Abstract
Mammalian cells contain activities that amplify the effects of activators on class II gene transcription in vitro. The molecular identity of several of these cofactor activities is still unknown. Here we identify poly(ADP-ribose) polymerase (PARP) as one functional component of the positive cofactor 1 activity. PARP enhances transcription by acting during preinitiation complex formation, but at a step after binding of transcription factor IID. This transcriptional activation requires the amino-terminal DNA-binding domain, but not the carboxyl-terminal catalytic region. In purified systems, coactivator function requires a large molar excess of PARP over the number of templates, as reported for other DNA-binding cofactors such as topoisomerase I. PARP effects on supercoiled templates are DNA concentration-dependent and do not depend on damaged DNA. The PARP coactivator function is suppressed by NAD+, probably as a result of auto-ADP-ribosylation. These observations provide another example of the potentiation of trancription by certain DNA-binding cofactors and may point to interactions of PARP with RNA polymerase II-associated factors in special situations.
Stimulation of class II gene transcription by activators requires the general factors and coactivators (1–4). Some of the coactivators are associated with TBP, the TATA box-binding protein, and RNA polymerase II, respectively (5–8). There is also another group of coactivators that appear not to be integral parts of the basal machinery. These soluble cofactors were identified on the basis of their capacity to enhance activation of transcription in vitro (9–12). In our initial study we had isolated a fraction from HeLa nuclear extracts, termed USA, for upstream factor stimulatory activity, that was essential for activator-dependent transcription in the presence of a complete set of general factors (9). Subsequent studies resolved the USA fraction into a minimum of four different positive cofactors, termed PC1 to PC4 (9–15). Several specific components of the USA fraction and related cofactors have been characterized in recent years. Examples include topoisomerase I, also called PC3 or Dr2 (13, 16), topoisomerase II (17), high mobility group proteins (18), and a protein called p15 or PC4 (14, 15). Although PC1 and PC2 were discovered earlier, their identities are yet unresolved. Here we have identified human poly(ADP-ribose) polymerase (PARP) as one active component of the PC1 fraction.
Mammalian PARP is a nuclear chromatin-associated protein of size 114 kDa that catalyzes the transfer of ADP-ribose units from NAD+ to nuclear protein acceptors (19–22). Up to several hundred ADP-ribose units are transferred to PARP itself. Subsequently PARP modifies cellular proteins that are located within the chromatin. Target proteins include topoisomerases I and II, histones, and high mobility group proteins (23). The activity of PARP is strongly stimulated by the presence of nicks and strand breaks in DNA. These observations have contributed to the idea that PARP mediates stress-induced signaling and functions in an NAD-dependent manner in certain DNA repair processes (19, 24, 25). There is convincing evidence for the binding of PARP to damaged DNA containing single-strand breaks and nucleotide excisions. Automodification releases PARP from DNA, thus providing a mechanism for rendering DNA more accessible to the DNA repair machinery (23). In the absence of NAD, PARP inhibits DNA repair through binding to damaged DNA (26). Other functions proposed for PARP include roles in cellular NAD depletion (27), antirecombination and genomic stability (28), and DNA replication (29). PARP also serves as a marker for the onset of apoptosis, after which it is cleaved by proteases into DNA-binding and catalytic fragments (30).
Earlier studies demonstrated that PARP suppresses nick-induced transcription in crude cell-free systems, whereas it was not required for basal transcription in systems reconstituted with purified factors (31). Moreover, inhibitors of PARP failed to demonstrate an essential role in transcription (32). However, PARP also proved to be nonessential in vivo for most of the functions suggested by the in vitro studies, with knock-out mice showing few defects in nucleotide excision and base excision-repair (33). PARP is the only nuclear ADP-ribosylating enzyme, and PARP homologues are evident in all eukaryotes except yeast (19). Although the enzyme has been investigated for more than 30 years, the physiological relevance of the heterologous protein modifications is generally unclear.
Here we uncover a novel property of PARP. We show that PARP is one active component of PC1, that it has an intrinsic capacity to enhance activator-dependent transcription in vitro, that its cofactor function requires the DNA binding domain (34) and a high PARP-template ratio, and that cofactor function is inhibited by auto-ADP-ribosylation. Our observations, though not supporting a role as a genuine commonly required transcription factor, demonstrate the capacity of PARP to enhance basal and activator-dependent transcription. This may point to a molecular connection between PARP and RNA polymerase II-associated factors in situations that remain to be defined in the cell.
