Abstract
Recently, we reported the isolation, cloning, and expression of a rat enzyme, fatty acid amide hydrolase (FAAH), that degrades bioactive fatty acid amides like oleamide and anandamide to their corresponding acids, thereby serving to terminate the signaling functions of these molecules. Here, we report the molecular characterization of both a mouse and a human FAAH and compare these enzymes to the rat FAAH. The enzymes are well conserved in primary structure, with the mouse and rat FAAHs sharing 91% amino acid identity and the human FAAH sharing 82% and 84% identity with the rat FAAH and mouse FAAH, respectively. In addition, the expressed human and rat FAAHs behave biochemically as membrane proteins of comparable molecular size and show similar, but distinguishable, enzymological properties. The identification of highly homologous FAAH proteins in rat, mouse, and human supports a general role for the fatty acid amides in mammalian biology.
Fatty acid amides represent a growing family of bioactive lipids with diverse cellular and physiological effects (1–5). Members of this molecular family include oleamide, a sleep-inducing lipid originally isolated from the cerebrospinal fluid of sleep-deprived cats (1, 6, 7), and anandamide, an endogenous ligand for the brain CB1 cannabinoid receptor (2). Both oleamide and anandamide possess intriguing physiological and cellular activities. In addition to its sleep-inducing properties in rats, oleamide has also been shown to potentiate the response of 5-HT2 receptors to serotonin (8). Anandamide has documented analgesic effects in rats (2) and, at the cellular and molecular level, has been demonstrated to regulate focal adhesion kinase activity in hippocampal slices (9), inhibit the 5-HT3 ion channel in rat nodose ganglion neurons (10), and arrest development of preimplantation mouse embryos (11). Also, both oleamide and anandamide have been found to block gap junction communication in glial cells (ref. 12 and unpublished observations). Recently, cultured cortical neurons were observed to synthesize and release anandamide along with a variety of other fatty acid ethanolamides in a regulated, calcium-dependent manner (13). The biosynthesis of oleamide and related fatty acid primary amides has been suggested to occur by a distinct route analogous to the biosynthesis of C-terminally amidated peptide hormones (14). In addition to oleamide and anandamide, other fatty acid amides have also been reported to possess biological activities (3–5, 15). In particular, palmitoyl ethanolamide has been shown to bind selectively over anandamide to the peripheral CB2 cannabinoid receptor, indicating perhaps that the CB1 and CB2 receptors recognize distinct fatty acid amides as their respective ligands (5, 15).
The neurophysiological effects of both oleamide and anandamide, in conjunction with the isolation of these compounds from cerebrospinal fluid and brain tissue, respectively, suggest that fatty acid amides may serve important neuromodulatory roles in the central nervous system. However, if neuroactive molecules like the fatty acid amides are to be generally accepted as in vivo participants in brain function, they must first fulfill several criteria (16). One expectation is that the molecules under consideration be closely linked to a mechanism for their expedient inactivation. The fatty acid amides appear to meet this requirement as membrane-bound enzymatic activities from brain tissue have repeatedly been observed to hydrolyze fatty acid amides to their corresponding acids (2, 17–21). A matter of some interest has concerned whether a single enzyme could account for the degradation of multiple fatty acid amides, in particular both oleamide and anandamide (22). Recently, we reported the isolation, cloning, and expression of a fatty acid amide hydrolase (FAAH) from rat liver plasma membranes and demonstrated that the expressed enzyme could indeed hydrolyze both oleamide and anandamide as well as several other fatty acid amides (23). We have since assessed the distribution of this FAAH in the rat central nervous system by in situ hybridization and have identified prominent FAAH expression in a variety of neuronal cells (E. A. Thomas, B.F.C., P. E. Danielson, N. B. Gilula, and J. G. Sutcliffe, unpublished work). We now describe the use of the rat FAAH cDNA in hybridization screening for the cloning of both human and mouse FAAHs from their respective liver cDNA libraries. The three FAAH cDNAs were comparatively characterized, and the human and rat FAAH enzymes were expressed in COS-7 cells for further enzymological and biochemical analyses.
EXPERIMENTAL PROCEDURES
Cloning of Mouse and Human FAAH cDNAs.
