Abstract
Translation initiation of the hepatitis C virus (HCV) RNA genome occurs through an internal ribosome entry site in a cap-independent manner. Here, we have examined the interaction between La antigen and the HCV 5′ noncoding region (5′NCR). In this analysis, competitor RNAs derived from HCV 5′NCR carrying deletions and a point mutation were used to identify the site(s) of La antigen binding during UV cross-linking assay. These studies suggest that La antigen recognizes the intact HCV 5′NCR structure. Further, these interactions occurred in the context of the initiator AUG. The latter view is supported by an analysis in which mutants of the HCV 5′NCR RNA with deletion or substitution in the initiator AUG codon failed to compete for La antigen binding to the wild-type 5′NCR. The evidence for the interaction between liver cell-derived La antigen and the HCV 5′NCR is provided by immunoprecipitation of a UV cross-linked species from the S100 fraction of Huh7 cell lysates. The functional relevance of this interaction was demonstrated by the stimulation of the HCV internal ribosome entry site-mediated translation in the presence of La protein. These results suggest an important functional role of La protein in the regulation of internal initiation of translation of the HCV RNA genome.
Keywords:
Human hepatitis C virus (HCV) causes chronic infection of the liver and has been linked to the development of hepatocellular carcinoma (1, 2). Based on genomic organization, biochemical properties, and molecular features, HCV has been classified in a separate genera of the family Flaviviridae (3, 4). The viral genome consists of a single-stranded, positive-sense RNA molecule of 9.4 kb. The 5′ noncoding region (5′NCR), which varies in length from 332 to 341 nucleotides, is followed by a long open reading frame encoding a polyprotein of about 3,000 amino acids that is processed into functionally active structural and nonstructural proteins. A relatively short noncoding region is located at the 3′ terminus (5). While there is considerable nucleotide heterogeneity among the clinical isolates of HCV, the 5′NCR displays a high degree of conservation (6).
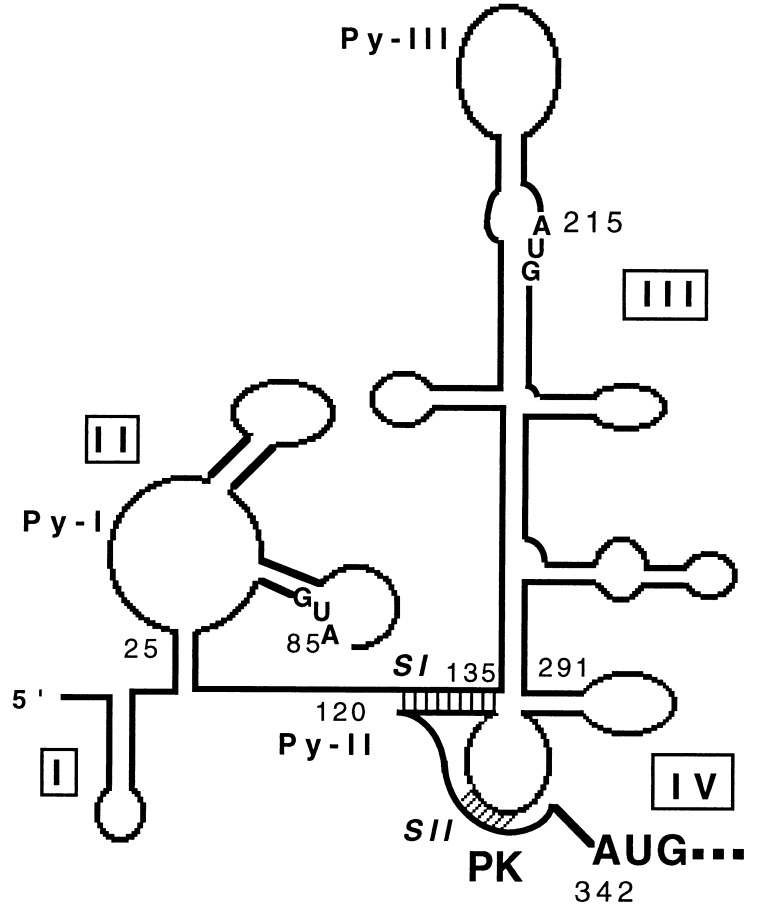
Translation by internal ribosome entry was first recognized as a scheme of translation initiation that was unique to picornaviral mRNA, but recently other viral and cellular mRNAs have been identified that use a similar translational strategy (7–9). Among other viruses, the RNA genomes of HCV and bovine viral diarrhea virus, another member of Flaviviridae, have been shown to contain an internal ribosome entry site (IRES) (10–16). Based on computer-assisted RNA folding and enzymatic probing, Brown et al. (17) proposed a model of the HCV 5′NCR. Consistent with the characteristic features of picornavirus IRES elements (18), the HCV 5′NCR contains multiple AUG codons and oligopyrimidine motifs (Fig. 1).
Figure 1.
Schematic representation of computer-generated RNA folding model as proposed by Brown et al. (17) with a modification in the vicinity of initiator AUG according to Wang et al. (19). The stem I (SI) and stem II (SII) of the pseudoknot (PK) structure are shaded.
Cellular transacting factors distinct from the canonical initiation (eIF) factors have been implicated in facilitating IRES-regulated translation (20). Among cellular proteins, polypyrimidine-tract binding protein and the La antigen (p52 or SS-B) are recognized for their important functional roles in stimulating IRES-mediated translation (see review ref. 21). The La protein was originally identified as an autoantigen that was recognized by sera from patients with systemic lupus erythematosus and Sjogren syndrome (see review ref. 22). La antigen binds to a variety of RNA structures via its RNA binding motif (23). The viral targets of the La protein include the 5′NCR of picornaviruses (18, 24), influenza virus (25), sindbis virus (26), and the HIV TAR element (27). In the case of poliovirus, the La protein plays a functional role in internal initiation of translation (28), where addition of purified La to rabbit reticulocyte lysates (RRL) inhibits the accumulation of aberrant translation products. This activity is accompanied by a modest stimulation of translation (28, 29). La binding to the HIV TAR structure alleviates the translational repression exerted by TAR on the downstream reporter gene (30). These studies together support the role of La antigen in the translational regulation of viral mRNAs.
