various growth factors, steroid hormones, and the vitamin A metabolite retinoic acid (RA). In this report, we present evidence that RA down-regulates MGP gene expression in different rat and human cell lines via endogenous retinoid receptors [RA receptor (RAR) and retinoid X receptor (RXR)]. Repression of the human MGP (hMGP) gene is specifically mediated by ligand-activated RAR and RXR. Deletion analysis led to the identification of a novel negative response element (NRE) within the hMGP promoter. DNA binding studies performed with bacterially expressed RAR/RXR reveal the formation of a specific heterodimer/NRE complex. Furthermore, electrophoretic mobility-shift assays performed with proteins from RA-treated cells show that endogenous RAR/RXR binds to the NRE. We demonstrate that the NRE contains a CCAAT box and that both RAR/RXR and CCAAT-binding proteins such as c/EBPβ recognize this common regulatory sequence in the hMGP promoter. Our results indicate that RA-mediated repression of the hMGP gene is due to binding of liganded RAR/RXR to a novel negative RA response element.
Keywords: transcriptional regulation, nuclear receptors, transrepression
Matrix Gla protein (MGP) is a 10-kDa, vitamin K-dependent protein that serves as a substrate for the enzyme γ-carboxylase, which converts glutamic acid to γ-carboxyl-glutamic acid (Gla). MGP expression has been detected in all vertebrate tissues, with the highest levels in cartilage, kidney, lung and heart (1). MGP is a secreted protein and accumulates to significant levels in the extracellular matrix of bone, cartilage, and calcified cartilage (2–4). Following the original isolation from bone (2, 3), MGP has been independently identified by differential cDNA screening as a gene that is overexpressed in primary human breast cancer cells (5), in prostate epithelial cells which undergo apoptosis (6), and in differentiated vascular smooth muscle cells (7). Although the exact function of MGP is presently unknown, there is evidence to suggest that the protein may play a role in cell growth and tumorigenesis. Interestingly, MGP gene expression is regulated by several growth factors and hormones. Especially, retinoic acid (RA) has been implicated in modulating MGP gene expression.
Retinoids play an important role in development, cellular proliferation, and differentiation and are well-known inhibitors of cell growth. Furthermore, retinoids can be effective therapeutic agents in treating certain human cancers (8). RA exerts biological effects by acting through at least two distinct classes of intracellular proteins including the RA receptors (RARs) and the retinoid X receptors (RXRs), both of which are members of the nuclear receptor superfamily. While both the RARs and RXRs are effective activators of some genes, RA is also known to repress gene expression. The inhibitory effects of RA on gene transcription can be exerted through various mechanisms such as competition between receptors and other transcription factors for overlapping binding sites (9, 10) or by competition for limiting cofactors (11, 12).
To further extend our understanding of RAR/RXR-mediated repression, we examined the effect of RA on the regulation of human (hMGP) gene expression. In this report, we show that RA is able to repress endogenous MGP gene expression in different cell lines. To identify cis-regulatory promoter elements that mediate RA-dependent repression we cloned and characterized the 5′ flanking region of the hMGP gene. Deletion analysis of the hMGP promoter identified a 37-bp region that is both necessary and sufficient to confer repression by RA. Electrophoretic mobility-shift assays (EMSAs) show that both RAR/RXR, expressed endogenously in NRK52E cells or recombinant receptors, bind to this novel negative response element (NRE). This NRE shows no sequence homology to other known RAR response elements, but contains a CCAAT box. Both RAR/RXR and the CAAT-binding protein c/EBPβ recognize the NRE, and c/EBPβ competes with RAR/RXR for binding to the NRE. Consequently, overexpression of c/EBPβ abrogates RA-mediated repression of the hMGP promoter. Finally, transient transfection analysis of RAR deletion mutants reveals that the DNA binding domain (DBD) and the activation domain (AF-2) of RAR are required for repression.
MATERIALS AND METHODS
Cell Culture and Transfection.
NRK52E cells were cultured in Ham’s F-12 medium. MCF-7 cells were cultured in Earl’s modified Eagle’s medium (EMEM). Both media were supplemented with 10% fetal calf serum. Transient transfection assays were carried out using the standard calcium phosphate coprecipitation technique (13) as described in ref. 14. NRK52E cells were exposed to the precipitate for 4 hr following a 2-min 10% dimethyl sulfoxide shock. Luciferase (LUC) activity was assayed as recommended by the manufacturer (Promega) in the Luminometer ML 3000 (Dynatech). Relative light units were normalized to β-galactosidase activity (15) and protein concentration using the Bradford dye assay (Bio-Rad). All experiments were repeated at least five times.
Reporter Plasmids.
The LUC reporter plasmids βRE2-TKLUC and (SPP)x2-TKLUC were described previously (15). The hMGP LUC reporter plasmid -3600LUC was generated by inserting a 3.6-kb hMGP promoter fragment at the KpnI–BglII restriction sites of pGL2LUC (Promega). The constructs -560LUC, -300LUC, -168LUC, -132LUC, and -560 Δ 168–132LUC were generated using PCR amplification followed by inserting the PCR products at the BglII–HindIII restriction sites of pGL2LUC.
Expression Vectors.
The eukaryotic expression plasmids CMX-hVDR, CMX-hRARα, CMX-hRXRα, CMX-RARα403*, CMX-RARα-DBD, CMX-RARα ΔDBD, CMX-RARα-N, CMX-RARα-C, and MSV-c/EBPβ were described previously (14–16).
DNA Binding Studies.