An 800-bp fragment of the rat FAAH cDNA was internally radiolabeled with [α-32P]CTP (Multiprime DNA Labeling kit, Amersham) and used as a probe to screen mouse and human liver 5′ Stretch Plus cDNA libraries (CLONTECH); screens and phage DNA isolation were conducted according to the manufacturer’s guidelines, except in the case of the human library, for which hybridization was conducted with 25% formamide rather than 50% formamide and 0.2× standard saline citrate washes were conducted at 50°C rather than 60°C. Five positive mouse clones were identified from a screening of 6 × 104 plaques, and two positive human clones were identified from a screening of 1.0 × 105 plaques. Two distinct 1.7-kb mouse clones were combined to provide a sequence covering the complete coding region for the mouse FAAH cDNA but lacking a portion of the 3′ untranslated region. A single 2.1-kb human clone provided the full-length human FAAH cDNA terminating in a poly(A) stretch. The mouse and human FAAH cDNAs were cloned into pBluescript II SK(+), and both strands of each clone were sequenced completely.
Expression Studies with Rat and Human FAAH in COS-7 Cells.
COS-7 cells were transiently transfected with either the human or the rat FAAH cDNA in the eukaryotic expression vector pcDNA3 (Invitrogen) as described (23). Cells were detached from their dishes with a cell scraper, washed twice in Hepes buffer (1 mM EDTA/100 mM NaCl/12.5 mM Hepes, pH 8.0), and Dounce-homogenized in Hepes buffer. FAAH enzymatic assays and Western blotting were conducted as described (23). Fractionation of the COS-7 cell extract into soluble and membrane fractions was conducted as follows. The cell homogenate was spun at 600 × g for 5 min and the supernatant was transferred to a Beckman airfuge and spun at 30 psi for 1 hr. The supernatant (soluble fraction) was removed, and the pellet (membrane fraction) was resuspended in Hepes buffer. The protein concentrations of each fraction were quantitated by Dc Protein Assay (Bio-Rad). Protein from each fraction was used for enzymatic assays and Western blotting (3.7 and 5.0 μg of protein, respectively).
Immunofluorescence Studies with FAAH-Transfected COS-7 Cells.
COS-7 cells were grown on 0.1% gelatinized 11 × 22 mm coverslips to 70% confluency and transfected as previously reported (23). The coverslips were washed in PBS and then fixed in 100% methanol for 5 min at −20°C. After fixation, the coverslips were washed in PBS, and a blocking solution (5% BSA in PBS) was applied for 1 hr at 25°C. The blocking solution was removed, and the coverslips were incubated with anti-FAAH rabbit polyclonal antibodies in blocking solution for 1 hr at 25°C. The anti-FAAH antibody solution was removed with PBS, and fluorescein isothiocyanate-labeled anti-rabbit goat antibody was applied in blocking solution for 1 hr at 25°C. The goat antibody was removed with PBS, and the coverslips were mounted on microscope slides with moviol.
Substrate Selectivity Analysis of Human and Rat FAAH.
14C-fatty acid amides were synthesized from their purchased acids (DuPont/NEN and Moravek Biochemicals, Brea, CA) as described (2, 23). FAAH activity was assayed in duplicate with 100 μM substrate (50 mCi/mmol; 1 Ci = 37 GBq) and 10 μg of COS-7 cell protein extract for 15 min at 37°C in 125 mM Tris·HCl, pH 9.0 (except in the case of stearamide, where owing to low solubility, 20 μM substrate was used). Reactions were worked up and analyzed as described (2, 23). Substrate hydrolysis in the presence of equal amounts of untransfected COS-7 cell protein served as a background control in all cases and was subtracted from transfected FAAH hydrolysis rates. Inhibitor studies were conducted in a similar manner with the addition of 10 μM 19-hydroxy-1,1,1-trifluoro-10(Z)-nonadecen-2-one (CF3-OH) to the reaction mixture just before enzyme addition.
Southern and Northern Blot Analyses of Human FAAH cDNA.