In this report, we investigated the possible interaction of La with the 5′NCR of the HCV RNA genome and its role in HCV IRES-dependent translation. Using a combination of UV cross-linking and competition assays, our studies demonstrate the interaction between La antigen and the HCV 5′NCR. While efficient binding by La antigen appears to require the intact 5′NCR, the specificity of this interaction is directed toward 3′ end of the 5′NCR. These studies further show that these interactions occurred in the context of the initiator AUG. Finally, the data presented here supports a functional role of La antigen during IRES-mediated translation of the HCV RNA in vitro.
MATERIALS AND METHODS
Construction of Plasmid DNA.
The construction of recombinant plasmids were carried out by standard protocols. The plasmid GEM5′NC contains the full-length 5′NCR of HCV-I subtype (11). The plasmid pNCR-C(AUG) is derived from GEM5′NC, but contains additional 12 nucleotides of the HCV core region. To construct this plasmid, antisense primer, NCR-C/AS/AUG (5′-GAGGAATTCAGGATTCGTGCTCATGGTGCACGG-3′), and T7 promoter primer (Promega) were used to PCR amplify from GEM5′NC DNA template. The amplified product was digested with HindIII and EcoRI, gel purified, and cloned in pGEM-4 at HindIII–EcoRI sites. The nucleotide sequence in these plasmids were confirmed by dideoxyribonucleotide sequencing method. The plasmid pNCR-C(AAG) is similar to pNCR-C(AUG) except that it contains a substitution (U to A) at nucleotide 343. The construction of plasmids, pNCR135–291, pT7Δ152–278, and pT7CΔ1–75/Δ142–268 has been previously described (31, 32).
In Vitro Transcription.
RNA transcripts were synthesized in vitro from linearized plasmid DNA that was purified by elution of the desired fragments from the agarose gels after digestion with an appropriate restriction endonuclease. The wild-type HCV 5′NCR RNA (NCR1–341) was transcribed from GEM5′NC DNA after digestion with EcoRI. NCR(-AUG) was transcribed from GEM5′NC, which was linearized by BspHI and digested with mung bean nuclease. To transcribe NCR(PKS) RNA, GEM5′NC digested with BspHI was used as template. NCR-C(AUG) and NCR-C(AAG) were transcribed from their respective plasmids linearized with EcoRI. Deletion mutants NCR1–83 or NCR1–131 were generated from GEM5′NC linearized by NcoI or SmaI, respectively. Plasmid pNCR135–291 was linearized by BamHI, and T7CΔ152–278 and T7CΔ1–75/Δ142–268 were digested with XbaI. T7LUC was digested with XbaI and transcribed to produce T7LUC(1–99) RNA. The monocistronic (T7C1–341) and dicistronic (T7DC1–341) RNAs were linearized by HpaI (11). All the linear DNAs were transcribed using T7 RNA polymerase under standard conditions. The radioactive RNA probes were synthesized under similar reaction conditions with 4-thio-UTP and [α-32P]CTP.
Purification of La Antigen.
A cDNA encoding the human La antigen in the pET-8c expression vector was a generous gift of G. J. M. Pruijn (University of Nijmegen, Nijmegen, The Netherlands). The expression of La antigen was induced by 1 mM isopropyl β-d-thiogalactoside in Escherichia coli [BL21(DE3)] cells. The cells were lysed after 5–6 h by repeated freeze-thaw, followed by sonication in buffer A [25 mM Tris·HCl, pH 8.0/75 mM NaCl/1 mM EDTA/1 mM DTT/0.2 mM phenylmethylsulfonyl fluoride (PMSF)/1 μM leupeptin]. The La protein was partially purified by DEAE-cellulose column chromatography. La protein-containing fractions were eluted between 150 and 200 mM NaCl in buffer A and dialyzed against buffer B [20 mM Hepes, pH 7.6/0.2 mM EDTA/0.5 mM DTT/0.1 M KCl/2 mM MgCl2/10% (vol/vol) glycerol/1 μM leupeptin/0.2 mM PMSF]. The sample was then mixed with poly(U)-Sepharose 4B for 2 h at 4 C and washed five times with the same buffer at 0.5 M KCl. The bound protein was eluted at 1.0 M KCl under similar conditions and dialyzed against buffer D [5 mM Hepes, pH 7.6/25 mM KCl/1 mM EDTA/1 mM DTT/10% (vol/vol) glycerol/0.2 mM PMSF/1 μM leupeptin].
UV Cross-Linking of Proteins with RNA.
4-Thio-UDP (Sigma) was phosphorylated with nucleoside 5′-diphosphate kinase to prepare 4-thio-UTP (33). RNA probes synthesized in the presence of 4-thio-UTP and [α-32P]CTP were UV cross-linked with protein samples in RNA binding buffer (buffer D plus 2 mM MgCl2) as described previously (32). For all the competition assays, competitor RNAs were added along with the components of the reaction mixture before UV cross-linking. The ribonucleoprotein complexes were treated with RNase A (10–20 units) (United States Biochemical) and analyzed by sodium dodecyl sulfate/polyacrylamide (12%) gel electrophoresis (SDS/PAGE) followed by autoradiography.
Immunoprecipitation of Huh7 La Antigen-HCV 5′NCR Complex.