MATERIALS AND METHODS
Purification of PC1.
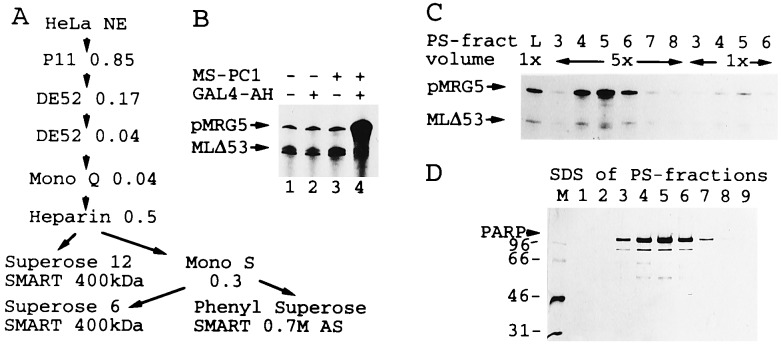
PC1 was purified from HeLa nuclear extract-derived USA fractions up to the heparin-Sepharose step as described previously (9). The specific activity of PC1 could not be determined during the first three to five chromatographic steps as it initially coeluted with other cofactor activities. The extract preparation used in this study contained mainly phoshorylated PC4, which is inactive and separated from PC1 on anion exchanger columns. In other preparations nonphosphorylated PC4 coeluted with PC1 through the initial five chromatographic steps but was separated on Mono S and Phenyl Superose columns (Fig. 1A). The 0.5 M KCl heparin-Sepharose eluate was dialyzed to 50 mM KCl in buffer T [20 mM Tris·HCl, pH 6.8 (room temperature)/0.2 mM EDTA/0.025% (vol/vol) Nonidet P-40/5 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/10% glycerol] and applied to a Mono S column. The column was washed with 200 mM KCl, and PC1 was eluted at 300 mM KCl in a linear gradient from 0.2 to 2 M KCl in buffer T. The peak fraction was adjusted to 1.3 M ammonium sulfate and loaded onto a Phenyl Superose column (SMART system, Pharmacia). PC1 was eluted with a linear gradient from 1.3 M to 0 M ammonium sulfate in buffer C [10% glycerol/0.2 mM EDTA/20 mM Tris·HCl, pH 7.3 (room temperature)/5 mM DTT/1 mM phenylmethylsulfonyl fluoride] with peak activity at 700 mM ammonium sulfate. Active fractions were dialyzed to buffer C containing 100 mM KCl and 20% glycerol. For size determination the Mono S fraction was applied to Superose 6 and Superose 12 columns (SMART system). The columns were developed with buffer P (200 mM NaPO4, pH 7.6/0.2 mM EDTA/10% glycerol). On the sizing columns, PC1 activity and PARP coeluted in a position corresponding to an approximate molecular mass of 400 kDa.
Figure 1.
(A) Schematic purification scheme for PC1 activity from HeLa nuclear extracts (NE). (B) PC1-induced activator-dependent transcription in a reconstituted class II gene transcription system. Saturating amounts of Mono S (MS) PC1 peak fractions (1 μl corresponding to approximately 2 μg of protein) were analyzed in the presence of saturating concentrations of general factors that included natural TFIIA (DE52 fraction), recombinant TFIIB, TFIIEα, TFIIEβ, natural Phenyl Superose TFIIF (8 μl), RNA polymerase II, and limiting amounts of TFIID (0.2 μl) and TFIIH under otherwise standard conditions and in combination with GAL4-AH activator as indicated. (C) Transcriptional stimulatory activity of purified Phenyl Superose (PS) PC1 fractions. The input (L) and two different concentrations of gradient fractions (numbers and relative volumes indicated) were analyzed in standard assays in the presence of GAL4-AH. Approximately 1 μg of total protein was present in peak fraction 5 (5 units). (D) SDS/PAGE analysis of the chromatographic fractions analyzed in C. The silver-stained gel contained 16 units of PC1 Phenyl Superose fractions. The 115-kDa band was identified as PARP by protein sequencing. Lower bands reflect degradation products.
Antibodies and Depletion Procedures.