Whole cell extracts from NRK52E cells were prepared as recommended (17). The glutathione S-transferase (GST) fusion proteins were expressed according to ref. 18. The purified GST fusion proteins or 2 μl of whole cell extracts were mixed with sample buffer resulting in a final concentration of 10 mM Tris (pH 8.0), 40 mM KCl, 0.05% Nonidet P-40, 6% (vol/vol) glycerol, 1 mM DTT, 1 μg poly(dI-dC). Approximately 0.5 ng of the 32P-labeled oligonucleotide probe was added to the reaction mixture and incubated on ice for 30 min. Subsequently, the samples were loaded on 5% nondenaturing polyacrylamide gels in 0.5 × TBE running buffer at 4°C. After electrophoresis, gels were dried and subjected to autoradiography at −70°C. The following oligonucleotides were used in gel retardation assays: NRE (5′-TGTTTGGGAAAAGTTCCAATGCTAGTTAAGTGCCAAC-3′), MT1 (5′-AGTTCCAATGCTAGTTAAGTGCCAAC-3′), MT2 (5′-AGTTCCAATGCTAGTTA-3′), DR2 (5′-AGCTTCAGGTCAGGAGGTCAGAG-3′), and DR5 (5′-AGCTTCAGGTCACCAGGAGGTCAGAG-3′). To determine the composition of various DNA/protein complexes the following antibodies were included in the reaction mixtures: α-RAR and α-RXR (Santa Cruz Biotechnology), α-Flag (Kodak), and α-c/EBPβ (generous gift from K.-H. Klempnauer, MPI für Immunobiologie, Freiburg, Germany).
Northern Blots.
Total RNA was prepared and subjected to Northern blot analysis using standard procedures (19). Hybridizations were performed by using hMGP, rat MGP, and mouse β-actin cDNA probes in a buffer containing 50% formamide at 42°C for 16 hr. The final washes were done in 0.1 × SSC at 60°C. The Northern blots were quantified by densitometry (Molecular Dynamics, Model 300A).
Radioimmunoassays.
MCF-7 and NRK52E cells were cultured until they had attained visual confluence. Culture medium was exchanged for fresh medium supplemented with ethanol vehicle or 10−6 M all-trans RA. After 24 hr, conditioned medium was collected from three to five individually treated dishes, centrifuged to remove any cellular debris, and aliquots of 0.2 ml were analyzed by radioimmunoassay in triplicate for MGP protein content as described (20).
RESULTS
RA Inhibits MGP Gene Expression.
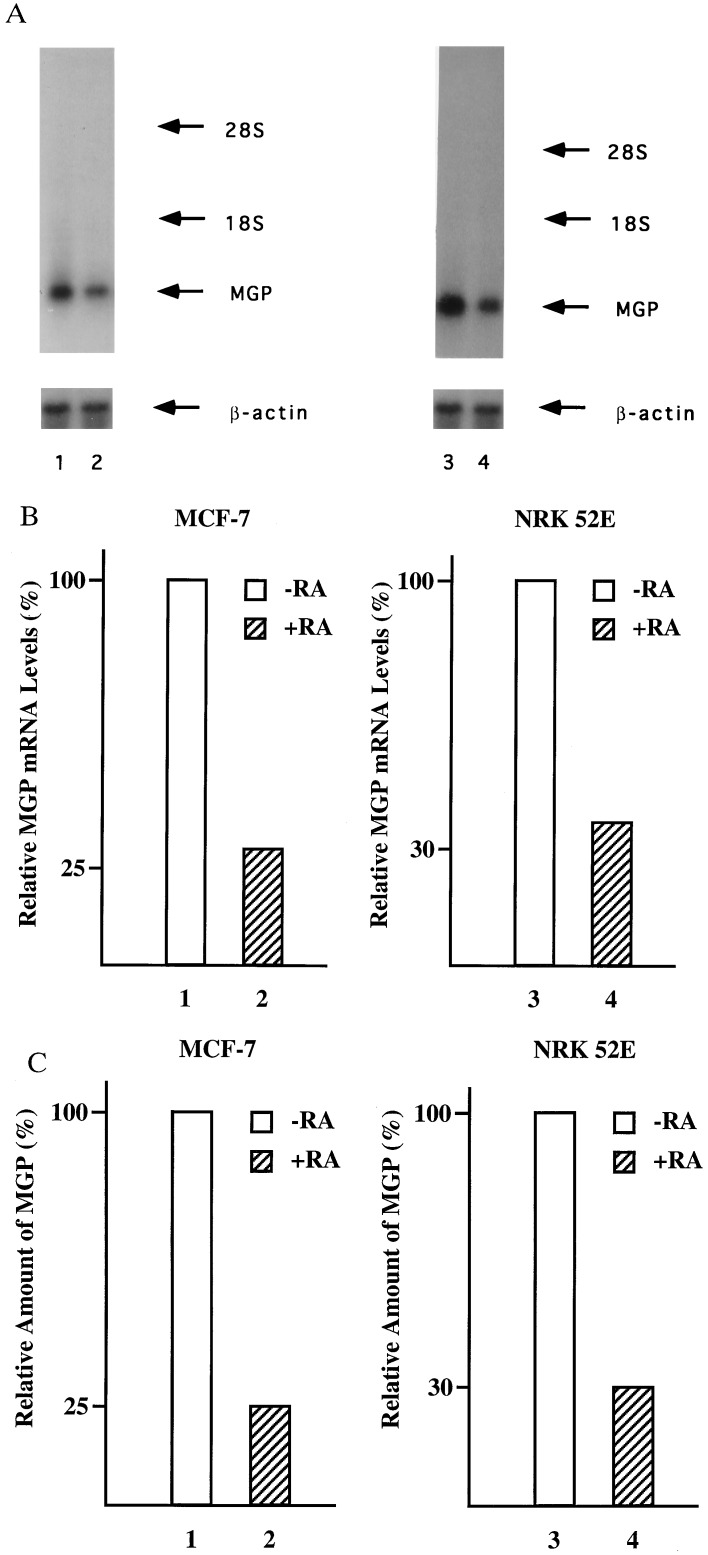
To evaluate the effects of RA on the expression of the endogenous MGP gene, the human breast cancer cell line MCF-7 and the rat kidney cell line NRK52E were treated for 24 hr with RA, and the levels of MGP mRNA were determined by Northern blot analysis. As shown in Fig. 1 A and B, RA decreased MGP mRNA levels in both the MCF-7 (lane 2) and NRK52E cells (lane 4) to ≈30% of the untreated control levels. RA-mediated repression is specific since RA treatment has no effect on the expression of the β-actin control gene (Fig. 1A, lanes 2 and 4). RA decreased MGP mRNA levels in the absence of new protein synthesis and as early as 10 hr after treatment (data not shown). To determine whether repression of MGP mRNA expression correlates with reduced levels of MGP protein, MCF-7 and NRK52E cells were treated for 24 hr with RA and the amounts of secreted MGP protein were determined by radioimmunoassays. In both cell lines, MGP protein levels are drastically reduced by RA treatment (Fig. 1C, lanes 2 and 4). Taken together, these results demonstrate that RA is able to repress MGP gene expression in rat and human cells under physiological conditions.