S100 cytoplasmic fraction from cultured Huh7 cells were prepared essentially as described by Dignam (34). S100 protein fraction (150 μg) maintained in RNA binding buffer was mixed with full-length HCV 5′NCR RNA probe in a total volume of 200 μl. After UV cross-linking and ribonuclease treatment, the sample was diluted to 500 μl with NETS buffer (50 mM Tris·HCl, pH 7.4/5 mM EDTA/1 mM DTT/100 mM NaCl/0.05% Nonidet P-40) and mixed with monoclonal anti-La antibody (SW5). The immunocomplexes were immobilized on protein A-Sepharose 4B beads saturated with anti-mouse IgG antibodies (Sigma). The unbound materials were washed five times with the same buffer. The bound protein(s) were analyzed by SDS/PAGE followed by autoradiography. A parallel reaction mixture was performed with normal serum and served as a control. Immunoprecipitation of the recombinant La-5′NCR complex was carried out as described above.
RESULTS
Mapping of La Antigen Binding Sites Within the HCV 5′NCR.
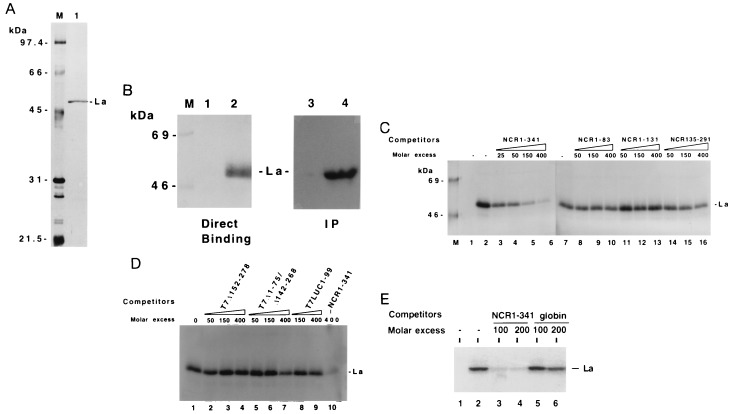
The human La antigen expressed in bacteria was purified by DEAE cellulose column chromatography followed by poly(U)-Sepharose affinity chromatography. An aliquot of purified La antigen was analyzed by SDS/PAGE, which revealed a single protein band migrating at an approximate molecular weight of 52,000 (Fig. 2A, lane 1). To determine RNA binding activity of La antigen in the purified fraction, the La preparation was UV cross-linked with thio-U containing 32P-labeled wild-type HCV 5′NCR RNA (NCR1–341). The result of the UV cross-linking experiment revealed a single band of radiolabeled RNA–protein complex that was fractionated on a denaturing polyacrylamide gel at the expected size of La antigen (Fig. 2B, lane 2). This RNA protein complex was recognized by a monoclonal anti-La antibody, SW5, (lane 4) during an immunoprecipitation assay. These results authenticated the La preparation and its UV cross-linking ability to the HCV 5′NCR. To identify the region(s) of HCV 5′NCR that interact with La antigen, a series of deletion and substitution mutants of the 5′NCR were used as unlabeled competitor RNAs during UV cross-linking experiments. First, a homologous competitor RNA (NCR1–341) was used during UV cross-linking assay. The intensity of binding signal due to cross-linking of La with the probe progressively decreased at increasing concentrations of the unlabeled homologous competitor RNA (Fig. 2C, lanes 3–6), thus confirming the specificity of La-5′NCR cross-linking. Next, three truncated RNAs derived from NCR1–341, all of which represent defined structural domains of the 5′NCR, were used in the competition assay. While the NCR1–83 and NCR1–131 RNAs are derived from the 5′-terminal sequences, NCR135–291 RNA represents the entire structural domain III of the 5′NCR. Both NCR1–83 and NCR1–131 competitor RNAs were unable to compete for the La binding to the RNA probe (lanes 8–13). These RNA fragments also failed to bind La antigen in a direct UV cross-linking assay (data not shown). The unlabeled NCR135–291 RNA competed to a negligible extent for La binding to the 5′NCR at a higher molar excess (lanes 14–16). Further competition experiments were carried out with the 5′NCR mutants devoid of domain III structure (Fig. 1). The mutant T7Δ152–278 RNA lacks the upper portion of domain III, including the apical loop containing Py-III motif. The mutant T7Δ1–75/Δ142–268 is similar to T7Δ152–278 except that it further lacks 75 nucleotides from the 5′ end. Both mutant RNAs did not compete for La binding (Fig. 2D, lanes 2–7). Similar results were obtained with an luciferase RNA fragment (nucleotides 1–99) that was synthesized from T7LUC plasmid (lanes 8 and 9). In a similar competition assay, rabbit globin mRNA, which does not contain an IRES element and is translated by a cap-dependent mechanism, was used as a control during the competition assay. While the binding of La to the probe was competed by the HCV 5′NCR RNA (Fig. 2E, lanes 3 and 4), globin mRNA competed poorly at 200-fold molar excess (lanes 5 and 6). Taken together, these results suggest that while segments of the structural domains of the 5′NCR are insufficient for La binding, the intact full-length 5′NCR binds La antigen efficiently.
Figure 2.