Western blots were performed by standard procedures. For depletion of PC1 activity 2 μl of Mono S PARP peak fractions were incubated with 1 μl PARP antisera in buffer C containing 150 mM KCl for 30 min at room temperature. Protein A-Sepharose (Sigma) equilibrated in the same buffer then was added, and the suspension was incubated for 30 min at room temperature. Reaction mixtures were centrifuged through 0.45-μm membranes (MC 0.45 tubes, Millipore), and flow-through fractions were used in transcription without dialysis. Control reactions containing preimmune sera were processed identically.
Cloning and Expression of PC1.
Approximately 100 μg of heparin-Sepharose PC1 fraction was subjected to SDS/PAGE, and the 115-kDa protein was excised and digested with trypsin. Derived peptides were separated on HPLC columns according to standard procedures. A peptide with sequence RKGDEVDGVDEVAK was sequenced (kindly performed by J. Kellermann at the Max Planck Institute for Biochemistry, Munich). To generate deletion proteins the complete PARP open reading frame was isolated by PCR from a first-strand HeLa cDNA that was synthesized with avian myeloblastosis virus reverse transcriptase by standard procedures. Upon cloning of the complete cDNA into Rc/CMV (Invitrogen), the DNA-binding region, comprising residues 1 to 450, was isolated by PCR and cloned into NdeI/BamHI sites of pET11d (Novagen). Proteins were expressed and purified on Ni-NTA and heparin-Sepharose columns as previously described for other His-tagged proteins (35).
In Vitro Transcription Assays.
Transcription reactions generally were conducted with supercoiled templates, pMRG5 and pMLΔ53 (9, 12). Nicked templates (Fig. 3C) were generated with amounts of DNAseI that completely converted supercoiled into relaxed templates without producing detectable double-strand DNA breaks. All transcription reactions were performed with purified natural and recombinant general transcription factors (ref. 35 and references therein). Standard reactions contained 50 ng of each transcription template, 20 ng of recombinant transcription factor II (TFII)B, 0.2–1.5 μl of TFIID (DE52-fraction, 0.35 mg/ml protein), 10 ng of recombinant TFIIEα, 5–10 ng of recombinant TFIIEβ, 1–8 μl of natural Phenyl Superose TFIIF (0.12 mg protein per ml), or 20 ng of baculovirus expressed RAP30/RAP74 (Superose 12 fraction), 1.0–1.5 μl of a DE52 TFIIH fraction (0.5 mg/ml protein) or 1 μl of a Phenyl Superose TFIIH fraction, 0.2 μl of calf thymus RNA polymerase II (DE52 fraction, 0.5 mg/ml protein) or 1.0 μl of a HeLa nuclear extract-derived RNA polymerase II fraction purified on phosphocellulose (0.5 M KCl step) and DE52 (0.1–0.3 M KCl step) columns, 30–50 ng of purified GAL4-AH or GAL4-TA1 (35), if not indicated otherwise in the figure legends. Activators were used in concentrations that were saturating for transcription and DNA binding. They were added to buffered transcription templates before addition of PC1 fractions and general factors. The partially purified TFIID contained TFIIA. Natural human or recombinant yeast TFIIA, present in some reactions, did not affect the activity of PARP. In order-of-addition experiments 1.0 μl of TFIIA (Mono Q fraction, 2.5 mg/ml) was added. Transcription reactions were conducted in a buffer containing 25 mM Hepes (pH 8.2), 10–20% glycerol, 5 mM MgCl2, 60–75 mM KCl, 5 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride, 200 ng/μl BSA, 0.002% Nonidet P-40, 100 μM each of UTP and ATP, 5 μM CTP, 20 μM 3′-O-methyl-GTP, 0.5 μM [α-32P]CTP (3,000 Ci/mmol; 1 Ci = 37 GBq), and 20 units of RNase-Block (Stratagene). NAD+ was purchased from Sigma and single-stranded DNA-binding protein (SSB, 1 mg/ml) from Stratagene. Transcription reactions were carried out for 1 h at 28–30°C and processed and analyzed on denaturing polyacrylamide gels as described (9, 35).