For Southern blotting, 4-μg samples of human genomic DNA (CLONTECH) were digested with the indicated restriction enzymes (100 units each) for 12 hr and then run on a 0.8% agarose gel. The DNA was transferred with a TurboBlotter downward capillary transfer system (Schleicher & Schuell) to a nylon membrane for use in Southern blot analysis. The blot was probed with a 1.0-kb fragment of the human FAAH cDNA and handled according to the manufacturer’s guidelines. Northern blot analysis was conducted using a Human MultiTissue Northern blot (CLONTECH) which was probed with the human FAAH cDNA fragment and handled according to the manufacturer’s guidelines.
RESULTS AND DISCUSSION
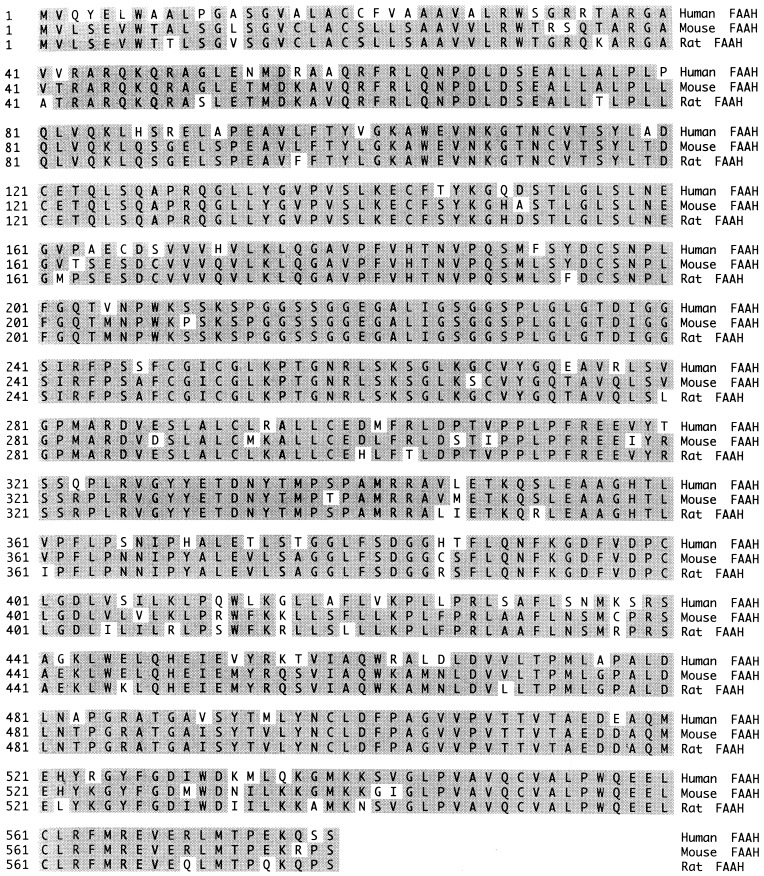
An 800-bp fragment of the rat FAAH cDNA was used as a probe for hybridization screening of mouse and human liver cDNA libraries. Two overlapping 1.7-kb positive clones isolated from the mouse library together encoded a complete ORF sequence for a mouse FAAH, whereas a single 2.1-kb clone isolated from the human library contained a complete ORF sequence for a human FAAH. Only the human FAAH cDNA ended in a poly(A) stretch, indicating that the mouse FAAH cDNA sequence lacked a portion of the 3′ untranslated region of the mouse FAAH gene. Comparison of the deduced amino acid sequences from the mouse and human FAAH ORFs to the rat FAAH sequence demonstrated that the three FAAH enzymes were quite homologous in primary structure (Fig. 1). The mouse and rat FAAHs shared 91% sequence identity, whereas the human FAAH shared 82% and 84% sequence identity with the rat and mouse FAAHs, respectively. In addition, all three FAAHs were 579 aa long with predicted molecular masses of 63.0, 63.4, and 63.2 kDa for the human, rat, and mouse FAAHs, respectively. Each FAAH possessed a putative transmembrane domain from amino acids 9–29 (tmpred program) and a completely conserved amidase consensus sequence from amino acids 215–246 (24). Finally, the putative Src homology 3-binding domain sequence, PPLPXR (25), identified previously in the rat FAAH (amino acids 310–315; see ref. 23) was completely conserved in the human and mouse FAAH sequences.
