Abstract
The sea urchin early histone repeating unit contains one copy of each of the five histone genes whose coordinate expression during development is regulated by gene-specific elements. To learn how within the histone repeating unit a gene-specific activator can be prevented to communicate with the heterologous promoters, we searched for domain boundaries by using the enhancer blocking assay. We focused on the region near the 3′ end of the H2A gene where stage-specific nuclease cleavage sites appear upon silencing of the early histone genes. We demonstrated that a DNA fragment of 265 bp in length, defined as sns (for silencing nucleoprotein structure), blocked the enhancer activity of the H2A modulator in microinjected sea urchin embryos only when placed between the enhancer elements and the promoter. We also found that sns silenced the modulator elements even when placed at 2.7 kb from the promoter. By contrast, the enhancer activity of the modulator sequences, located downstream to the coding region, was not affected when sns was positioned in close proximity to the promoter. Finally, the H2A sns fragment placed between the simian virus 40 regulative region and the tk promoter repressed chloramphenicol acetyltransferase expression in transfected human cell lines. We conclude that 3′ end of the H2A gene contains sequence elements that behave as functional barriers of enhancer function in the enhancer blocking assay. Furthermore, our results also indicate that the enhancer blocking function of sns lacks enhancer and species specificity and that it can act in transient assays.
Genetic and biochemical evidence supports the notion that chromatin is organized in discrete functional domains and that each domain defines a functionally autonomous unit of gene expression (1–3). Such an organization infers the existence of topological barriers, flanking the domain, that would preserve gene units in which they reside from the influence of regulatory sequences located in neighboring domains. Domain boundary elements can take the form of specialized chromatin structures or are the binding sites for regulators. The best characterized are as follows: the scs elements of the Drosphila 87A1 hsp 70 locus (4–6), the 5′ HS4 element of the chicken β-globin locus control region (LCR) locus (7), and the cluster of binding sites for the Drosphila suppressor of Hairy-wing [su(Hw)] protein of the gypsy retrotrasposon (8, 9). These elements are chromatin insulators in that when placed at the borders of a transcription unit they reduce the position effect at the integration sites in gene transfer experiments (7, 10–14).
Another element thought to be associated with domain structures is the matrix or scaffold-associated region (MAR or SAR). However, while some SARs attach to a nuclear matrix, few elements, such as the A elements of the chicken lysozyme gene and the MAR elements of the human Apolipoprotein gene, confer position-independent expression to linked reporter genes in stable transformants (15, 16). SAR sequences have been found in close association with regulative elements and are thought to define the basis of a chromatin loop (17). The mutually exclusive binding of histone H1 and high mobility group I/Y has led to the suggestion that SAR sequences are involved in the condensation and decondensation of chromatin domains and hence in the regulation of gene expression (18).
Scaffold-associated region elements have been mapped in the intergenic H1–H3 spacer of the Drosophila histone repeat unit (17). By analogy, it might be postulated that chromosomal boundary elements subdivide the tandem repeated arrays of the sea urchin early histone genes (19) into discrete functional domains, each domain containing one repeat. Topological constraining of the repeating unit in one functional domain might give the advantage of placing the five histone genes under the control of master regulators, so that their transient expression in the early cleavage embryos (20) can be regulated in a coordinated fashion. However, extensive investigations on the molecular mechanism underlying the timing of transcription in the sea urchin embryo indicate that each of the five early histone genes, within a tandem repeat, constitutes an independent transcription unit with different promoter organization and gene-specific regulative elements (21–25). Hence, if we assume that the five histone genes are confined in a chromosomal domain, then some mechanisms must operate to prevent a gene specific enhancer to activate the other promoters of the repeating unit.
The modulator sequence element of the early H2A gene, first described by Birnstiel and collaborators (26, 27), is the only transcriptional enhancer identified in the early histone repeating unit. Its binding factor, the modulator binding factor 1 (MBF-1), is required for maximal activity of the early H2A promoter at morula stage but is not involved in the silencing of the early H2A gene at mesenchyme blastula/gastrula stage (28). Since the modulator, like enhancers, communicates with its target promoter in an orientation- and position-independent manner (28), what then prevents the MBF-1 activator, once bound to the modulator, to contact downstream and upstream promoters in the histone repeating unit?
We have started addressing these issues by looking for enhancer blocking elements. In this paper we present evidence for one sequence element located near the 3′ end of the sea urchin H2A early subtype histone gene that insulates homologous and heterologous enhancers from the basal promoter.
MATERIALS AND METHODS
Constructions of Plasmids.
The DNA constructs, schematically drawn in Figs. 2, 4, and 5 were constructed from the tk-70 pBL2 and the pBL3-CAT vectors (29). Multiple copies of the modulator oligonucleotide were cloned upstream the tk and downstream the chloramphenicol acetyltransferase (CAT) coding region as described (28). The simian virus 40 (SV40) enhancer fragment that included the 21-bp repeats was cloned upstream the tk promoter of the tk-70 pBL2 vector. The sns fragment (for silencing nucleoprotein structure) was isolated by HpaII and Sau3A restriction enzyme digestion of the SacI subclone of PH70 histone DNA (20). In some constructs the 265-bp-long sns DNA or a pUC18 fragment of an equivalent size were cloned between the modulator or the SV40 enhancer and the tk promoter. In other constructs the sns fragment was cloned in the downstream multiple cloning sites, either 3′ of the CAT coding region or 3′ of the modulator sequences. The orientation of the DNA inserts was determined by sequence analysis.