Figure 1.
Inhibition of MGP gene expression by RA. (A) Cell lines MCF-7 and NRK52E were grown in the absence or presence of 10−6 M all-trans RA. MGP mRNA levels were analyzed by hybridization with cDNA probes encoding the human (lanes 1 and 2) or the rat MGP (lanes 3 and 4) gene, respectively. The amount of RNA loaded in each lane was controlled by hybridization with a mouse β-actin cDNA. (B) Densitometric quantitation of the MGP mRNA levels shown in A. The relative ratios of MGP/β-actin RNA in untreated cells are set as 100%. (C) RA treatment represses the level of MGP antigen in conditioned medium of MCF-7 and NRK52E cells. Cells were incubated with ethanol vehicle (open bars) or 10−6 M all-trans RA (hatched bars) for 24 hr. The levels of MGP antigen secreted into the conditioned medium were assayed by radioimmunoassay. The amount of MGP antigen secreted by the ethanol-treated control cells is set as 100%.
RA-Mediated Repression of hMGP Promoter Activity.
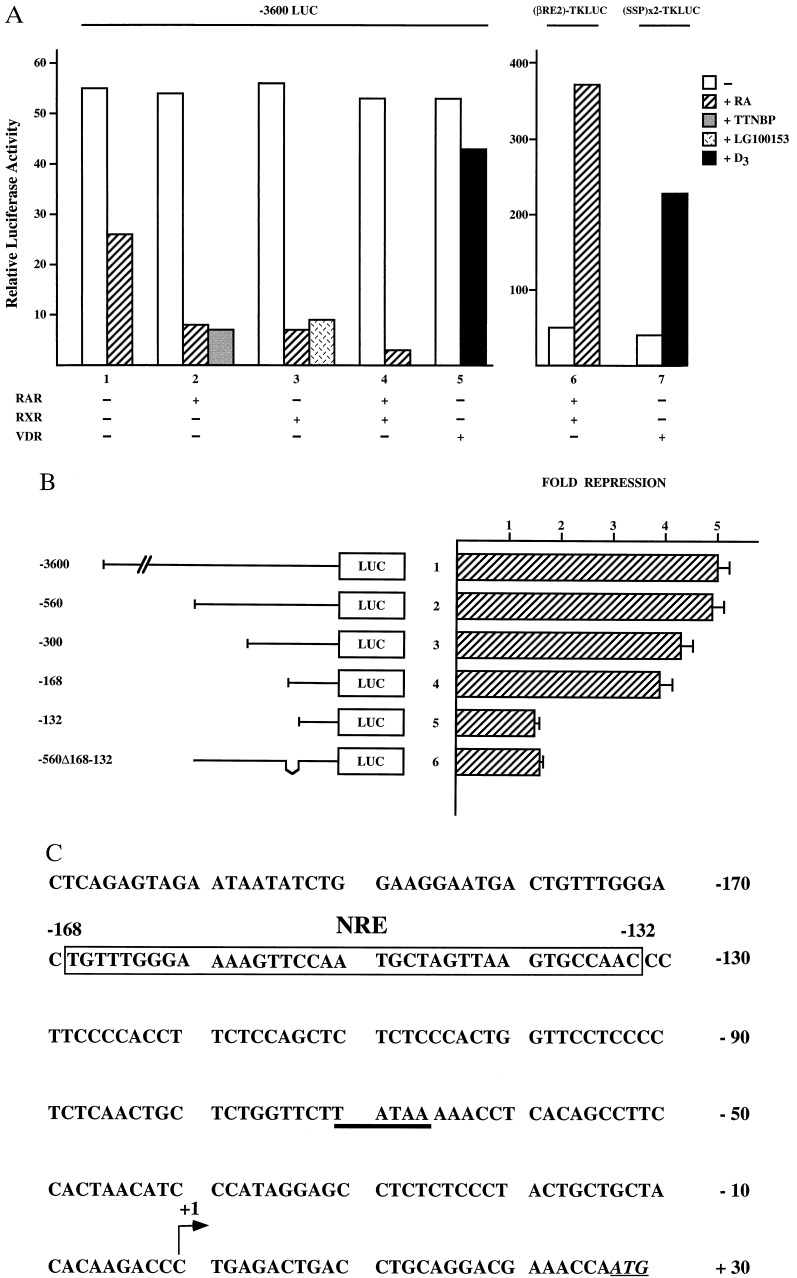
To investigate the mechanism of repression, we fused a 3600-bp fragment of the hMGP promoter to the firefly LUC reporter gene. Transcriptional activity of the hMGP reporter construct was monitored following transient transfection in NRK52E cells. As shown in Fig. 2A, activity of the -3600LUC reporter is significantly repressed in RA-treated cells (Fig. 2A, bar 1). Cotransfection of either RAR or RXR expression plasmids results in a even more dramatic RA-dependent repression (Fig. 2A, bars 2 and 3). Interestingly, the RAR- or RXR-specific ligands TTNPB and LG100153 also inhibit -3600LUC reporter activity ≈10-fold (Fig. 2A, bars 2 and 3). Cotransfection of both RAR and RXR expression plasmids further enhances the ligand-mediated repression (Fig. 2A, bar 4). In contrast, the control plasmid βRE2-TKLUC, which contains a known RA response element is activated by RA, indicating that RA-mediated repression is specific for the hMGP promoter (Fig. 2A, bar 6). In addition, cotransfected vitamin D receptor (VDR) fails to repress the hMGP promoter in NRK52E cells (Fig. 2A, bar 5), whereas the control plasmid (SSP)x2-TKLUC, which contains a well-known VDR element (15) is activated by D3 (Fig. 2A, bar 7). Taken together, these data demonstrate that RA-induced repression of hMGP promoter activity is specifically mediated by hormone-activated RAR/RXR, whereas VDR and several other nuclear receptors fail to repress hMGP promoter activity.