Purification of bacterially expressed human La protein and competition assay for the binding of La antigen with the full-length wild-type HCV 5′NCR. (A) SDS/PAGE and silver staining of poly(U)-Sepharose 4B affinity purified La antigen. Seventy-five nanograms of protein was loaded in lane 1. Lane M, molecular weight markers. (B) Direct UV cross-linking of the purified La preparation (shown in Fig. 2A, lane 1) with the wild-type 5′NCR RNA probe followed by 12% SDS/PAGE. The probe contained 4-thio-U and was labeled with [32P]CTP. The binding reactions were carried out with 15 ng of purified La (lane 2). Lane 1, probe alone. A similar UV cross-linking assay was carried out as shown in lane 2 followed by immunoprecipitation (IP) with monoclonal anti-La antibody, SW5 (lane 4), or normal serum (lane 3). The samples were fractionated on 12% SDS/PAGE and autoradiographed. (C) Effect of unlabeled competitor RNAs (homologous and deletion mutants) during the interaction of La protein with the thio-U-containing 32P-labeled NCR1–341 RNA probe. Lane 1, probe alone; lane 2, no competitor RNA; lanes 3–6, increasing amounts of unlabeled homologous RNA. The UV cross-linked samples presented from lanes 7 to 16 were fractionated on a separate SDS/polyacrylamide gel. Sample in lane 7 is same as in lane 2. Lanes 8–16 represent deletion mutants of the 5′NCR RNA used as competitors. The numbers indicate the length of nucleotide sequences of the 5′NCR. (D) Competition assay with the HCV 5′NCR mutants lacking domain III structure. UV cross-linking of the NCR1–341 RNA probe with La antigen was carried out in presence of unlabeled competitor RNA as described in the legend to Fig. 2B. Lane 1, no competitor RNA. Lanes 2–7, unlabeled competitor RNAs derived from 5′NCR as indicated. The T7LUC1–99 RNA used as a competitor (lanes 8, 9) represents 99 nucleotides derived from the luciferase gene. (E) Effect of a heterologous competitor RNA on the La binding to the 5′NCR RNA probe. The competitor RNAs, globin mRNA (GIBCO) (lanes 5 and 6), and homologous RNA (lanes 3 and 4), were included in the reaction mixture during UV cross-linking as described above. Lane 1, probe alone. Lane 2, no competitor RNA.
La Protein Binds HCV 5′NCR in the Context of the Initiator AUG.
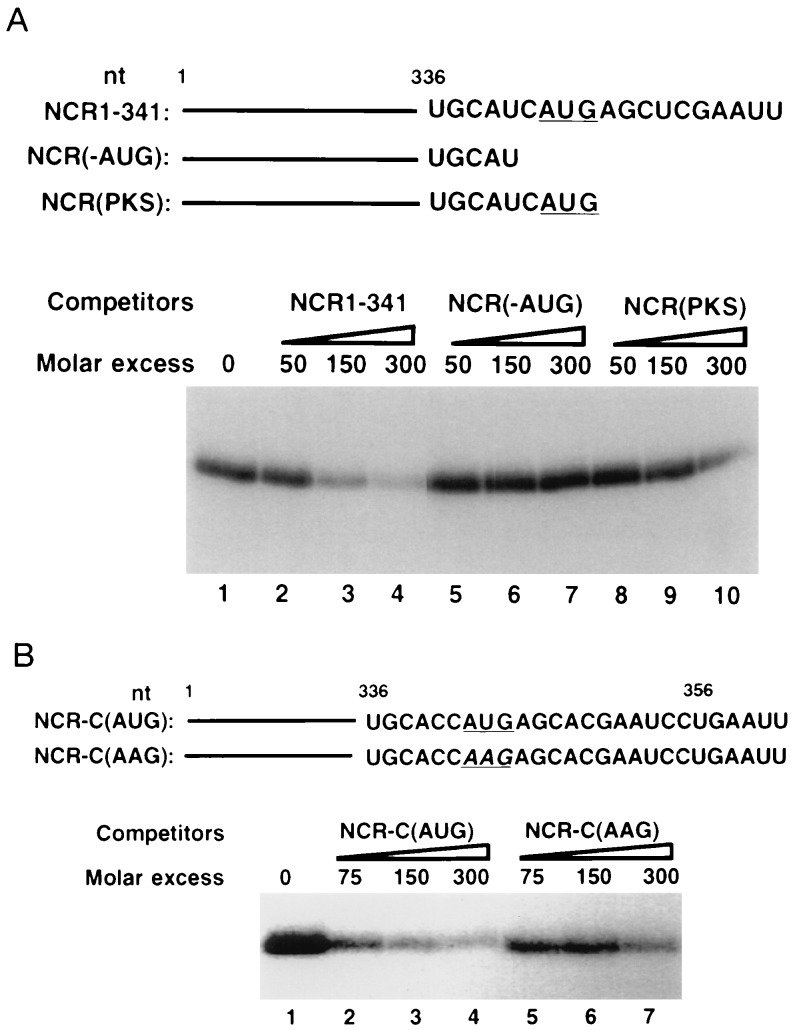
From the data described in Fig. 2, it is evident that various structural domains of the 5′NCR RNA when used independently failed to interact with La antigen, suggesting that this interaction occurs in the context of the entire 5′NCR. Because the 3′ border of the 5′NCR contains critical cis-elements required for the HCV IRES activity, we investigated the role of these motifs in La binding. Several pseudoknot mutants were tested in this analysis, and the results showed that the integrity of the 3′ border was essential for La binding (data not shown). During these investigations, we observed that one mutant RNA [NCR(-AUG)] in which the initiator AUG was deleted, failed to compete for La binding to wild-type RNA (Fig. 3A, lanes 5–7). In the subsequent analysis, initiator AUG was included in the 5′NCR [NCR(PKS)], but lacked the complete Kozak consensus. The RNA again did not compete efficiently (lanes 8–10). These results indicated that one of the La recognition motifs is located at the translation initiation site of the HCV RNA, and the initiator AUG may be directly involved in La interaction with the 5′NCR. This hypothesis was tested using two additional RNA constructs shown in Fig. 3B. These RNAs contain the full-length HCV 5′NCR and extend to 15 nt of the HCV open reading frame that encodes capsid protein. These capsid-encoding sequences have been recently shown to form a hairpin structure in which initiator AUG resides within a loop (35). The NCR-C(AUG) RNA, which represents wild-type HCV RNA, and NCR-C(AAG) RNA are identical except that the latter contains a base substitution at the initiator AUG (AUG to AAG) (Fig. 3B). These RNAs were used as competitors during UV cross-linking of La with NCR-C(AUG) RNA probe. The results of this experiment are described in