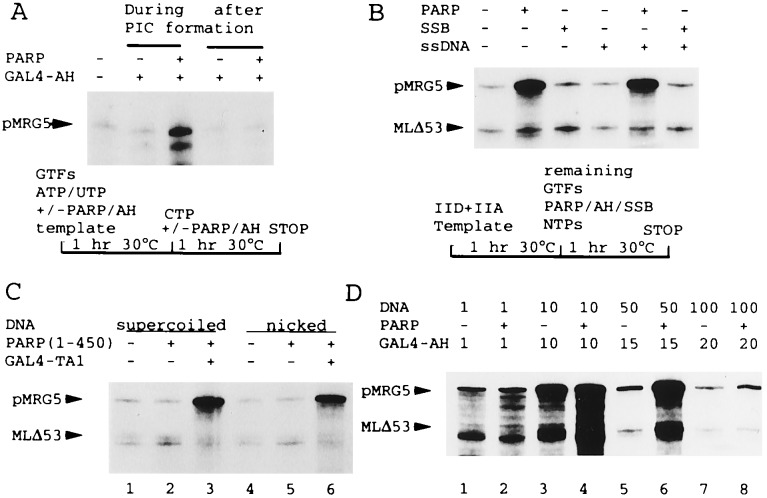
Figure 3.
(A) A combination of PARP and GAL4-AH is inactive when added after PIC formation. GAL4-AH and PARP (Mono S fraction) were added, as indicated, either during a 1-h preincubation of GTFs, ATP, and UTP, and 100 ng of pMRG5 template (lanes 3 and 4) or during the following 1-h incubation under standard transcription conditions with added CTP. A schematic protocol is shown below. (B) PARP and GAL4-AH function when added after formation of a TFIID-TFIIA-DNA complex. After preincubation of DNA, TFIID, and TFIIA according to the protocol indicated, transcription was initiated by addition of other GTFs, NTPs, GAL4-AH (all lanes) along with PARP (Mono S fraction), E. coli single-stranded-DNA-binding protein (SSB) (200 ng), or a 60-bp single-stranded DNA promoter-derived oligonucleotide (ssDNA, present at twice the concentration of the template) as indicated. (C) PARP does not act through binding to nicked templates. Standard transcription reactions contained supercoiled (lanes 1–3) or nicked (lanes 4–6) DNA templates and GAL4-TA1 and purified recombinant PARP (500 ng of a deletion protein containing amino acids 1–450) as indicated. (D) PARP function is strictly DNA concentration-dependent. Standard transcription reactions contained variable amounts of template DNA (indicated in ng), GAL4-AH (units indicated, with 10 units the standard amount), and PARP (natural Mono S fraction) as indicated.
RESULTS
Purification of a Cofactor from PC1 Fractions.
PC1 activity was isolated from HeLa nuclear extracts that were fractionated as schematically indicated in Fig. 1A. PC1 activity originated from the USA precursor fraction (corresponding to the P11 0.85 M KCl fraction in Fig. 1A). It was separated from other PC activities such as PC2 (9, 11), PC3 (12), and PC4 (14, 15) on subsequent chromatographic steps (see Materials and Methods). The cofactor activity was monitored using a reconstituted class II gene transcription system with GAL4 fusion proteins carrying an AH (GAL4-AH) or the NFκB TA1 (GAL4-TA1) activation domain. A TATA-containing HIV promoter carrying five GAL4 sites (pMRG5) and a basal adenovirus major late promoter as an internal control (MLΔ53) served as reporters. GAL4-AH enhanced transcription in the presence of PC1 but failed to affect transcription in the absence of cofactors (Fig. 1B), consistent with previous studies of the activator-dependent transcription process (9–15). GAL4-TA1 activity also was potentiated by PC1, though it moderately stimulated transcription in the apparent absence of PC activities (Fig. 2). Although PC1 can markedly enhance activator-dependent transcription (Fig. 1B), it can also moderately stimulate basal transcription (e.g., Fig. 2A). The relative strengths of the individual effects depend upon PC1 concentrations, DNA concentrations (Fig. 3D) and the specific reconstituted transcription system (Fig. 1B versus Fig. 2A; data not shown).