Figure 2.
RA induced down-regulation of the hMGP promoter. (A) Reporter plasmids (1.25 μg) -3600LUC, βRE2-TKLUC, and (SPP)x2-TKLUC were transfected in NRK52E cells. Transcriptional activity was analyzed either by taking advantage of endogenous retinoid receptor activity present in NRK52E cells (bar 1) or by cotransfection of 125 ng CMX plasmids expressing RAR, RXR, or VDR. Cells were either untreated (open bars) or treated with 10−6 M all-trans RA (hatched bars), 10−7 M TTNPB (shaded bars), 10−7 M LG100153 (cross-hatched bars), or with 10−6 M D3 (solid bars). (B) Indicated reporter plasmids (1.25 μg), together with 125 ng of CMX-RAR and CMX-RXR expression plasmids, were cotransfected into NRK52E cells. Transcriptional repression is presented as “fold repression.” (C) Nucleotide sequence of the first 209 bp of the hMGP 5′ flanking region. The NRE located from position −168 to −132 bp is marked by the open rectangle. The TATA box is underlined. Arrow indicates the transcription initiation site. The ATG start codon of the hMGP coding region is underlined and shown in italics.
To further delineate the sequences responsible for repression of hMGP promoter activity, we generated a series of promoter deletion mutants and tested these constructs in transient transfection assays in NRK52E cells. Subsequent deletions of 5′ flanking sequences up to the position −168 bp do not alter repression by RAR and RXR significantly (Fig. 2B, bars 1–4). However, a reporter construct containing only the promoter region up to position −132 bp is no longer down-regulated by liganded RAR/RXR (Fig. 2B, bar 5). Importantly, the activity of the internal mutant -560Δ168–132 LUC is also not repressed by ligand-activated RAR/RXR (Fig. 2B, bar 6). These results demonstrate that the DNA sequences located between −168 and −132 bp contain a NRE that is necessary and sufficient to confer RA-mediated repression (Fig. 2C).
Both the AF-2 and the DBD of RAR Are Necessary for Repression.
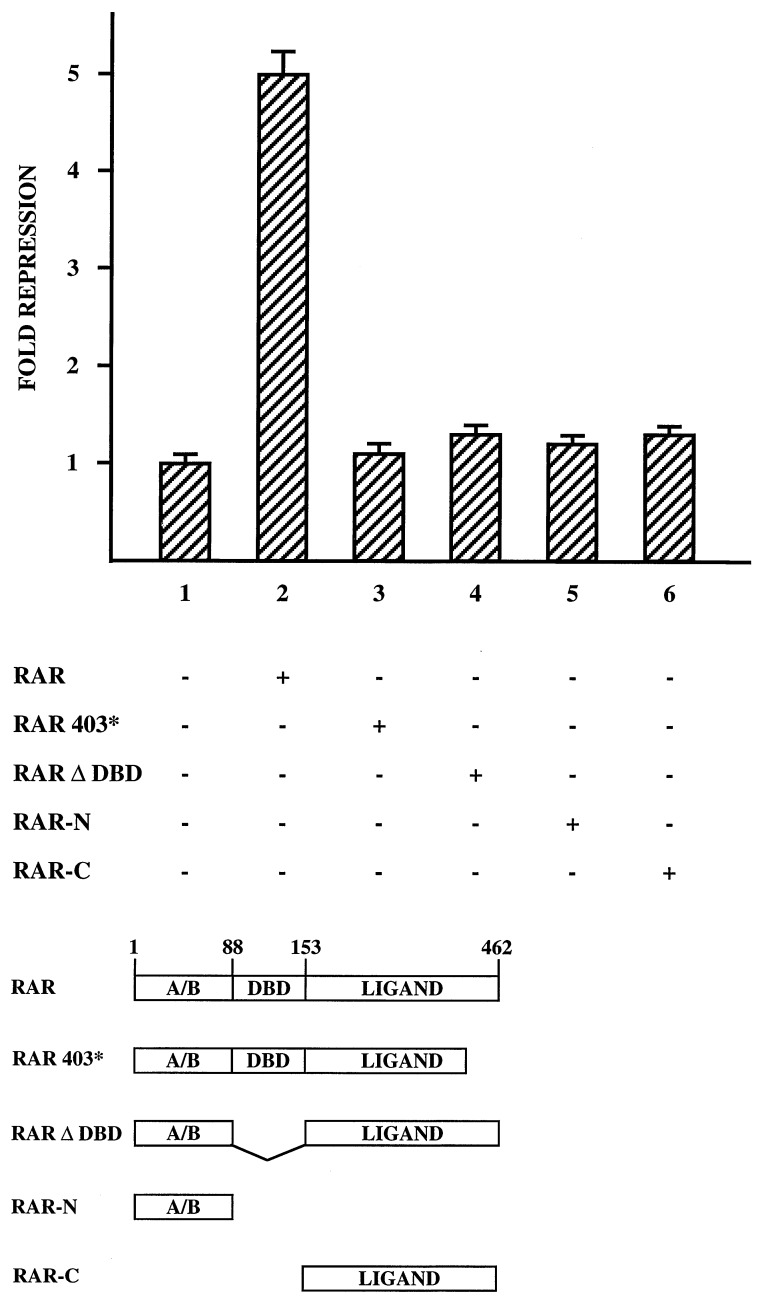