Figure 1.
Comparison of deduced amino acid sequences from human, mouse, and rat FAAH cDNAs. Sequence identity shared by at least two of the three FAAH proteins is shaded.
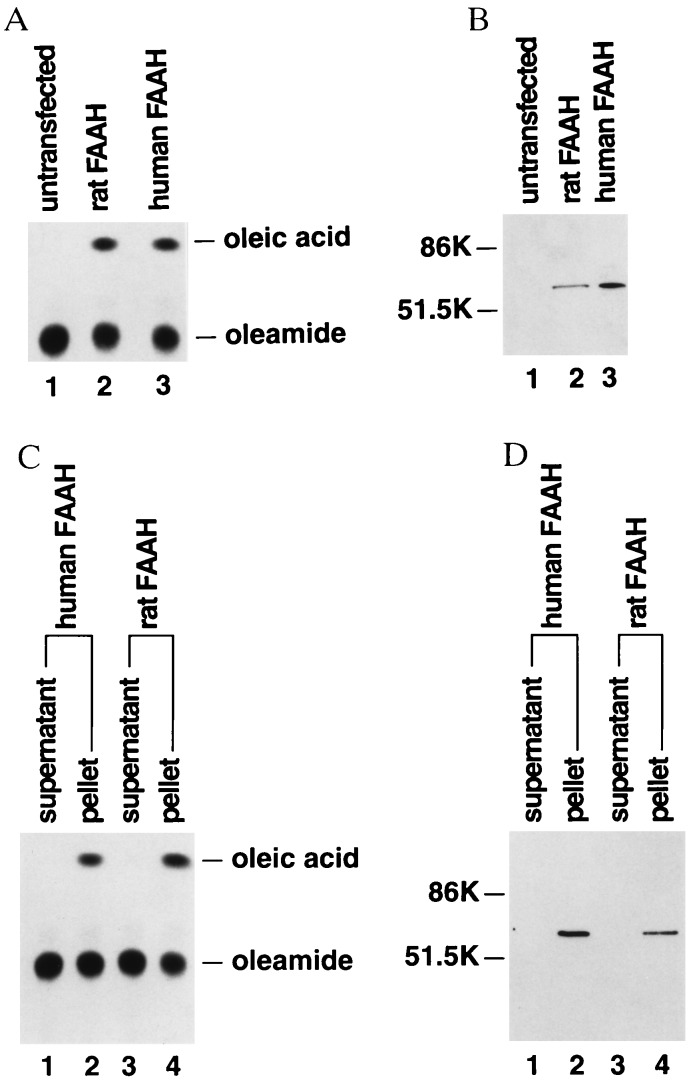
To investigate further the properties of the human and rat FAAHs, their respective cDNAs were cloned into the eukaryotic expression vector pcDNA3 and transiently transfected into COS-7 cells. COS-7 cells transfected with either the human or the rat FAAH cDNA expressed high levels of oleamide hydrolase activity, whereas untransfected COS-7 cells did not effectively hydrolyze oleamide (Fig. 2A). Western blotting of COS-7 cell extracts with polyclonal antibodies generated against the well conserved FAAH sequence VGYYETDNYTMPSPAMR identified a single 60- to 65-kDa protein in both rat and human FAAH-transfected COS-7 cell extracts but not in untransfected COS-7 cell extract (Fig. 2B). Given that the FAAH enzymes were predicted integral membrane proteins, further biochemical fractionation of the COS-7 cell extracts was conducted to determine whether the expressed FAAH enzymes associated with membranes. High speed centrifugation separated the soluble and membrane fractions of COS-7 cell extracts, and each fraction was assayed for FAAH activity. Both rat and human FAAH enzyme activities completely segregated with the pelleted membrane fraction of COS-7 cell extracts (Fig. 2C), indicating that the expressed enzymes were indeed attached to membranes. Likewise, Western blotting identified the FAAH protein exclusively present in these same membrane fractions (Fig. 2D).
Figure 2.