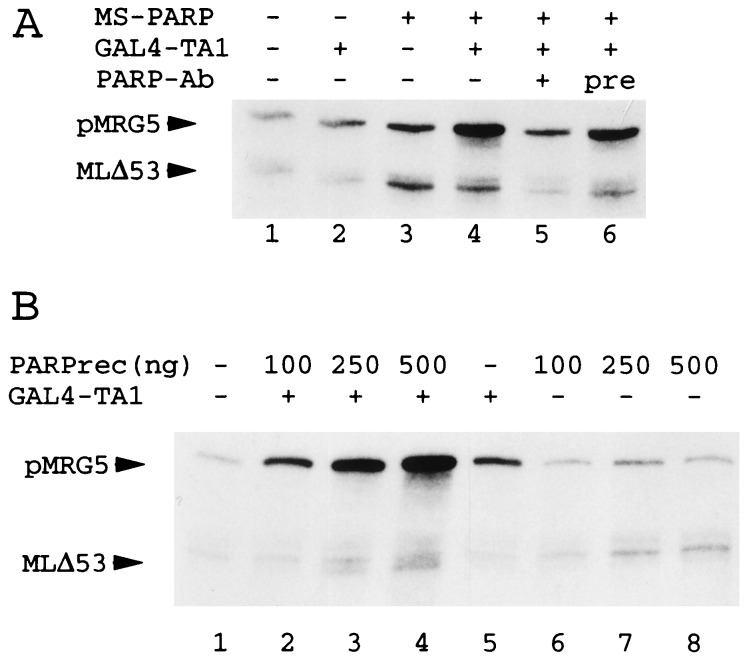
Figure 2.
(A) Anti-PARP polyclonal antibodies (PARP-Ab) deplete PC1 basal and activator-dependent activity. Standard transcription assays contained untreated (lanes 1–4), anti-PARP-treated (lane 5), and preimmune serum-treated (lane 6) Mono S PC1 fractions and the GAL4-TA1 activator as indicated. (B) Purified full-length recombinant PARP enhances activator-dependent transcription. Standard transcription reactions contained GAL4-TA1 and variable levels of PARP as indicated.
PC1 activity coeluted with a 115-kDa peptide at the final Phenyl Superose gradient step (Fig. 1 C and D). Minor contaminants of sizes 55 to 90 kDa did not cofractionate with PC1 activity on Superose 6 and Superose 12 gel filtration columns on which PC1 eluted with an apparent molecular weight of 400 kDa (Fig. 1 and data not shown). Sequencing of a tryptic peptide of the 115-kDa protein identified the latter as PARP. Anti-PARP antibodies (Fig. 2A, lane 5), but not pre-immune sera (lane 6), eliminated effects of partially purified PC1 on both basal- and activator-dependent transcription (lanes 2 to 5). Recombinant PARP, expressed in and purified from Escherichia coli, behaved indistinguishably from its natural counterpart at comparable concentrations and conditions (compare Fig. 2 A and B). Altogether, these data provide strong evidence that PARP is at least one active component of transcription cofactor PC1 (9).
PARP Acts in a DNA Concentration-Dependent Manner During Preinitiation Complex (PIC) Formation.
Because PARP appeared to act specifically on transcription we reasoned that it might affect PIC formation. Indeed, order-of- addition experiments demonstrated that PARP acts during PIC formation at a time after binding of TFIID to the TATA box but before complete PIC formation. Thus, addition of activator and PARP after preincubation of all general transcription factors (GTFs) with the template abolished transcriptional activation, whereas concommitant addition with GTFs revealed the previously described effect of PARP (Fig. 3A). In contrast to the lack of an effect when added after PIC formation, PARP enhanced transcription when added together with GAL4-AH after prebinding of TFIID (and TFIIA) to the promoter (Fig. 3B). We conclude that PARP is required at some point during the assembly of TFIIB, TFIIE, TFIIF, TFIIH, and RNA polymerase II, whereas it appears not to support binding of TFIID to the promoter. Consistent with this conclusion, PARP effects were not dependent upon TFIID concentrations (data not shown). In contrast to these observations for PARP, the potent coactivator PC4 stimulates TFIID binding when added simultaneously with TFIID but not when added after TFIIA-TFIID promoter complex formation (35).
PARP is a DNA-binding protein that preferentially binds to damaged DNA containing nicks or excised nucleotides. It also recognizes supercoiled DNA in a cooperative manner (34, 36, 37). Indeed, we also could monitor PARP activity during chromatography (e.g. in Mono S gradient fractions) with single-stranded DNA oligonucleotides in gel mobility shift assays (data not shown). However, the cofactor activity appears not to be mediated via binding to damaged DNA; and although other cofactors such as PC4 also bind single-stranded DNA (14, 15), this property, while potentially involved, is not sufficient for activity (35). Thus, the E. coli single-stranded-DNA-binding protein does not enhance transcription at comparable concentrations (Fig. 3B, lane 3) and PARP functions indistinguishably on nicked versus supercoiled templates (Fig. 3C).