The above experiments demonstrate that the hMGP promoter is efficiently repressed by RAR/RXR in a ligand-dependent manner. To define the domains of RAR involved in repression, several receptor mutants were analyzed in cotransfection studies in NRK52E cells for their ability to repress -560LUC reporter activity. All mutants were expressed in similar amounts in the tested cells (data not shown). The C-terminal truncation mutant RAR403* is unable to repress (Fig. 3, compare bars 2 and 3). Also, deletion of the DBD results in a complete loss of repression (Fig. 3, bar 4). Further analysis confirm that RAR mutants, which express either the C terminus (RAR-C) or the N-terminus (RAR-N), also fail to repress (Fig. 3, bars 5 and 6). In conclusion, these data suggest that RAR-mediated repression requires both the DBD and an intact ligand binding domain of RAR.
Figure 3.
Functional analysis of RAR mutants. Transcriptional repression of 1.25 μg of the reporter plasmid -560LUC was assayed in NRK52E cells cotransfected as indicated (+) with 125 ng expression plasmids coding for either the full-length RAR, RAR403*, RARΔDBD, RAR-N, or RAR-C. Full-length RAR protein is composed of the N-terminal A/B domain (amino acids 1–87), the DBD (amino acids 88–152), and the C-terminal ligand binding domain (amino acids 153–462). RA-dependent repression of -560LUC reporter activity obtained without cotransfection of RAR expression plasmids was set as 1.
RAR/RXR Bind to the hMGP Promoter.
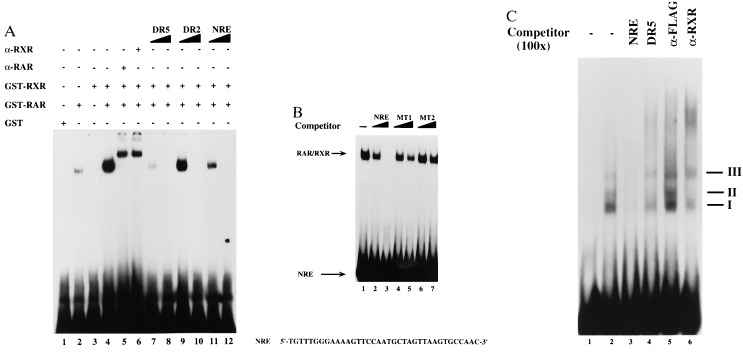
To investigate the molecular basis of RA repression, we examined the potential of RAR and RXR to bind to the NRE in vitro. Therefore, we performed EMSAs with bacterially expressed GST-RAR and GST-RXR proteins. GST-RAR forms a specific complex with an oligonucleotide that includes a single copy of the NRE (Fig. 4A, lane 2). Addition of equal amounts of RXR enhances the binding of RAR (Fig. 4A, lane 4), whereas RXR alone does not bind (Fig. 4A, lane 3). The RAR/RXR complex is supershifted by both α-RAR and α-RXR antibodies. Oligonucleotides containing known RAR/RXR binding sites (DR5 and DR2) are able to compete for RAR/RXR binding (Fig. 4A, lanes 7–10). As expected, the DR5 element competes more efficiently than the DR2-containing oligonucleotide. Importantly, binding of RAR/RXR is also efficiently competed by the addition of unlabeled NRE (−168 to −132 bp)-containing oligonucleotide (Fig. 4A, lane 12). In contrast, the 5′-truncated mutant oligonucleotide MT1 (−157 to −132 bp) competes with greatly reduced affinity (Fig. 4B, lanes 4 and 5), whereas the mutant oligonucleotides MT2 (−154 to −137 bp) is unable to compete (Fig. 4B, lanes 6 and 7).
Figure 4.