Fig. 3B. While binding of La antigen to the wild-type NCR-C(AUG) RNA probe was efficiently competed by homologous RNA (lanes 2–4), the mutant RNA, NCR-C(AAG), containing the base substitution at the AUG, competed poorly at 75–150 molar excess (compare lanes 2 and 3 with lanes 5 and 6). At 300 molar excess, however, a moderate competition by this RNA was observed (lane 7). Thus, a single nucleotide substitution at the initiator AUG dramatically reduced the La interaction to the 5′NCR. Deletion of the initiator AUG in the NCR-C(AUG) RNA showed similar reduction in La binding (data not shown). It should be noted that C or U residues at nucleotide 340 are found in natural HCV isolates. We have used wild-type HCV 5′NCR containing either of these nucleotides at position 340. For instance, NCR1–341 RNA contains U, whereas NCR-C(AUG) RNA had C at this position. Both RNAs interact efficiently with La antigen. Thus interchange of the pyrimidine residues at nucleotide 340 probably do not affect La binding. In conclusion, deletion and substitution mutagenesis of the HCV 5′NCR unambiguously support the notion that while La binding to the 5′NCR occurs in the context of other structural domains in that region, the initiator AUG is one of the probable recognition motifs.
Figure 3.
Effect of deletion and substitution of initiator AUG codon (iAUG) on the binding of La antigen to the 32P-labeled HCV 5′NCR. (A) Competition assay using HCV 5′NCR derived mutant RNA lacking nucleotide sequences at the translation initiation site. (Upper) Nucleotide sequences at the 3′ end of each competitor RNA. The iAUG is underlined. (Lower) Results of competition assay. The NCR(1–341) (lanes 2–4), NCR(-AUG) (lanes 5–7), and NCR(PKS) (lanes 8–10) unlabeled RNAs were used as competitors during UV cross-linking of the wild-type RNA probe (NCR1–341) with La antigen. Lane 1, no competitor RNA. (B) Effect of substitution (U to A) at the initiator AUG on La binding. (Upper) Nucleotide sequences at the 3′ end of the unlabeled RNAs used during competition assay. (Lower) The NCR-C(AUG) RNA probe was UV cross-linked with La antigen in the presence of unlabeled homologous (lanes 2–4) and the mutant NCR-C(AAG) (lanes 5–7) RNAs. Lane 1, no competitor RNA. The UV cross-linked products were fractionated by SDS/PAGE and autoradiographed.
Interaction of Cellular La with the HCV 5′NCR.
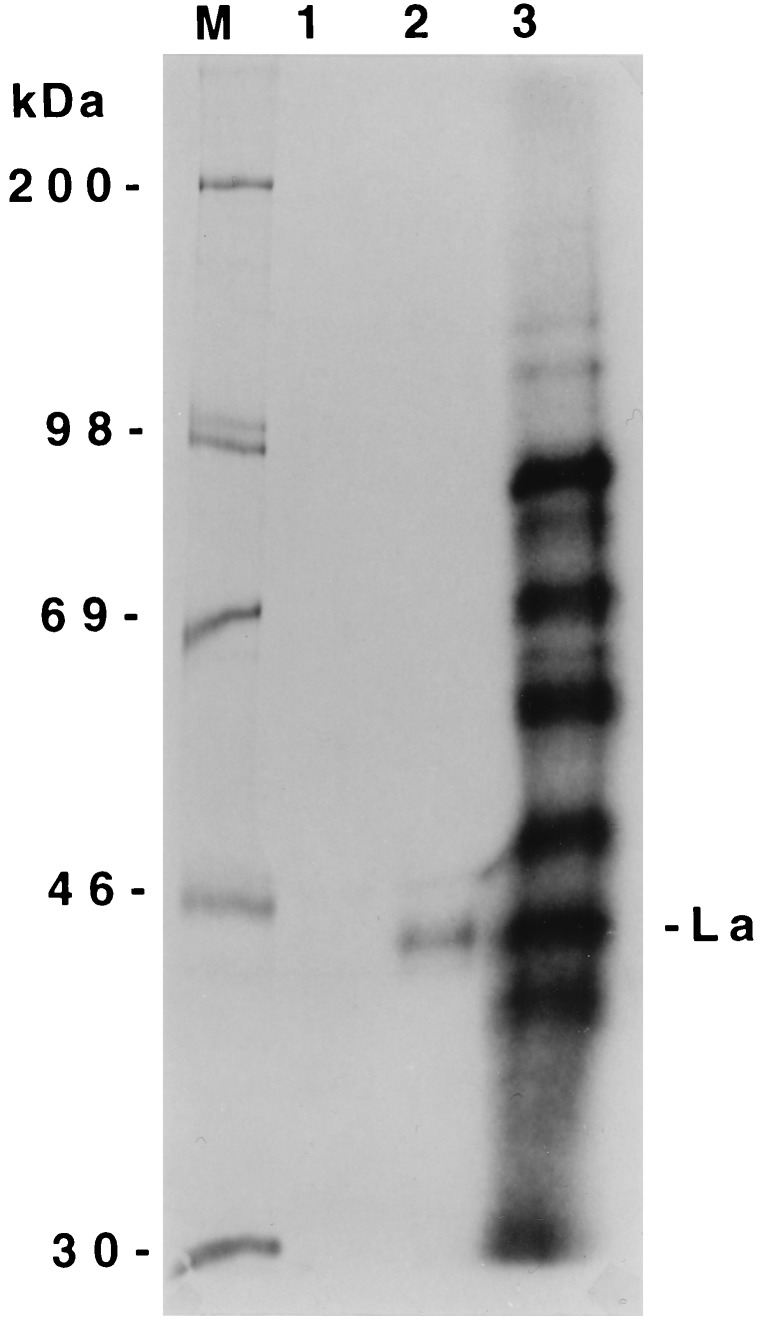
All the characterizations of La protein binding to the HCV 5′NCR described here were carried out with a bacterially expressed purified La protein. To determine whether La protein-HCV 5′NCR interaction occurs in liver cell-derived cytoplasmic extracts, a S100 fraction of Huh7 cell lysates was prepared and UV cross-linked with the full-length wild-type NCR1–341 RNA (Fig. 4, lane 3). An aliquot of UV cross-linked sample was immunoprecipitated with a monoclonal anti-La antibody, SW5. A single RNA-protein complex migrating at approximately 45 kDa was observed after immunoprecipitation with the SW5 anti-La antibody (Fig. 4, lane 2), but not with an unrelated serum (lane 1). The SW5 antibody has been shown to recognize 45–48 kDa isomers of La from several cell lines (36). In addition, this antibody also immunoreacted with recombinant La antigen used here in the binding studies (Fig. 2B). These results are consistent with the previously documented specificity of SW5 antibody (37). Further, the results also suggest that at least one of the La polypeptides that is capable of binding the HCV 5′NCR is present in the cytosolic fraction of the hepatocytes. Among various other proteins of the Huh7 S100 fraction that were UV cross-linked with 5′NCR RNA (Fig. 4, lane 3), a 50- to 52-kDa protein complex was also observed migrating just above the SW5-reactive La isomer. This ribonucleoprotein complex could be one of the isomers of La protein, known to bind poliovirus 5′NCR (24, 28). It is highly likely that the SW5 may not have recognized the epitopes of this isomer after UV cross-linking or cellular protein(s) in the lysates might have masked the epitope. Indeed, a 52-kDa polypeptide in Huh7 lysates was immunoblotted with SW5 antibody (data not shown).