As previously found for other DNA-binding general coactivators (12), high concentrations of PARP are required to stimulate transcription, and this activity depends strictly on the concentration of double-stranded DNA templates. A titration analysis revealed a narrow optimum for coactivator function at intermediate DNA concentrations with little activity at very low or at higher DNA concentrations (Fig. 3D). The capacity of PARP to modulate basal transcription was also strongly influenced by DNA concentration (compare lanes 3–8). The narrow activity window explains the sometimes weak stimulatory effects of PARP and variations in basal versus activator-dependent effects in different experiments. We estimated that reactions in which optimal PARP coactivator function was observed contained up to 80 molecules of PARP per template. At these PARP/template ratios, supercoiled plasmids were completely shifted by PARP in agarose gels (data not shown). Thus, PARP is in excess over nicks and probably binds to double-stranded supercoiled templates to induce transcription. Consistent with this, high concentrations of single-stranded oligonucleotide competitors failed to eliminate effects of PARP on activator-dependent transcription (Fig. 3B, lanes 4–6). These findings suggest that PARP binds to double-stranded DNA and, consequently, modulates activity of the preinitiation complex.
NAD Inhibits Stimulation of Transcription by PARP.
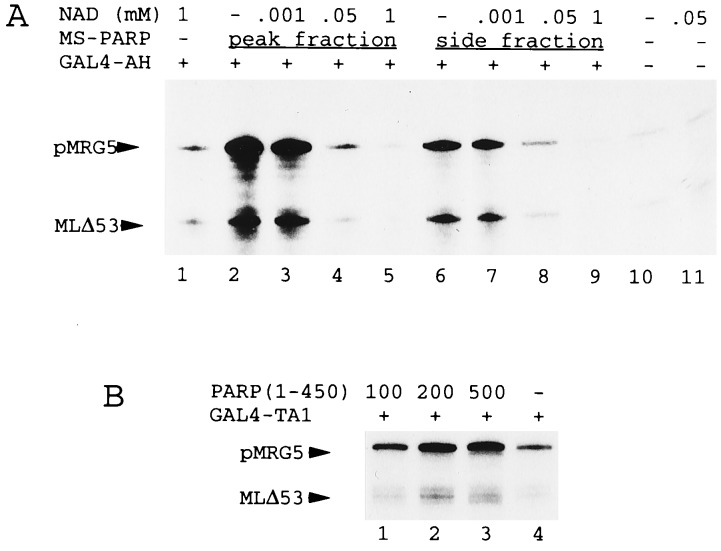
Auto-ADP-ribosylation by PARP is mediated through carboxyl-terminal catalytic regions that contain two putative NAD-binding sites. The central part of PARP functions as a receptor for the poly(ADP) chains (19). To determine whether ADP ribosylation influences PARP coactivator functions, we analyzed effects of natural PARP (Mono S peak and side fractions) on transcription in the presence of increasing concentrations of NAD+ (Fig. 4A). NAD+ completely inhibited all transcription (basal and activator-mediated) that was dependent upon PARP (Fig. 4A, lanes 2–9), while it did not affect basal transcription in the absence of PARP (lanes 1 and 11 versus lane 10). In control experiments the activity of other cofactors such as PC2 (12) was not affected by NAD (data not shown). A recombinant PARP fragment containing the amino-terminal 450 amino acids and lacking the catalytic region stimulated transcription indistinguishably from the intact protein (Figs. 3C and 4B), demonstrating that neither ADP ribosylation nor the NAD-binding region are required for cofactor activity. These experiments suggest that ADP-ribosylation abolishes cofactor function.
Figure 4.
(A) NAD+ inhibits PARP stimulatory effects on transcription. Standard transcription reactions contained NAD+, GAL4-AH, and either Mono S PARP peak (lanes 2–5) or side (lanes 6–9) fractions as indicated. (B) Carboxyl-terminal catalytic domains of PARP are not required for transcription. Standard transcription reactions contained GAL4-TA1, and variable levels (amounts in ng) of recombinant purified PARP(1–450) as indicated.