Expression and comparison of human and rat FAAHs in COS-7 cells. (A) FAAH activity as measured by oleamide hydrolysis was present in COS-7 cells transiently transfected with either rat FAAH or human FAAH (lanes 2 and 3, respectively) but not in untransfected COS-7 cells (lane 1). (B) Western blot analysis of COS-7 cell extracts with polyclonal anti-FAAH antibodies (see text) identified a single immunoreactive 60- to 65-kDa protein present in COS-7 cells transfected with either rat or human FAAH (lanes 2 and 3, respectively) but not in untransfected COS-7 cells (lane 1). (C and D) Association of FAAH activity and immunoreactivity (lanes 1 and 2, rat FAAH; lanes 3 and 4, human FAAH) with COS-7 cell membranes upon cellular fractionation (supernatant, soluble protein fraction; pellet, membrane protein fraction).
Preliminary immunofluorescence studies with COS-7 cells transiently transfected with either the human or the rat FAAH cDNA showed a surprising difference in the apparent cellular distribution of the two proteins (Fig. 3). In COS-7 cells expressing the rat FAAH, a relatively diffuse staining pattern was detected, with the highest intensity in regions near, but not including, the nucleus (Fig. 3 C′ and D′). This staining is most compatible with predominate association of the rat FAAH protein with the endoplasmic reticulum and/or Golgi apparatus. In contrast, in COS-7 cells expressing the human FAAH, a staining pattern consisting of intense, fibrous or rod-like structures was observed (Fig. 3 A′ and B′). These fibrous arrays were usually disorganized throughout the cell body of the cells and also appeared to organize around the nucleus in several cases. Given that the human FAAH behaved as a membrane protein in our biochemical assays (Fig. 2), it is perhaps intriguing to speculate that this enzyme may somehow be involved in attaching the intracellular membranes of the COS-7 cells to a portion of the cytoskeletal network. Along this line, a membrane-bound form of glutamate dehydrogenase was recently reported to mediate the association of lysosomes to microtubules (26). Whether the different apparent distributions of the rat and human FAAHs can be attributed to fundamental differences in the proteins themselves or simply to distinct expression levels of the two proteins is unclear at this time. However, in this regard, transient transfection experiments usually provide a population of cells with various protein expression levels, and although COS-7 cells expressing very low levels of the human FAAH did appear void of the aforementioned fibrous structures, COS-7 cells expressing the rat FAAH were never found to contain such immunoreactive fibrous structures regardless of their level of FAAH expression.
Figure 3.
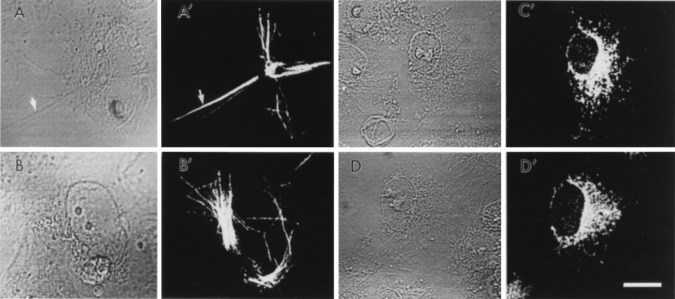
Immunofluorescence microscopy of COS-7 cells transfected with either human FAAH (A and B, cell phase; A′ and B′, anti-FAAH antibody staining) or rat FAAH (C and D, cell phase; C′ and D′, anti-FAAH antibody staining). Note the fibrous, rod-like staining pattern of the human FAAH (arrow) and, in contrast, the more diffuse staining pattern of the rat FAAH. No significant immunostaining was observed in untransfected COS-7 cells, and both the rat and the human FAAH immunoreactivities were effectively competed away by excess peptide antigen.
The expressed rat FAAH has been demonstrated to hydrolyze a variety of fatty acid amides with a selectivity dependent on both chain length and degree of unsaturation of the substrate (23). The substrate preference of the expressed human FAAH was therefore examined and compared with that of the rat FAAH (Table 1). The human and rat FAAHs shared a similar qualitative substrate selectivity, with anandamide, oleamide, and myristic amide serving as the preferred substrates, and palmitic and stearic amide being hydrolyzed at significantly lower rates. However, the human FAAH did appear quantitatively somewhat less selective than the rat FAAH as it hydrolyzed myristic amide at a rate comparable to oleamide, and palmitic amide at a rate only 2-fold slower than oleamide. Still, both FAAH enzymes were quite sensitive to the degree of unsaturation of the substrate, as exemplified by their nearly 20-fold preference for oleamide over stearic amide, two fatty acid amides that differ only by the presence of a single degree of unsaturation in oleamide.