Figure 4.
Immunoprecipitation of La antigen from liver cell-derived (Huh7) S100 fraction after UV cross-linking with the 32P-labeled HCV 5′NCR RNA probe. UV cross-linked S100 lysates (lane 3) were subjected to immunoprecipitation with monoclonal anti-La antibody (SW5) (lane 2) or normal rabbit serum (lane 1). The samples were fractionated by SDS/PAGE.
Stimulation of HCV IRES-Mediated Translation in the Presence of La Antigen.
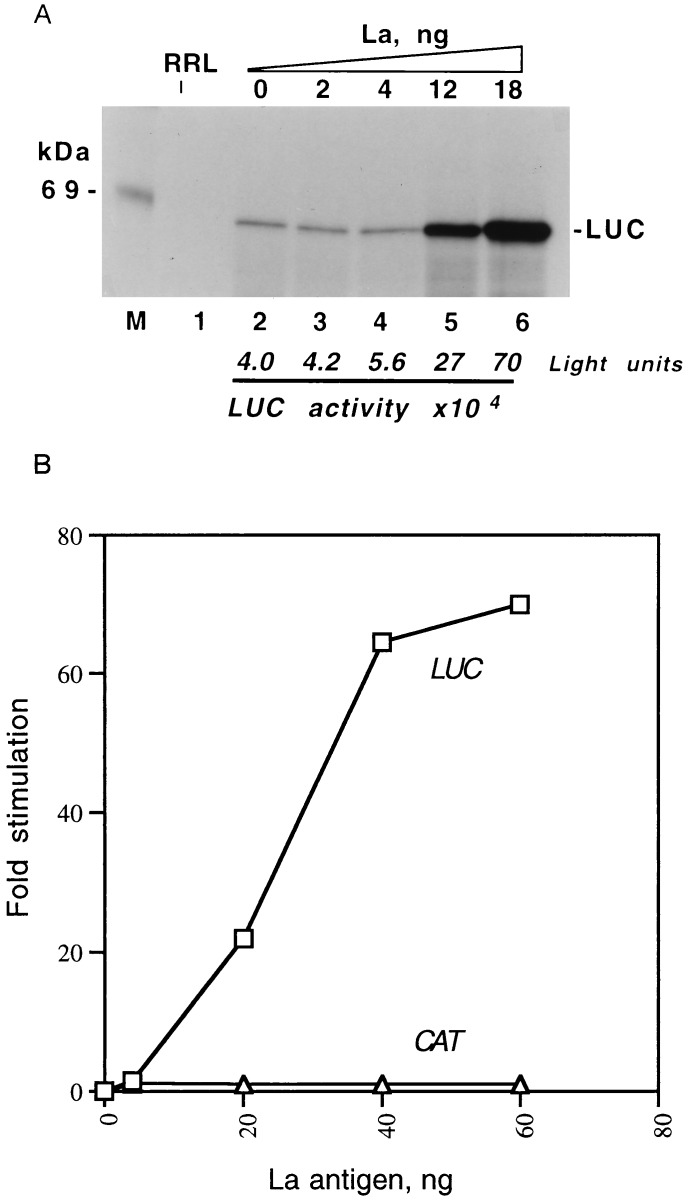
Next, we investigated the functional role of La-HCV 5′NCR interaction. The functional relevance of La interaction was examined using both the monocistronic (T7C1–341) and dicistronic (T7DC1–341) uncapped RNAs synthesized in vitro. In both constructs, the translation of luciferase is dictated by the HCV 5′NCR (11). RRL, which contain sub-optimal level of La (24), were used for the translation of these RNAs. In the first analysis, translation of T7C1–341 RNA was carried out in RRL with increasing amounts of poly(U)-Sepharose purified La antigen. The translational efficiency of the lysates in expressing HCV IRES-controlled luciferase RNA increased from 5- to 15-fold in the presence of La (Fig. 5A, lanes, 3–6) compared with the translational efficiency in the absence of added La (lane 2). To rule out the possibility that the enhancement of translational efficiency observed in the presence of La was not due to a nonspecific effect of protein(s), increasing amounts of BSA were added. Addition of BSA to the lysates did not result in any stimulation of translation of the template RNA (data not shown). To further confirm the transactivating effect of La antigen on the HCV IRES-mediated translation, a dicistronic template RNA (T7DC1–341) was used in which the downstream cistron (luciferase) is translated by internal ribosome entry. The RNA was translated in the presence of varying amounts of La and fractionated on denaturing polyacrylamide gel. The band intensities representing the actual luciferase and chloramphenicol acetyltransferase (CAT; first cistron) expression were quantitated and plotted against La concentration (Fig. 5B). Under the in vitro translation assay conditions used in these studies, the expression of luciferase was significantly stimulated with increasing amounts of La antigen (Fig. 5B). In contrast, the stimulation of CAT expression was negligible under similar concentrations of La. Addition of nonspecific proteins, such as host bacterial lysates not expressing La antigen or normal rabbit serum, to the translation mixture did not show any stimulatory effect on luciferase or CAT expression (data not shown). These data support the stimulatory role of La binding in the HCV IRES function.
Figure 5.
La antigen stimulates translation of the HCV IRES-controlled luciferase RNA in RRL. (A) Translation of monocistronic RNA (T7C1–341) with increasing amounts of purified La antigen (lanes 3–6) or without added La antigen (lane 2). The translation was carried out with (lanes 2–6) or without (lane 1) T7C1–341 RNA (0.2 μg) in RRL. The translation products were radiolabeled with [35S]methionine. The final volume (20 μl) of the reaction mixtures was adjusted with buffer D. Five microliters of translation mixtures was fractionated by SDS/PAGE. The gels were treated with Fluoro-Hance (Research Products International) before autoradiography. Luciferase activity was measured with a 2 μl aliquot of the translation mixture according to de Wet et al. (38). (B) Stimulation of HCV IRES activity in the presence of recombinant La antigen. The dicistronic RNA template (T7DC1–341) used for translation as described in A contained HCV 5′NCR between upstream chloramphenicol acetyltransferase (CAT) and downstream luciferase (LUC) cistrons. Increasing amounts of purified La antigen were added to the translation mixture. The translation mixtures were fractionated by SDS/ PAGE. The band intensities of the LUC and CAT expression were measured by densitometry using PhosphorImager. The fold-stimulation of translation was calculated using these data and plotted against the recombinant La antigen added to the translation mixtures.