Figure 2.
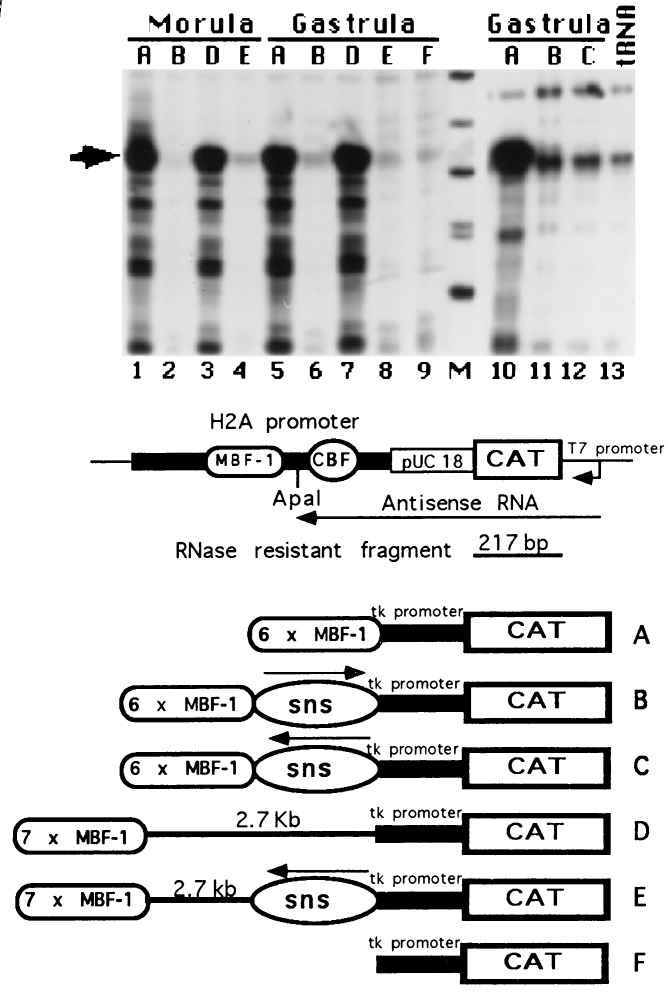
Modulator blocking activity by sns sequences in microinjected sea urchin embryos. (Top) CAT RNA transcripts were detected by RNase protection assay. 32P-labeled antisense RNA, transcribed in vitro from the H2A-CAT DNA containing a pUC18 insert (28) and schematically drawn (Middle), was hybridized with total RNA extracted from 50 sea urchin embryos at the indicated developmental stage, microinjected with the constructs A-F depicted (Bottom) or with 100 μg of yeast tRNA. One half of the RNase-resistant products were run on denaturing polyacrylamide gel along with labeled HpaII-digested end labeled pUC18 fragments (lane M). Only fragments ranging from 353 bp to 110 bp are shown. Gels were dried and exposed overnight at −80°C with an intensifying screen. The expected 217-nucleotide resistant fragment is indicated by an arrow.
Figure 4.
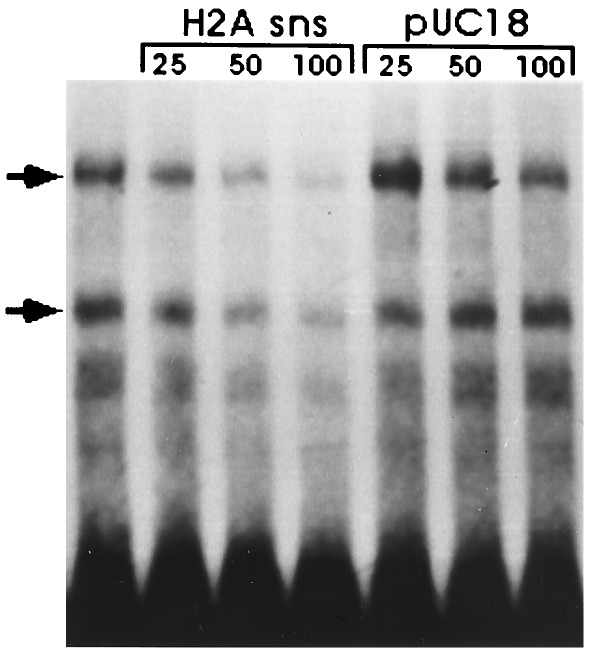
Nuclear protein binding at the sns sites. The HpaII–Sau3A fragment (Fig. 1) was 3′ end labeled with [α-32P]dATP and the large fragment of DNA polymerase I. The probe (1.5 ng) was incubated with 5 μg of nuclear extract from gastrula embryos in the absence or in the presence of 25, 50, and 100 ng of unlabeled homologous and plasmid fragments, respectively. Nuclear protein complexes were fractionated by acrylamide gel electrophoresis.