DISCUSSION
The fractionation of nuclear extracts has led to the identification of an increasing number of coactivators that modulate transcription in vitro. Examples are high mobility group 1/2 (18), topoisomerases I/II (13, 17), and PC4 (14, 15). Here we have isolated PARP as an active component of PC1 and demonstrated its capacity to stimulate activator-dependent and basal transcription. PARP itself is neither an integral part of the basal transcription machinery nor essential for transcription. Although the identities of several cofactors that have similar effects on transcription are now known, sequence comparisons have not yet revealed any underlying common structural principle. Indeed, PARP and other transcription cofactors show little sequence homology, although they are related with respect to their DNA-binding properties (35). With the characterization of PARP as a cofactor we provide yet another example of a DNA-binding protein that stimulates transcription.
Several mechanisms by which cofactors stimulate transcription have been proposed. These include enhancement of TFIID binding and activation of the preinitiation complex (refs. 14–16 and 18, reviewed in ref. 10). In the case of PARP we demonstrated that its presence is required during assembly of RNA polymerase II and other general factors with a preformed complex consisting of TFIID and possibly TFIIA. We have no indication that PARP stimulates formation of a TFIID-promoter complex, as was reported for PC4 (14, 15, 35). The inability to stimulate transcription after formation of a complete PIC containing only basal factors points to a role in activation of the preinitiation complex during assembly. This might be achieved through a number of different mechanisms, which include recruitment of GTFs or conformational changes within components of the basal machinery (reviewed in ref. 4). Although the precise mechanism of activation remains to be specified, it would appear that the complete PIC (or an intermediate) formed in the absence of the activator/coactivator is in a distinct stable conformation that is both refractory to activator/coactivator and unable to function efficiently in factor recruitment and/or initiation or reinitiation. An understanding of the molecular mode of action of transcription cofactors could be further complicated by recent observations demonstrating that certain members of the DNA-binding cofactors function synergistically with others (K. Kaiser and M.M., unpublished observations).
Transcriptional activation is correlated with template-recognition by PARP. The DNA-binding domain of PARP, comprised of two Zn fingers within the amino-terminal 165 amino acids and additional carboxy-terminal regions that may be involved in nonspecific binding to DNA (34, 36, 37), suffices for stimulation of transcription. PARP effects are strictly dependent on template concentrations. Auto-ADP-ribosylation, which releases PARP from DNA, abolishes cofactor activity. Earlier investigations demonstrated that PARP prevents initiation of transcription from damaged DNA through binding to nicked DNA (31). In agreement with this analysis we could observe PARP-dependent repression of incorporation of label into high molecular weight nucleic acids in the presence of nicked templates, though this was restricted to cruder systems and usually not observed in purified transcription systems. However, coactivator effects do not rely on binding of PARP to damaged DNA. Enhancement of activator-dependent transcription required high concentrations (approximately 0.2 μM) of PARP, and the number of PARP molecules exceeded by far the number of nicks in the templates. In fact, the concentrations of PARP necessary for coactivator function probably suffice to cover transcription templates.
If we tentatively assume that all molecules are bound to templates, approximately one PARP molecule will be localized every 40 base pairs. Although PARP is very abundant in mammalian cells (19), we believe it unlikely that PARP performs similarly in the natural genomic environment. Consistent with this judgment, none of the earlier characterizations were able to document a role for PARP in transcription either in living cells or in animals (33). Nonetheless, our data demonstrate the capacity of PARP to alter the efficiency of transcription in vitro. Minimally, this suggests that PARP may simply be mimicking the action of, and substituting for, another bona fide transcriptional coactivator. At this time, the redundancy generated through many functionally equivalent cofactor activities in nuclear extracts makes it difficult to test the role of individual components. For this reason, it also is difficult to exclude the formal possibility that PARP functions occasionally as a transcription cofactor on specific genes or in certain situations. The coactivator function of PARP can be observed with activators that play a role in many biological processes. Thus, PARP also could act in other processes, such as DNA replication (29), that are subject to regulation by site-specific DNA-binding proteins that include bonafide transcriptional activators. It remains to be tested whether binding of PARP to nicked DNA could induce similar events at significantly lower PARP concentrations, leading, for example, to recruitment of factors associated with the basal machinery to damaged DNA in transcriptionally active regions. Future experiments could help to elucidate whether PARP interacts with basal factors or with functionally distinct components, such as DNA repair proteins, that have been reported to associate with the basal machinery (38, 39).