DISCUSSION
There is a growing body of evidence that suggests that the translation of the HCV RNA genome occurs by internal ribosome entry through the 5′NCR in a cap-independent fashion (see reviews refs. 9 and 39). Although, the HCV 5′NCR contains multiple AUG codons, the translation of the polyprotein is initiated from an internal AUG triplet located at nucleotide 342. Studies from this laboratory suggest that the sequences in the vicinity of the initiator AUG maintain a higher-order structure that includes RNA pseudoknot (19). It was further demonstrated that maintenance of these structural domains was crucial for the HCV IRES function (19, 31). Here we show that the integrity of these structures also plays a significant role in the interaction of La with the 5′NCR. Competition assays show that full-length wild-type HCV 5′NCR binds La protein efficiently, whereas the deletion mutants in domain III and/or 3′ terminal sequences of the 5′NCR abrogated binding. The most surprising finding was the requirement of an intact initiator AUG codon for La binding. The 5′NCR fragment (NCR1–131), which contains a highly conserved noninitiator AUG at nucleotide position 85, failed to efficiently interact with La protein in both direct UV cross-linking (data not shown) and competition assays. Thus, these results suggest a role of La antigen in the selection of the initiator AUG in the context of the structural elements of the HCV 5′NCR.
The mechanism by which La antigen recognizes diverse RNA sequences or structural motifs and plays a significant role in disparate biological pathways remains elusive. On the basis of the data presented here and those reported for other viruses (28–30), La antigen enhances the overall translational capacity of the reticulocyte lysates to translate RNAs in an IRES-dependent manner by the following mechanisms. First, the protein may increase the fidelity of the initiation of HCV IRES-dependent translation by binding in the context of the initiator AUG codon. This interaction appears to be dependent on the integrity of the higher-order superstructure of the 5′NCR in the immediate vicinity of the initiator AUG (19, 31). Because mutation in the initiator AUG codon will lead to alteration in translation initiation, the functional analysis of the La protein binding in the context of mutated AUG codon is not feasible. Indeed, Reynolds et al. (14) observed that a mutation in the initiator AUG to AAG severely reduced the HCV IRES-mediated translation. Second, the flanking sequences of the initiator AUG codon constitute highly structured region (19). The binding of La alone or in cooperation with the initiation factors such as eIF-4B and eIF-4F, which also modestly enhance internal initiation (40), may facilitate unwinding of the 5′NCR by virtue of its helicase property, mediating base pairing of the AUG codon with the anticodon of the Met-tRNAi. In fact, La protein has been shown to bind both synthetic and naturally occurring reovirus double-stranded RNA and exhibit RNA-RNA as well as RNA-DNA unwinding activity (41, 42). RNA helicase activity of La could further facilitate smooth translocation of the ternary complex in 5′ –> 3′ direction. The third possibility could be that La sequesters other cellular factor(s) that may be important for stimulation of translation by protein-protein interactions. Svitkin et al. (29) have shown that a truncated version of La antigen (amino acids 1–194), which still contains the RNA recognition motif, binds poliovirus 5′NCR, but fails to correct or stimulate the translation initiation. Similar studies showed that immunodepletion of La from HeLa cell lysates inhibit the translation of poliovirus RNA. However, supplementation with exogenous La did not restore the translational capacity of the lysate (21). These observations suggest that La protein may function as a chaperone in stabilizing higher-order RNA structure and also be involved in protein–protein interactions.