Table 1.
Substrate selectivity of human FAAH expressed in COS-7 cells and comparison to rat FAAH
| Substrate | Human FAAH rate of hydrolysis, nmol·min−1·mg−1 | Human FAAH % hydrolysis rate | Rat FAAH % hydrolysis rate |
|---|---|---|---|
| Anandamide, 100 μM | 30.7 ± 1.5 | 100 | 100 |
| Oleamide, 100 μM | 21.5 ± 0.9 | 70 | 73 |
| Myristic amide, 100 μM | 19.8 + 1.4 | 65 | 24 |
| Palmitic amide, 100 μM | 10.2 ± 0.4 | 33 | 10 |
| Oleamide, 20 μM | 5.5 ± 0.2 | 100 | 100 |
| Stearic amide, 20μM | 0.32 + 0.01 | 5.8 | 5.8 |
FAAH activity measured in transfected COS-7 cells with various 14C-fatty acid amides as substrates. Anandamide and oleamide hydrolysis rates were considered to be 100% of FAAH activity where indicated, to which other fatty acid amide hydrolysis rates were compared. Previously measured relative rates of fatty acid amide hydrolysis for the rat FAAH (23) are listed in the final column for comparison.
Trifluoromethyl ketone derivatives of oleamide have previously been shown to potently inhibit the rat FAAH (23, 27). Therefore, the relative sensitivities of the human and rat FAAHs to the trifluoromethyl ketone inhibitor CF3-OH were examined (Table 2). Both FAAH enzymes were inhibited by CF3-OH to remarkably similar degrees, supporting further the notion that the human and rat FAAHs share several common enzymological features.
Table 2.
Inhibition of rat and human FAAH by CF3-OH
| Enzyme | Rate of hydrolysis, nmol·min−1·mg−1 | % |
|---|---|---|
| Rat FAAH | 30.7 ± 1.5 | 100 |
| Rat FAAH + CF3-OH | 5.2 + 0.8 | 16.8 |
| Human FAAH | 21.5 + 0.9 | 100 |
| Human FAAH + CF3-OH | 4.3 + 0.5 | 19.8 |
Relative inhibition of rat and human FAAH activity by 10 μM CF3-OH. Oleamide (100 μM) was used as substrate in each assay.
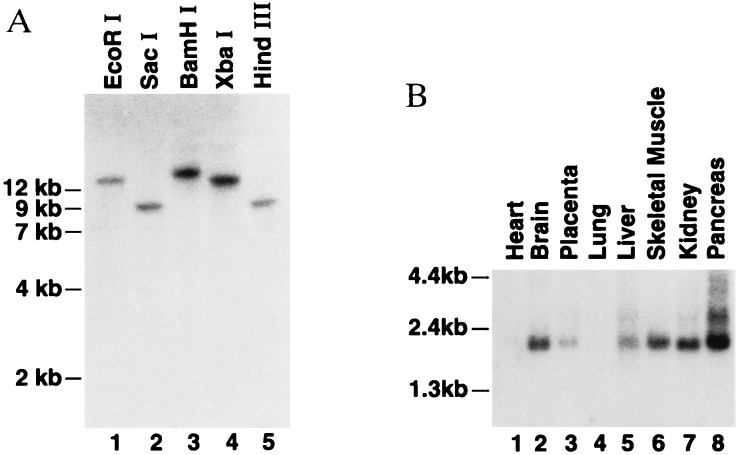
Taken as a whole, this preliminary biochemical and enzymological comparison of the rat and human FAAH proteins suggests that the essential features of the FAAH enzyme have been well conserved from rat to human. Another means of comparing the human and rat FAAHs would be with regards to their respective gene copy number and tissue distribution. Therefore, Southern and Northern blot analyses of human FAAH were conducted using a 1.0-kb fragment from the human FAAH cDNA as a probe. Southern blots indicated that the human FAAH probe hybridized to single DNA fragments in several different restriction enzyme digests of the human genome (Fig. 4A). These results are similar to those obtained from Southern analysis of the rat FAAH (23) and are most consistent with the notion that the human FAAH, like the rat FAAH, is derived from a single copy gene. Northern blot analysis with the human FAAH cDNA probe and mRNA from various human tissues identified a single major mRNA transcript of approximately 2.1 kb that was most abundant in pancreas, brain, kidney, and skeletal muscle, with lesser amounts present in liver and placenta. This transcript was not detected in either heart or lung. The tissue distribution of the human FAAH message was markedly distinct from the distribution of the rat FAAH transcript (23), which was most abundant in liver and was not detected in skeletal muscle. Still, both the human and the rat FAAH transcripts were expressed at strong levels in brain and appeared absent from heart tissue, indicating that the human and rat FAAH transcripts also shared significant features of their tissue distribution profiles.