Figure 5.
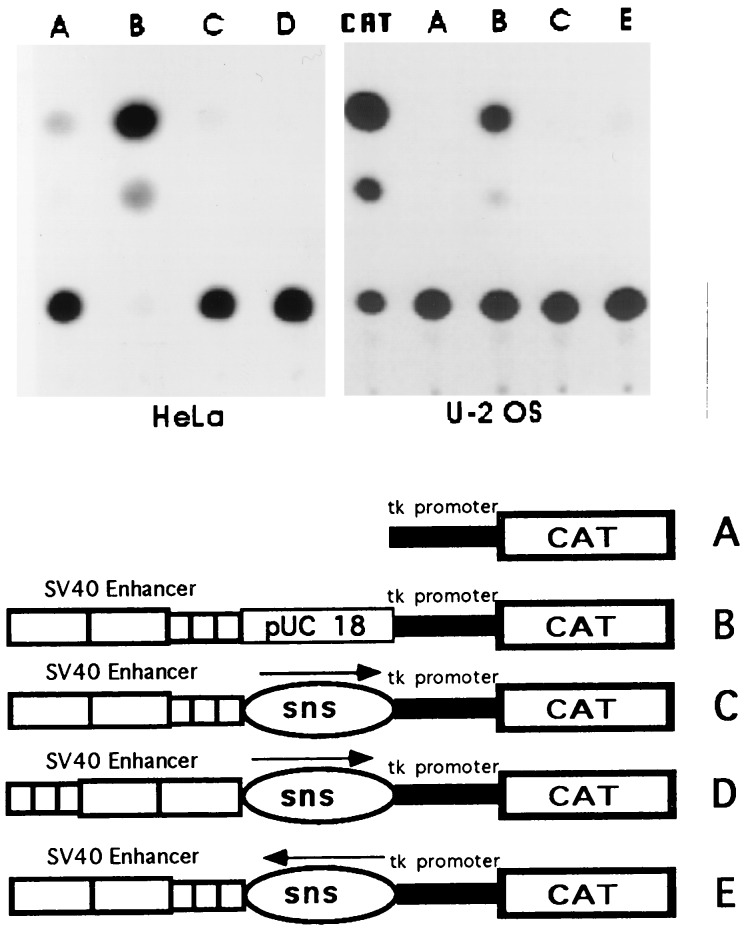
SV40 enhancer blocking function by the sns fragment in human cell lines. The plasmid constructs A–E were transfected into HeLa cells and in the osteosarcoma cell line U-2 OS. Detection of the CAT enzymatic activity on cellular extracts were performed 48 h after transfection. CAT refers to an enzymatic reaction carried out by using 1.2 units of a commercial chloramphenicol acetyltransferase. The autoradiogram shows the thin-layer chromatographic separation of [14C]chloramphenicol and its acetylated derivatives.
Gene Transfer and CAT Expression.
Plasmids were linearized by digestion with either HindIII or ClaI restriction enzymes whose sites are located in the polylinker regions 5′ and 3′ to the CAT sequences, respectively. Linearized plasmids were brought to a total concentration of 200 μg/ml with HindIII digested Paracentrotus lividus sperm DNA of roughly 5 kb and were microinjected into sea urchin P. lividus eggs from a mature female as described (28). Embryos were raised and processed to monitor CAT gene expression by RNase protection experiments. Nucleic acid samples from a pool of 50–70 microinjected embryos, at morula (5 h of development) or early gastrula (20 h of development) stages, were digested with RNase-free DNase I and hybridized with 32P-labeled antisense CAT RNA transcribed in vitro from the H2A- INSERT-CAT plasmid (28). Hybridization conditions, RNase digestion, and gel fractionation of the RNase-resistant hybrids were as described (30). Transfection was carried out by incubating the human cell lines, HeLa and U-2 OS, with calcium phosphate precipitates containing 20 μg of recombinant SV40 CAT plasmids and 2 μg of the internal control β-galactosidase expression plasmid pON1 (31). Cell lysates were assayed for β-galactosidase activity (31), normalized, and then assayed for CAT enzymatic activity (32).
Electrophoretic Mobility Shift Assays.
Nuclear extracts from P. lividus embryos at the gastrula stage were prepared as described (33). Nuclear extracts (5 μg) were preincubated with 7 μg of poly(dA-dT)·(dA-dT) and O.3 μg of Escherichia coli DNA in 15 μl of 10 mM Hepes (pH 7.9), 60 mM KCl, 1 mM DTT, 1 mM EDTA, and 4% Ficoll for 5 min on ice before the addition of 1.5 ng of end labeled HpaII–Sau3A H2A fragment. After 20 min incubation on ice, binding reactions were loaded onto 4% polyacrylamide gel in 50 mM Tris, 50 mM H3BO3, and 2 mM EDTA (pH 8.3) and electrophoresed at 10 V/cm for 2 h at room temperature. For competition experiments unlabeled homologous or plasmid DNA fragments in the amounts described in the legend to Fig. 3 were added to the preincubation mixture prior to the addition of the extract.