In summary, the studies described here show that the AUG at nucleotide 342, which serves as an initiator codon for the long single open reading frame of the HCV RNA genome, is one of the recognition motifs for La protein binding. This AUG, which is in translationally favorable context (43), and the surrounding structural elements are highly conserved among all the HCV subtypes. Our studies further show that the La binding to the HCV 5′NCR leads to a substantial stimulation of translation. Whether La recognizes initiator AUG codon as one of the motifs or the initiator AUG is being recognized as a component of the superstructure in that region needs to be determined. These studies are currently under investigation. Interestingly, McBratney and Sarnow (44) using synthetic RNA molecules as substrate, recently have shown that La antigen interacted directly with the AUG triplet, and the binding was influenced by the Kozak sequence context. These observations together suggest a novel role of La protein in start-site selection in translation initiation.
Acknowledgments
We thank G. J. M. Pruijn for human La cDNA clone and monoclonal anti-La antibody (SW5). We also thank Peter Sarnow for constant help and fruitful discussions throughout the course of this study. This work was supported by grants from American Cancer Society (VM-193) and Lucille P. Markey Charitable Trust. N.A. received support from the Colorado Advanced Technology Institute.
ABBREVIATIONS
- HCV
hepatitis C virus
- IRES
internal ribosome entry site
- 5′NCR
5′ noncoding region
- RRL
rabbit reticulocyte lysates
References
- 1.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo Q-L, Houghton M, Kuo G. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukuma H, Hiyama T, Tanaka S, Nakao M, Yabuuchi T, Kitamura T, Nakanishi K, Fujimoto I, Inoue A, Yamazaki H, Kawashima T. N Engl J Med. 1993;328:1797–1801. doi: 10.1056/NEJM199306243282501. [DOI] [PubMed] [Google Scholar]
- 3.Miller R H, Purcell R H. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo Q L, Richman L K, Han J H, Berger K, Lee C, Dong C, Gellegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 6.Bukh J R, Purcell R H, Miller R H. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macejak D G, Sarnow P. Nature (London) 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 8.Oh S-K, Scott M P, Sarnow P. Genes Dev. 1993;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 9.Jackson R J, Kaminski A. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 10.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Sarnow P, Siddiqui A. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 13.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds J E, Kaminski A, Kettinin H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rijnbrand R, Brenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 16.Lu H-H, Wimmer E. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown E A, Zhang H, Ping L-H, Lemon S M. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellen C U T, Wimmer C. In: Cap-Independent Translation. Sarnow P, editor. Heidelberg: Springer; 1995. pp. 31–63. [Google Scholar]
- 19.Wang C, Le S-Y, Ali N, Siddiqui A. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson R J, Howell M T, Kaminski A. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- 21.Belsham G J, Sonenberg N, Svitkin Y V. In: Cap-Independent Translation. Sarnow P, editor. Heidelberg: Springer; 1995. pp. 85–98. [Google Scholar]
- 22.Tan E M. Adv Immunol. 1989;44:93–159. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- 23.Kenan D J, Query C C, Keene J D. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 24.Meerovitch K, Pelletier J, Sonenberg N. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 25.Park Y W, Katze M G. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 26.Pardigon N, Strauss J H. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y-N, Kenan D J, Keene J D, Gatignol A, Jeang K-T. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svitkin Y V, Meerovitch K, Lee H S, Dholakia J N, Kenan D J, Agol V I, Sonenberg N. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svitkin Y V, Pause A, Sonenberg N. J Virol. 1994;68:7001–7008. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Sarnow P, Siddiqui A. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali N, Siddiqui A. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stade K, Rinke-Appel J, Brimacombe R. Nucleic Acids Res. 1989;17:9889–9908. doi: 10.1093/nar/17.23.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dignam J D. In: Guide to Protein Purification. Deutscher M P, editor. New York: Academic; 1990. pp. 194–203. [Google Scholar]
- 35.Honda M, Ping L-H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 36.Smith P R, Williams D G, Venables P J W, Maini R N. J Immunol Methods. 1985;77:63–67. doi: 10.1016/0022-1759(85)90184-x. [DOI] [PubMed] [Google Scholar]
- 37.Pruijn G J M, Thijssen J P H, Smith P R, Williams D G, Van Venrooij W J. Eur J Biochem. 1995;232:611–619. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- 38.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramaniam S. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Siddiqui A. In: Cap-Independent Translation. Sarnow P, editor. Heidelberg: Springer; 1995. pp. 99–115. [Google Scholar]
- 40.Anthony D D, Merrick W C. J Biol Chem. 1991;266:10218–10226. [PubMed] [Google Scholar]
- 41.Bachmann M, Pfeifer K, Shroder H-C, Muller W E G. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Q, Sharp T V, Jeffrey I W, James M C, Pruijn G J M, van Venrooij W J, Clemens M J. Nucleic Acids Res. 1994;22:2512–2518. doi: 10.1093/nar/22.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBratney S, Sarnow P. Mol Cell Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]