Figure 4.
Southern (A) and Northern (B) blot analyses of human FAAH. (A) Southern blot of human genomic DNA digested with several restriction enzymes (4 μg of digested DNA per lane); restriction enzymes used are indicated at the top of each lane. (B) Northern blot of human mRNA from different human tissues (CLONTECH; 2 μg of mRNA per lane).
Given the increasing number of reports documenting biological activities for the fatty acid amides (1–12), a detailed understanding of the means by which these lipids are degraded in vivo has become imperative. The cloning of mouse and human FAAH enzymes described herein, in conjunction with previous work detailing the cloning of a rat FAAH (23), has allowed for the present molecular comparison of FAAHs from three different organisms. The strong degree of sequence homology shared by the rat, mouse, and human FAAHs suggests that the enzymatic mechanism for fatty acid amide inactivation has been well conserved throughout mammalian evolution. In further support of this notion, the expressed rat and human FAAHs were found to share many biochemical and enzymological features, including membrane association, apparent molecular size, substrate selectivity, and inhibitor sensitivity. On the other hand, the rat and human FAAHs were distinguishable in terms of their immunostaining in COS-7 cells and their respective tissue distributions as judged by Northern blot analysis. A more detailed investigation into the unique and common features of the rat, mouse, and human FAAH enzymes is presently underway, with a fundamental goal being a deeper understanding of the mechanisms by which fatty acid amides exert their profound biological effects.
Acknowledgments
We thank Dr. K. Sullivan and G. Klier for their assistance and advice regarding immunofluorescence; G. Klier for his assistance with Fig. 3; and Drs. D. Boger, N. Gilula, and R. Lerner for their advice and insights. This work was supported by the Lucille P. Markey Charitable Trust and The B. G. Corbin Foundation for Molecular Biology Research.
ABBREVIATIONS
- FAAH
fatty acid amide hydrolase
- CF3-OH
19-hydroxy-1,1,1-trifluoro-10(Z)-nonadecen-2-one
Footnotes
References
- 1.Cravatt B F, Prospero-Garcia O, Siuzdak G, Gilula N B, Henriksen S J, Boger D L, Lerner R A. Science. 1995;268:1506–1509. doi: 10.1126/science.7770779. [DOI] [PubMed] [Google Scholar]
- 2.Devane W A, Hanus L, Breuer A, Pertwee R G, Stevenson L A, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 3.Facci L, Dal Toso R, Romanello S, Buriani A, Skaper S D, Leon A. Proc Natl Acad Sci USA. 1995;92:3376–3380. doi: 10.1073/pnas.92.8.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wakamatsu K, Masaki T, Itoh F, Koichi K, Sudo K. Biochem Biophys Res Commun. 1990;168:423–429. doi: 10.1016/0006-291x(90)92338-z. [DOI] [PubMed] [Google Scholar]
- 5.Barg J, Fride E, Hanus L, Levy R, Matus-Leibovitch N, Heldman E, Bayewitch M, Mechoulam R, Vogel Z. Eur J Pharmacol. 1995;287:145–152. doi: 10.1016/0014-2999(95)00487-4. [DOI] [PubMed] [Google Scholar]