Figure 3.
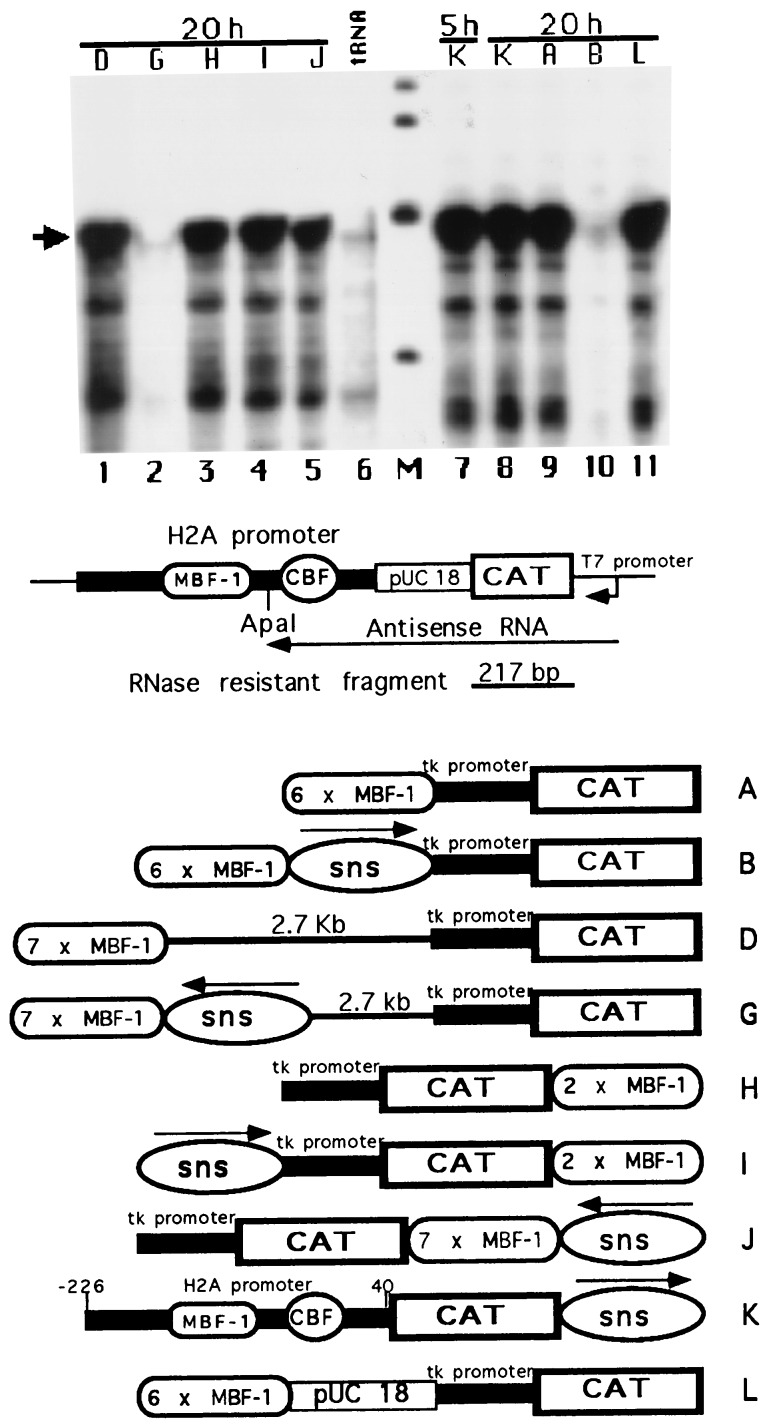
Detection of the enhancer blocking function of the sns element placed at different locations with respect to the promoter. Sea urchin embryos were microinjected with the constructs drawn in the lower part of the figure. RNase protection to monitor CAT transcription was carried out as in Fig. 2. Developmental stages expressed as hours of postfertilization are indicated on top of the figure. The lanes marked 5 h and 20 h corresponded to morula and early gastrula stages, respectively. The marker DNA fragments (lane M) range in size from 344 to 154 bp and are derived from pBR322 cut with HinfI restriction enzyme and end labeled. Fluorograms were exposed overnight. Arrow refers to the 217-nucleotide protected RNA band.
RESULTS
Modulator Blocking Sequences Are Located Near the 3′ End of the H2A Gene.
Chromatin boundary elements are often flanked by micrococcal nuclease cutting sites or are embedded in nuclease hypersensitive domains (4, 7). As previously shown, the early histone gene chromatin undergoes structural rearrangements during development that can be summarized as follows. First, the regular nucleosomal packaging that is lost when the early histone genes are transcriptionally activated, is regained at mesenchyme blastula stage upon silencing (34). Second, at mesenchyme blastula stage the DNA region located in proximity of the 3′ palindromic sequence of the early H2A is preferentially cleaved by the micrococcal nuclease (35).