Acknowledgments
We are grateful to E. Ackerman and A. Spoonde for providing bacterially expressed and affinity-purified PARP proteins and J. Kellermann for performing the protein sequencing of PARP. We further thank P. Halle and S. Malik for their help with reagents and I. Gander for critical reading of the manuscript. This work was supported by grants from the Bundesministerium für Bildung, Forschung, und Technologie (BMBF), and the Deutsche Forschungsgemeinschaft (SFB 190) to M.M. and by grants from the National Institutes of Health to R.G.R.
ABBREVIATIONS
- TFII
transcription factor II
- PARP
poly(ADP-ribose) polymerase
- GTFs
general transcription factors
- PIC
preinitiation complex
- PC
positive cofactor
- USA
upstream factor stimulatory activity
References
- 1.Kornberg R. Trends Biochem Sci. 1996;21:325–326. [PubMed] [Google Scholar]
- 2.Kim Y-J, Björklund S, Li Y, Sayre M H, Kornberg R. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 3.Orphanides G, Lagrange T, Reinberg D. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 4.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 5.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. Nature (London) 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 6.Dynlacht B D, Hoey T, Tjian R. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 7.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 8.Burley S, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser K, Meisterernst M. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 11.Liebermann P. Mol Cell Biol. 1994;14:8365–8375. doi: 10.1128/mcb.14.12.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kretzschmar M, Stelzer G, Roeder R G, Meisterernst M. Mol Cell Biol. 1994;14:3927–3937. doi: 10.1128/mcb.14.6.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kretzschmar M, Meisterernst M, Roeder R G. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge H, Roeder R G. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 15.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 16.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. Nature (London) 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 17.Brou C, Kuhn A, Staub A, Chaudhary S, Grummt I, Davidson I, Tora L. Nucleic Acids Res. 1993;21:4011–4018. doi: 10.1093/nar/21.17.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shykind B M, Kim J, Sharp P A. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl T, Satoh M S, Poirier G G, Klungland A. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 20.Althaus F R, Richter C. Mol Biol Biochem Biophys. 1987;37:1–126. [PubMed] [Google Scholar]
- 21.de Murcia G, Menissier-de Murcia J M, Schreiber V. BioEssays. 1991;13:455–462. doi: 10.1002/bies.950130905. [DOI] [PubMed] [Google Scholar]
- 22.Boulikas T. Toxicol Lett. 1993;67:129–150. doi: 10.1016/0378-4274(93)90051-x. [DOI] [PubMed] [Google Scholar]
- 23.Satoh M S, Lindahl T. Nature (London) 1992;356:356–359. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee S, Berger N A. Mol Cell Biochem. 1994;138:61–69. doi: 10.1007/BF00928444. [DOI] [PubMed] [Google Scholar]
- 25.Stevnsner T, Ding R, Smulson M E, Bohr V A. Nucleic Acids Res. 1994;22:4620–4624. doi: 10.1093/nar/22.22.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smulson M E, Istock N, Ding R, Cherney B. Biochemistry. 1994;33:6186–6191. doi: 10.1021/bi00186a018. [DOI] [PubMed] [Google Scholar]
- 27.Berger N A. Radiat Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 28.Ding R, Smulson M E. Cancer Res. 1994;54:4627–4634. [PubMed] [Google Scholar]
- 29.Simbulan-Rosenthal C M, Rosenthal D S, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson M E. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- 30.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 31.Slattery E, Dignam J D, Matsui T, Roeder R G. J Biol Chem. 1983;258:5955–5959. [PubMed] [Google Scholar]
- 32.Griffin R J, Curtin N J, Newell D R, Golding B T, Durkacz B W, Calvert A H. Biochemie. 1995;77:408–422. doi: 10.1016/0300-9084(96)88154-5. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 34.Gradwohl G, Menissier-de Murcia J M, Molinete M, Simonin F, Koken M, Hoeijmakers J H, de Murcia G. Proc Natl Acad Sci USA. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaiser K, Stelzer G, Meisterernst M. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gradwohl G, Mazen A, de Murcia G. Biochem Biophys Res Commun. 1987;148:913–919. doi: 10.1016/s0006-291x(87)80219-x. [DOI] [PubMed] [Google Scholar]
- 37.Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill D M, Miwa M. J Biol Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- 38.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J-P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J-M. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 39.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]