- 6.Lerner R A, Siuzdak G, Prospero-Garcia O, Henriksen S J, Boger D L, Cravatt B F. Proc Natl Acad Sci USA. 1994;91:9505–9508. doi: 10.1073/pnas.91.20.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cravatt B F, Lerner R A, Boger D L. J Am Chem Soc. 1996;118:580–590. [Google Scholar]
- 8.Huidobro-Toro J P, Harris R A. Proc Natl Acad Sci USA. 1996;93:8078–8082. doi: 10.1073/pnas.93.15.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derkinderen P, Toutant M, Burgaya F, Le Bert M, Siciliano J C, de Franciscis V, Gelman M, Girault J-A. Science. 1996;273:1719–1722. doi: 10.1126/science.273.5282.1719. [DOI] [PubMed] [Google Scholar]
- 10.Fan P. J Neurophysiol. 1995;73:907–910. doi: 10.1152/jn.1995.73.2.907. [DOI] [PubMed] [Google Scholar]
- 11.Paria B C, Das S K, Dey S K. Proc Natl Acad Sci USA. 1995;92:9460–9464. doi: 10.1073/pnas.92.21.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venance L, Piomelli D, Glowinski J, Giaume C. Nature (London) 1995;376:590–594. doi: 10.1038/376590a0. [DOI] [PubMed] [Google Scholar]
- 13.Di Marzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz J-C, Piomelli D. Nature (London) 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 14.Merkler D J, Merkler K A, Stern W, Fleming F F. Arch Biochem Biophys. 1996;330:430–434. doi: 10.1006/abbi.1996.0272. [DOI] [PubMed] [Google Scholar]
- 15.Skaper S D, Buriani A, Dal Toso R, Petrelli L, Romanello S, Facci L, Leon A. Proc Natl Acad Sci USA. 1996;93:3984–3989. doi: 10.1073/pnas.93.9.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel G J, Agranoff B W, Albers R W, Molinoff P B. Basic Neurochemistry. New York: Raven; 1994. pp. 182–183. [Google Scholar]
- 17.Deutsch D G, Chin S A. Biochem Pharmacol. 1993;46:791–796. doi: 10.1016/0006-2952(93)90486-g. [DOI] [PubMed] [Google Scholar]
- 18.Desarnaud F, Cadas H, Piomelli D. J Biol Chem. 1995;270:6030–6035. doi: 10.1074/jbc.270.11.6030. [DOI] [PubMed] [Google Scholar]
- 19.Hillard C J, Wilkison D M, Edgemond W S, Campbell W B. Biochim Biophys Acta. 1995;1257:249–256. doi: 10.1016/0005-2760(95)00087-s. [DOI] [PubMed] [Google Scholar]
- 20.Ueda N, Kurahashi Y, Yamamoto S, Tokunaga T. J Biol Chem. 1995;270:23823–23827. doi: 10.1074/jbc.270.40.23823. [DOI] [PubMed] [Google Scholar]
- 21.Omeir R L, Chin S, Hong Y, Ahern D G, Deutsch D G. Life Sci. 1995;56:1999–2005. doi: 10.1016/0024-3205(95)00181-5. [DOI] [PubMed] [Google Scholar]
- 22.Maurelli S, Bisogno T, De Petrocellis L, Di Luccia A, Marino G, Di Marzo V. FEBS Lett. 1995;377:82–86. doi: 10.1016/0014-5793(95)01311-3. [DOI] [PubMed] [Google Scholar]
- 23.Cravatt B F, Giang D K, Mayfield S M, Boger D L, Lerner R A, Gilula N B. Nature (London) 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 24.Mayaux J-F, Cerbelaud E, Soubrier F, Faucher D, Petre D. J Bacteriol. 1990;172:6764–6773. doi: 10.1128/jb.172.12.6764-6773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng S, Chen J K, Yu H, Simon J A, Schreiber S L. Science. 1994;266:1241–1246. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 26.Rajas F, Gire V, Rousset B. J Biol Chem. 1996;271:29882–29890. doi: 10.1074/jbc.271.47.29882. [DOI] [PubMed] [Google Scholar]
- 27.Patterson J E, Ollmann I R, Cravatt B F, Boger D L, Wong C-H, Lerner R A. J Am Chem Soc. 1996;118:5938–5945. [Google Scholar]