In light of these observations, we explored the possibility that chromatin boundary elements reside in the DNA region containing the nuclease hypersensitive cutting sites located near the 3′ end of the H2A gene. Previously, the micrococcal nuclease cleavage sites were erroneously mapped downstream to the 3′ palindromic sequence because of the partial DNA sequence that was available when the experiments were performed. With the completion of the DNA sequence gaps we relocated these sites in the H2A stop codon and at 200 bp from it (Fig. 1). To search for chromatin boundary elements we performed the enhancer blocking assay (5). The DNA fragment containing the micrococcal cleavage sites flanking the 3′ H2A palindromic sequence was isolated by cutting a PH70 (20) subclone with the HpaII and Sau3A restriction enzymes (Fig. 1). The DNA fragment was defined as sns. The sns fragment was cloned in both orientations between multiple copies of the modulator sequence element and the tk promoter fused to a CAT reporter gene. The constructs depicted in Figs. 2 and 3 were microinjected into sea urchin eggs, embryos raised and processed to determine CAT expression by the RNase protection assay. The results of four different microinjection experiments are presented in Figs. 2 and 3. The tk-CAT vector, construct F, is not expressed (Fig. 2, lane 9). In agreement to our previous evidence (28), the presence of multiple copies of the MBF-1 binding sites, as in construct A, activated CAT expression from the tk promoter at morula (Fig. 2, lane 1) and gastrula (Fig. 2, lanes 5 and 10; Fig. 3, lane 9) stages. Placing the H2A sns element between the enhancer and the promoter, constructs B and C, drastically reduced the transcription of the reporter gene (Fig. 2, lanes 2, 6 and 11; Fig. 3, lane 10). This modulator blocking activity was independent of the orientation (Fig. 2, lane 12). To test whether the absence of detectable CAT transcripts in embryos microinjected with constructs B and C was not due to the increased distance of the modulator sequence elements from the tk promoter, we cloned seven copies of the modulator sequence elements in the polylinker of the tk-CAT vector (29) located downstream of the CAT coding sequence. When this plasmid was cut with the ClaI restriction enzyme, whose site is located between the CAT gene and the modulator elements, the linear construct D was generated (Figs. 2 and 3). In this construct 2.7 kb of plasmid DNA separates the modulator sequences from the tk promoter. Nevertheless, the extent of CAT transcription in embryos at morula and gastrula stages microinjected with construct D (Fig. 2, lanes 3 and 7) is identical to that observed in embryos at the same developmental stages raised from the same batch of eggs microinjected with construct A (Fig. 2, lanes 1 and 5). These results confirm that the enhancer activity of the modulator sequence elements is independent of the transcriptional state of the early histone genes (28). Again, placing the H2A sns fragment in the inverted orientation between the enhancer and promoter, as in constructs E, silenced the enhancer function of the modulator elements, and no CAT expression was detected in the microinjected embryos (Fig. 2, lanes 4 and 8).
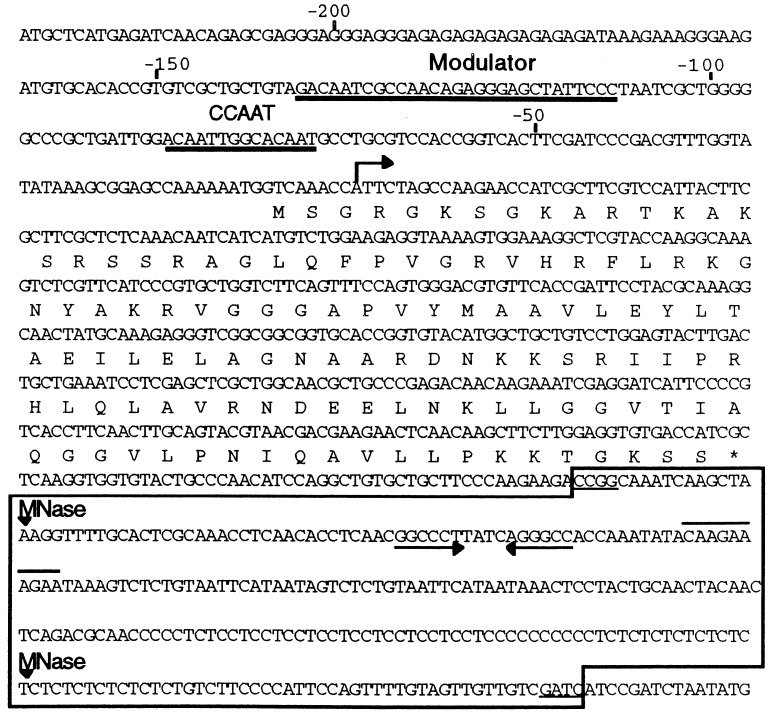
Figure 1.