(A) RAR/RXR binds to the hMGP promoter in vitro. EMSAs were performed by incubating a 32P-labeled oligonucleotide containing the NRE sequence (−168 to −132 bp) with purified recombinant GST-RAR (lane 2) and GST-RXR (lane 3) or both proteins (lane 4). The RAR/RXR heterodimer shift is specifically supershifted by an α-RAR antibody (lane 5) as well as by an α-RXR antibody (lane 6). Competition experiments were performed by using the indicated competitors in a 50- or 200-fold molar excess. The retarded receptor/DNA complex is competed by the unlabeled DR5 oligonucleotide (lanes 7 and 8), DR2 oligonucleotide (lanes 9 and 10), and NRE oligonucleotide (lanes 11 and 12). Bacterially expressed GST-protein was also added as a control (lane 1). (B) Purified GST-RAR/GST-RXR were incubated with a 32P-labeled oligonucleotide containing the NRE sequence (−168 to −132 bp). The RAR/RXR shift is specifically competed by a 200-fold molar excess of NRE oligonucleotide (lanes 2 and 3). Oligonucleotide MT1 (−157 to −132 bp) competes only very little (lanes 4 and 5), whereas oligonucleotide MT2 (−154 to −137 bp) fails to compete (lanes 6 and 7). (C) RXR binds to the hMGP promoter in vivo. Samples (2 μl) of whole cell extracts prepared from RA-treated NRK52E cells were incubated with the 32P-labeled NRE probe. The DNA/protein complexes are specifically competed by a 100-fold molar excess of NRE oligonucleotide (lane 3). In contrast, oligonucleotide DR5 competes only complex II (lane 4). To establish the composition of the DNA/protein complexes, the EMSAs were challenged with α-RXR (lane 6) or unrelated α-Flag antibodies (lane 5).
To establish that the RAR/RXR endogenously expressed in NRK52E cells binds to the NRE sequence, we performed additional EMSAs. Proteins present in whole cell extracts from RA-treated NRK52E cells are able to form the DNA/protein complexes I, II, and III (Fig. 4C, lane 2). These complexes are competed by a 100-fold molar excess of unlabeled NRE-containing oligonucleotide (Fig. 4C, lane 3). Complex II is competed by a 100-fold molar excess of unlabeled DR5 oligonucleotide, indicating that RAR/RXR is part of complex II (Fig. 4C, lane 4). Finally, we challenged the EMSAs with an α-RXR antibody. Complex II is specifically supershifted by the α-RXR antibody showing that RXR is part of this DNA/protein complex (Fig. 4C, lane 6). Incubation with an unrelated α-Flag control antibody has no influence on the mobility of complex II (Fig. 4C, lane 5). Taken together, these data reveal that retinoid receptors present in NRK52E cells bind to the NRE element.
The NRE Is a Common Binding Site for RAR/RXR and c/EBPβ.
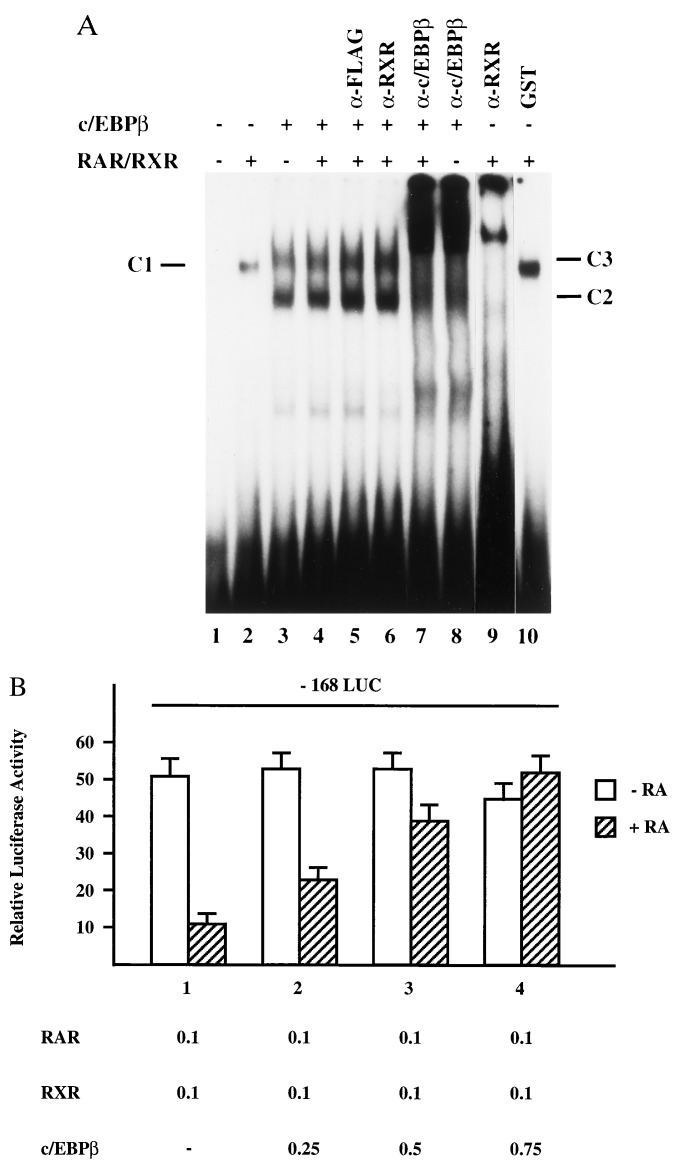
The NRE conferring RA repression to the hMGP promoter does not show any sequence homology to other known RAR response elements (Fig. 2C). Interestingly, a CCAAT box can be mapped within the NRE. To investigate the molecular basis of hormonal repression in more detail, we examined the ability of the CAAT-binding protein c/EBPβ and RAR/RXR heterodimers to bind to the NRE. Accordingly, we performed gel mobility-shift experiments using bacterially expressed GST-RAR, GST-RXR, and GST-c/EBPβ proteins. RAR/RXR heterodimers form a retarded DNA/protein complex C1 (Fig. 5A, lane 2). Incubation of the NRE oligonucleotide with GST-c/EBPβ alone results in the formation of two DNA/protein complexes (C2 and C3), which migrate with different mobilities than the RAR/RXR/NRE complex C1 (Fig. 5A, lane 3). Incubation of the NRE oligonucleotide with equal amounts of both c/EBPβ and RAR/RXR fusion proteins results only in the formation of complexes C2 and C3 (Fig. 5A, lane 4). To establish the composition of the different DNA/protein complexes, we challenged the EMSAs with specific antibodies directed against RXR, c/EBPβ, and an unrelated α-Flag control antibody. Complex C1 is specifically supershifted by the RXR antibody (Fig. 5A, lane 9), whereas complexes C2 and C3 are supershifted by the c/EBPβ antibody (Fig. 5A, lane 8). However, EMSAs containing both RAR/RXR and c/EBPβ proteins are supershifted by the c/EBPβ antibody only (Fig. 5A, lane 7). The α-RXR antibody fails to supershift complexes C2 and C3 (Fig. 5A, lane 6). Hence, RAR/RXR is not present in complexes C2 and C3. Incubation with an unrelated α-Flag control antibody has no influence on the mobility of the complexes (Fig. 5A, lane 5). These in vitro binding studies show that RAR/RXR and c/EBPβ do not physically interact on the NRE. Instead, the data suggest that c/EBPβ competes with RAR/RXR for binding to a common binding site.