Nucleotide sequence of early H2A gene and flanking regions. Thick underlines indicate the binding sites for the MBF-1 and CCAAT binding factor (CBF) factors to, respectively, the modulator and the CCAAT sequences of the H2A promoter. Arrow refers to the transcription starting site. The deduced amino acid sequence of the H2A coding region is reported above the DNA sequence. Arrowheads pointing to the sequence refer to the mesenchyme blastula-specific micrococcal nuclease (MNase) cutting sites. The 3′ palindrome is indicated by arrows while the conserved motif for the RNA processing is overlined. The sns sequence is boxed, and the HpaII and Sau3A recognition sites flanking the sns fragment are underlined.
The H2A sns Sequences Insulate the Modulator from the Basal tk Promoter.
To provide further insights regarding both the specificity and the mechanism of action of the antimodulator element, we monitored CAT expression in sea urchin embryos injected with the constructs depicted in Fig. 3. In one construct (L), the H2A sns element was substituted with a plasmid fragment of an equivalent size. Equivalent amounts of CAT transcripts were detected by RNase protection experiments in microinjected embryos at 20 h of development (Fig. 3, lane 11), indicating that placing the plasmid fragment between the modulator and the tk promoter did not block the enhancer function, as it did the H2A sns fragment. In the construct K, the H2A sns fragment was cloned downstream to the CAT coding region whose expression is under the control of the early H2A promoter. We showed previously (28) that a H2A 5′ flanking region which included the binding sites for the modulator (MBF-1) and the CCAAT (CBF) factors (see Fig. 1 for the DNA sequence) can promote reporter gene expression even in embryos at the gastrula stage (20 h postfertilization) when the early histone genes are transcriptionally silent. As expected, placing the H2A sns element downstream the CAT coding region had no effect on the level of CAT gene expression at both early (5 h postfertilization) and late (20 h postfertilization) developmental stages (Fig. 3, lanes 7 and 8). A similar result was obtained with construct J in which the sns fragment was moved downstream to the heptamerized modulator sequences placed 3′ to the coding region (Fig. 3, lane 5). Finally, to distinguish between enhancer blocking and promoter interference, two further constructs were microinjected in the sea urchin embryo. In construct G (Fig. 3) the sns fragment was moved 2.7 kb away from the tk promoter and placed in closed association with the heptamerized MBF-1 binding site. Construct I (Fig. 3) had the sns sequences placed immediately upstream the basal tk promoter, whereas a dimer of the modulator sequence was located downstream to the CAT coding region. As shown in Fig. 3, placing the sns fragment away from the tk promoter efficiently repressed CAT expression (compare lanes 1 and 2). By contrast, the abundance of the reporter gene transcripts in gastrula embryos injected with construct I (lane 4) was similar to that of embryos injected with construct D (lane 1). From these results, we conclude that (i) the sns sequences abolished CAT transcription by affecting the function of the modulator elements and not by interfering with the activity of the basal promoter, and (ii) the repression of the enhancer activity of the modulator is not due to the binding of negative regulators to the sns fragment, but rather to a mechanism by which one or more elements function as chromatin boundaries that insulate the enhancer from the promoter.
Nuclear Protein Binding to the sns Sequences.
As a first effort on the characterization of the proteins that bind to the H2A sns boundary element we performed an electrophoretic mobility shift assay. Two major DNA binding activities were detected by gel shift with crude nuclear extracts from sea urchin gastrula embryos (Fig. 4). The two proteins seem to interact specifically with the H2A sns sequences since addition of molar excess of unlabeled homologous DNA reduced the formation of the labeled complexes, whereas plasmid sequences competed very poorly. However, attempts to identify the interacting sequences by DNase I footprinting showed no clear protection pattern on the H2A fragment (not shown). This would imply that either the two complexes recognize structural features rather that specific sequences on the sns DNA, or the protein complexes bind with low affinity to the DNA.
The Enhancer-Blocking Activity of H2A sns Element Is Not Enhancer- and Species-Specific.
To try to understand more of the function of the H2A sns element, we tested whether this element was capable to insulate an heterologous enhancer from the basal promoter and in a different species. We chose the SV40 enhancer because it activates transcription from heterologous promoters of linked reporter genes in a variety of vertebrate systems. The H2A sns fragment or plasmid sequences were cloned between the SV40 enhancer and the basal tk promoter fused to the reporter CAT gene. These constructs, schematically drawn in Fig. 5, were transiently transfected in two human cell lines, Hela and U-2 OS. The expression of the reporter gene was monitored 2 days after transfection by detecting the CAT enzymatic activity on total cellular extracts. As shown in Fig. 5, the H2A sns fragment insulated the SV40 enhancer from the tk promoter in either orientation. Substituting the H2A sns element with plasmid sequences no blocking effect was detected.
DISCUSSION
We have demonstrated that a DNA fragment located near the 3′ end of the early histone H2A gene, defined as sns, contains sequence elements that repress enhancer function in the enhancer blocking assay. As functional tests in the sea urchin embryo, we microinjected CAT expression vectors whose promoters were under the influence of multiple copies of the 30-bp-long modulator element. As previously demonstrated, the modulator can be equated to an enhancer since one copy of this element was sufficient to elicit transcription from an heterologous promoter in a position and orientation manner (26, 28). Here we confirmed these data and further show (Figs. 2 and Fig. 3) that moving the heptamerized modulator sequences at almost 3 kb from the basal tk promoter did not reduce the extent of transcription of the linked reporter gene. Similar results were obtained with two copies of the modulator (data not shown). Despite the presence of multiple copies of the modulator in several of the constructs we tested, the sns fragment when inserted between the modulator elements and the tk promoter prevented transcriptional activation of the reporter gene. This enhancer blocking function was position-dependent because placing the sns fragment in any other location did not affect transcription of the CAT gene. From these results we may suggest that sns represses transcription by an active silencing mechanism with the binding of one or more repressor molecules.
The sns fragment, concomitantly with the repression of the early histone genes repeating unit (20), becomes most probably associated with a positioned nucleosome (35). Since a positioned nucleosome nearby a promoter might affect transcription, constructs in which either element, the modulator and the sns, were placed at different locations were microinjected into sea urchin embryos and tested for CAT gene expression. Two lines of evidence indicate that the sns element blocks the activity of enhancers rather than interfering with the promoter. First, repression of CAT expression was observed in sea urchin embryos microinjected with construct G (Fig. 3) in which the sns fragment was cloned downstream the modulator sequences at 2.7 kb from the basal promoter. Second, when the sns fragment was placed immediately upstream to the tk promoter, while the modulator elements was moved downstream to the reporter coding sequences (construct I of Fig. 3), the abundance of CAT transcripts did not change significantly.
The sns region contains different sequences features: the last four and the termination codons of the H2A gene, the G+C-rich inverted repeat and the CAAGAAAGA motif, and the homopyrimidine tract. The palindrome and the CAAGAAAGA sequence are highly conserved features of all five histone genes (36) and are required for the 3′ processing of the histone mRNA precursors (37). Similar hairpin–loop structures and downstream sequence elements are needed for the production of mature histone mRNA analyzed in other systems (38). Interestingly, a highly conserved homopyrimidine sequence, where a S1 nuclease cleavage site has been mapped (39), is also located in similar position in the early H2A histone gene of the closely related sea urchin Psammechinus miliaris (M. Birnstiel, personal communication). It remains to be seen whether all these sequence features are needed for the repression of the enhancer function.
Our results suggest, but do not prove, that the H2A sns contains a chromatin insulator. Consistent with this hypothesis is the evidence that enhancer activity is not affected when the sns fragment is placed 3′ to the coding region. If the H2A sns element is a chromatin insulator, then what is its function in the histone repeating unit? Given that the H2A modulator is the only transcriptional enhancer identified among the regulatory regions of the early histone genes (26–28), we may postulate the following model. The H2A sns element, in its normal location, would create a chromosomal functional domain containing the five genes, whose promoters should then all be under the influence of the H2A modulator. Alternatively, the sns element could block the H2A-specific MBF-1 transcription factor in the activation of the heterologous promoter of the neighboring transcriptional units.
The elucidation of the mechanisms by which the H2A sns sequences blocks enhancer function could help the clarification of the mechanism of action of the modulator element. We may speculate that nucleoprotein complexes at the sns sequences, when inserted between an enhancer and a promoter, interfere with DNA looping or with the tracking of the MBF-1 transcription factor bound at the enhancer. Alternatively, as it has been shown for the β-globin locus control region insulator (7), they could block MBF-1 from displacing a nucleosome over the modulator. A mechanism of action consisting in the alteration of the nuclear compartmentalization of the transcription unit is less likely. In fact, we observed repression of the SV40 enhancer by the sea urchin H2A sns sequences in transient assays, in the human cell lines, where the transfected DNA might not be properly packed into chromatin. This result is not surprising because it has been shown that the binding sites for the suppressor of Hairy-wing [su(Hw)] protein can insulate the heat shock elements from the basal promoter also in a transient assay (40). Therefore, our results would indicate that enhancer blocking by the sns complex does not require integration in the genome. In addition, they would imply an evolutionary conservation for the function of the sea urchin H2A sns sequences.
Finally, the lack of species and enhancer specificity of action of the H2A sns element has some interesting practical implications. Integration of exogenous DNA in gene transfer experiments and in transgenic animals occurs randomly into chromatin; therefore, the constructs are subjected to position effects at the integration site that cause differences in expression (41). When placed flanking a transcriptional unit the H2A sns element could drastically reduce this chromosomal position effect at the integration site. The demonstration of a chromatin insulator function for the sea urchin H2A sns sequences should clarify this issue.
Acknowledgments
We are grateful to Drs. A. Giallongo and A. Di Leonardo for providing us the human cell lines, and to Dr. M. Sollazzo for revising the manuscript. The technical assistance of Mr. D. Cascino and Mr. A. Oliva and the photography assistance of Mr. S. Buccoleri is acknowledged. This work was supported in part by the Progetto Finalizzato Ingegneria Genetica and by funds of the Ministero dell’ Università e della Ricerca Scientifica e Tecnologica (40% and 60%).