Figure 5.
(A) RAR/RXR and c/EBPβ recognize an overlapping binding site. The EMSAs were performed by using the 32P-labeled NRE oligonucleotide and purified recombinant GST-RAR, GST-RXR, GST-c/EBPβ, and GST. To determine binding of these proteins to the NRE sequence, equal amounts of GST (lane 1), GST-RAR and GST-RXR (lanes 2 and 9), GST-c/EBPβ (lanes 3 and 8), or a combination of GST-RAR/GST-RXR and GST-c/EBPβ (lanes 4–7) were incubated with the probe. To establish the composition of the different complexes, the EMSAs were challenged with α-RXR (lane 6) or α-c/EBPβ antibodies (lanes 7 and 8). The RAR/RXR/NRE complex is denoted C1, the c/EBPβ/NRE complexes are denoted C2 and C3. (B) c/EBPβ is able to abrogate RAR/RXR-mediated repression. The reporter plasmid -168LUC was cotransfected in NRK52E cells together with constant amounts of plasmids expressing RAR, RXR, and various amounts of expression vector encoding c/EBPβ. The cells were either untreated (open bars) or treated with 10−6 M all-trans RA (hatched bars). Numbers indicate the amount of cotransfected plasmid DNA in micrograms.
To investigate the effect of c/EBPβ on RAR/RXR-mediated repression, we cotransfected RAR/RXR and c/EBPβ expression vectors together with the -168LUC reporter plasmid into NRK52E cells. As shown in Fig. 5B, increasing amounts of c/EBPβ expression vectors abolishes RAR/RXR-mediated repression (Fig. 5B, compare bar 1 with bars 2–4). In summary, these data suggest competition between RAR/RXR and CCAAT-binding proteins, such as c/EBPβ, for a common binding site as a mechanism for the RA-mediated repression of the hMGP promoter.
DISCUSSION
In this manuscript, we demonstrate that RA functions as a potent inhibitor of MGP gene expression. To assess the physiological relevance of RA regulation, we examined the RA-dependent expression level of the endogenous MGP gene in different cell lines. Importantly, we demonstrate by Northern blot analysis and radioimmunoassays that RA is able to down-regulate MGP gene expression via endogenous receptors present in the cell lines tested. Repression of hMGP gene transcription is further increased by enhanced expression of ligand-activated RAR and RXR. Furthermore, our results show that MGP gene repression is specifically mediated by RAR/RXR, whereas VDR and several other nuclear hormone receptors fail to repress hMGP promoter activity. In this respect, it is interesting to note that induction of F9 cell differentiation, as well as inhibition of F9 cell proliferation, is achieved by saturating concentrations of an RAR-specific ligand, in the absence of an RXR ligand, whereas ligand-mediated activation of both RAR and RXR is required to obtain efficient apoptosis in F9 cells (21). In contrast, repression of hMGP gene activity can be achieved independently by ligand-activated RAR or RXR.
To delineate the cis-regulatory sequences that are responsible for RA-mediated repression, we performed functional analysis with various MGP promoter constructs containing 5′ as well as internal deletions. We identified a novel NRE located between nucleotides −168 and −132 bp. Activity of a mutant promoter, lacking this region, is no longer repressed by ligand-activated RAR/RXR. DNA binding studies, performed with bacterially expressed RAR and RXR, reveal the formation of a specific RAR/RXR/NRE complex. In addition, EMSAs performed with extracts from RA-treated cells demonstrate the formation of a specific NRE/protein complex. The NRE/protein complex is recognized by an α-RXR antibody, clearly demonstrating that RXR, present in NRK52E cells, binds to the NRE. Importantly, the NRE shows no sequence homology to previously identified negative RA response elements like the previously reported NRE in the rat oxytocin promoter (22).
Mechanistically, negative gene regulation mediated by nuclear receptors can be achieved by the competition between receptors and positive transacting factors for common or overlapping binding sites (9, 10). Alternatively, repression might also occur through direct protein–protein interactions between receptors and transcriptional activators. Such protein–protein interactions might interfere with the DNA binding and/or transactivation properties of the transcriptional activator (14). In addition, recent reports describe that the cofactors CBP/p300 directly interact with the ligand binding domain of multiple nuclear receptors. CBP/p300 might serve as a common cofactor for many transcription factors and competition for limiting amounts of CBP/p300 might account for many of the inhibitory actions of RAR and glucocorticoid receptor on AP-1 activity (11, 12).