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- SV40
simian virus 40
- sns
silencing nucleoprotein structure
- MBF-1
modulation binding factor
- CBF
CCAAT binding factor
Footnotes
References
- 1.Eissenberg J C, Elgin S C R. Trends Genet. 1991;7:335–340. doi: 10.1016/0168-9525(91)90424-o. [DOI] [PubMed] [Google Scholar]
- 2.Dillon N, Grosveld F. Curr Opin Genet Dev. 1994;4:260–264. doi: 10.1016/s0959-437x(05)80053-x. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe A P. Curr Biol. 1994;4:85–87. doi: 10.1016/s0960-9822(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 4.Udvardy A, Maine E, Schedl P. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 5.Kellum R, Schedl P. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vazquez J, Schedl P. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J H, Whiteley M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 8.Geyer P K, Corces V G. Genes Dev. 1993;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 9.Harrison D A, Gdula D A, Coyne R S, Corces V G. Genes Dev. 1992;7:1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- 10.Kellum R, Schedl P. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 11.Roseman R R, Pirrotta V, Geyer P K. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai H, Levine M. Nature (London) 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 13.Gerasimova T, I, Gdula D A, Gerasimov D V, Simonodova O, Corces V G. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 14.Scott S K, Geyer P K. EMBO J. 1995;24:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonifer C, Vidal M, Grosveld F, Sippel A E. EMBO J. 1990;9:2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalos M, Fournier R E K. Mol Cell Biol. 1995;15:198–207. doi: 10.1128/mcb.15.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli U K, Käs E, Poiljak L, Adachi Y. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhao K, Käs E, Gonzales E, Laemmli U K. EMBO J. 1993;8:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentschel C, Birnstiel M. Cell. 1981;25:301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- 20.Spinelli G, Gianguzza F, Casano C, Acierno P, Burckhardt J. Nucleic Acids Res. 1979;6:545–560. doi: 10.1093/nar/6.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Liberto M, Lai Z C, Fei H, Childs G. Genes Dev. 1989;3:973–985. doi: 10.1101/gad.3.7.973. [DOI] [PubMed] [Google Scholar]
- 22.Lee I J, Tung L, Bumcrot D A, Weinberg E S. Mol Cell Biol. 1991;11:1048–1061. doi: 10.1128/mcb.11.2.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell J, Char B R, Maxson R. Dev Biol. 1992;150:363–371. doi: 10.1016/0012-1606(92)90248-f. [DOI] [PubMed] [Google Scholar]
- 24.Fei H, Childs G. Dev Biol. 1993;155:383–395. doi: 10.1006/dbio.1993.1037. [DOI] [PubMed] [Google Scholar]
- 25.Palla F, Casano C, Albanese I, Anello L, Gianguzza F, Di Bernardo M G, Bonura C, Spinelli G. Proc Natl Acad Sci USA. 1989;86:6033–6037. doi: 10.1073/pnas.86.16.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosschedl R, Birnstiel M L. Proc Natl Acad Sci USA. 1980;77:7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosschedl R, Machler M, Rohrer U, Birnstiel M L. Nucleic Acids Res. 1983;11:8123–8136. doi: 10.1093/nar/11.23.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palla F, Bonura C, Anello L, Di Gaetano L, Spinelli G. Proc Natl Acad Sci USA. 1994;91:12322–12326. doi: 10.1073/pnas.91.25.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckow B, Schütz G. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Bernardo M, Russo R, Oliveri P, Melfi R, Spinelli G. Proc Natl Acad Sci USA. 1995;92:8180–8184. doi: 10.1073/pnas.92.18.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spaete R R, Mocarski E S. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spinelli G, Ciliberto G. Nucleic Acids Res. 1985;13:8065–8081. doi: 10.1093/nar/13.22.8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palla F, Bonura C, Anello L, Casano C, Spinelli G. Proc Natl Acad Sci USA. 1993;90:6854–6858. doi: 10.1073/pnas.90.14.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spinelli G, Albanese I, Anello L, Ciaccio M, Di Liegro I. Nucleic Acids Res. 1982;10:7977–7992. doi: 10.1093/nar/10.24.7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anello L, Albanese I, Casano C, Palla F, Gianguzza F, Di Bernardo M G, Di Marzo R, Spinelli G. Eur J Biochem. 1986;156:367–374. doi: 10.1111/j.1432-1033.1986.tb09592.x. [DOI] [PubMed] [Google Scholar]
- 36.Hentschel C C, Irminger J, Bucher P, Birnstiel M L. Nature (London) 1980;285:147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- 37.Birnstiel M L, Busslinger M, Strub K. Cell. 1985;41:349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- 38.Mowry L, K, Steitz J A. Trends Biochem. 1988;13:447–451. doi: 10.1016/0968-0004(88)90220-4. [DOI] [PubMed] [Google Scholar]
- 39.Hentschel C C. Nature (London) 1982;295:714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- 40.Holdridge C, Dorsett D. Mol Cell Biol. 1991;11:1894–1900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson G, Garrick D, Wu W, Kearns M, Martin D, Whitelaw E. Proc Natl Acad Sci USA. 1995;92:5371–5375. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]