To better understand the mechanism of RAR-mediated MGP gene repression, deletion mapping studies were used to identify domains in the RAR responsible for repression. Nuclear receptor function depends on a conserved motif within the C terminus, referred to as the AF-2 domain. Several lines of evidence indicate that nuclear receptors such as RAR or TR interact with certain cofactors in an AF-2- and ligand-dependent fashion. These interactions release the binding of corepressors like N-CoR/SMRT and allow the receptors to be transcriptional activators (23, 24). Accordingly, the deletion mutant RAR403*, in which the AF-2 core is lacking, functions as an RA-independent, constitutive repressor. In clear contrast, our data show that RAR403* is unable to repress hMGP promoter activity. Furthermore, overexpression of SMRT does not influence RA-mediated MGP repression (data not shown). In addition, the DBD deletion mutant RARΔDBD, which cannot bind DNA, fails to repress hMGP promoter activity. In summary, our results indicated that the DBD and the AF-2 function of RAR are necessary for repression.
Interestingly, the NRE in the hMGP promoter contains a CCAAT box. To test for mutual binding of RAR/RXR heterodimers or CCAAT-binding factors such as c/EBPβ, we tested the ability of RAR/RXR and c/EBPβ to bind the NRE. DNA binding studies reveal that RAR/RXR and c/EBPβ can recognize this common regulatory sequence in the hMGP promoter. Additionally, our data indicate that RAR/RXR does not interact with c/EBPβ bound to the NRE. Indeed, GST-pulldown and mammalian two-hybrid assays reveal no heteromeric protein–protein interactions between c/EBPβ and RAR/RXR (data not shown). These results suggest that RA-mediated repression of the hMGP gene is dependent on the binding of RAR/RXR to overlapping c/EBPβ binding sites. Consequently, overexpression of c/EBPβ can overcome RA-mediated repression of the hMGP promoter.
Acknowledgments
We thank Drs. Michael Wegner and Karl-Heinz Klempnauer for the generous gift of c/EBPβ expression plasmids and c/EBPβ antibodies and Drs. Rich Heyman and Michael Klaus for the RAR/RXR specific ligands TTNPB and LG100153. We also thank Corina Schüle for providing the art work and administrative assistance and Drs. Heike L. Pahl and Erich F. Greiner for discussion and critical reading of the manuscript. This work was supported in part by Grant AR25921 from the U.S. Public Health Service to P.A.P. and by the Deutsche Forschungsgemeinschaft Grant Schu 688/2-1 to R.S.
ABBREVIATIONS
- RA
retinoic acid
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- VDR
vitamin D receptor
- MGP
matrix Gla protein
- hMGP
human MGP
- LUC
luciferase
- NRE
negative response element
- EMSA
electrophoretic mobility-shift assay
- GST
glutathione S-transferase
- DBD
DNA binding domain
References
- 1.Rannels S R, Cancela M L, Wolpert E B, Price P A. Am J Physiol. 1993;265:L270–L278. doi: 10.1152/ajplung.1993.265.3.L270. [DOI] [PubMed] [Google Scholar]
- 2.Price P A, Urist M R, Otawara Y. Biochim Biophys Res Commun. 1983;117:765–771. doi: 10.1016/0006-291x(83)91663-7. [DOI] [PubMed] [Google Scholar]
- 3.Hale J E, Fraser J D, Price P A. J Biol Chem. 1988;263:5820–5824. [PubMed] [Google Scholar]
- 4.Rice J S, Williamson M K, Price P A. J Bone Miner Res. 1994;9:567–576. doi: 10.1002/jbmr.5650090417. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, O’Bryan J P, Smith H S, Lui E. Oncogene. 1990;5:1391–1395. [PubMed] [Google Scholar]
- 6.Briehl M M, Miesfeld R L. Mol Endocrinol. 1991;5:1381–1388. doi: 10.1210/mend-5-10-1381. [DOI] [PubMed] [Google Scholar]
- 7.Shanahan C M, Weissberg P L, Metcalfe J C. Circ Res. 1993;73:193–204. doi: 10.1161/01.res.73.1.193. [DOI] [PubMed] [Google Scholar]
- 8.Sporn M B, Roberts A B, Goodman D S. The Retinoids. New York: Raven; 1994. pp. 443–520. [Google Scholar]
- 9.Schüle R, Umesono K, Mangelsdorf D J, Bolado J, Pike J W, Evans R M. Cell. 1990;61:497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- 10.Diamond M I, Miner J N, Yoshinaga S K, Yamamoto K R. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 11.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 13.Gorman C M, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfitzner E, Becker P, Rolke A, Schüle R. Proc Natl Acad Sci USA. 1995;92:12265–12669. doi: 10.1073/pnas.92.26.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Z, Umek R M, McKnight S L. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 17.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlmann T, Rangarajan P N, Umesono K, Evans R M. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Otawara Y, Price P A. J Biol Chem. 1986;261:10828–10832. [PubMed] [Google Scholar]
- 21.Clifford J, Chiba H, Sobieszczuk P, Metzger D, Chambon P. EMBO J. 1996;15:4142–4155. [PMC free article] [PubMed] [Google Scholar]
- 22.Lipkin S M, Nelson C C, Glass C K, Rosenfeld M G. Proc Natl Acad Sci USA. 1992;89:1209–1213. doi: 10.1073/pnas.89.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hörlein A J, Näär A M, Heinzel T, Torchia J, Gloss G, Kurokawa R, Ryan A, Kamei Y, Sönderström, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–403. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 24.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]