Abstract
The biological activities of the retinoids are mediated by two nuclear hormone receptors: the retinoic acid receptor (RAR) and the retinoid-X receptor (RXR). RXR (and its insect homologue ultraspiracle) is a common heterodimeric partner for many other nuclear receptors, including the insect ecdysone receptor. As part of a continuing analysis of nuclear receptor function, we noticed that, whereas RXR can be readily expressed in Escherichia coli to produce soluble protein, many of its heterodimeric partners cannot. For example, overexpression of RAR results mostly in inclusion bodies with the residual soluble component unable to interact with RXR or ligand efficiently. Similar results are seen with other RXR/ultraspiracle partners. To overcome these problems, we designed a novel double cistronic vector to coexpress RXR and its partner ligand-binding domains in the same bacterial cell. This resulted in a dramatic increase in production of soluble and apparently stable heterodimer. Hormone-binding studies using the purified RXR–RAR heterodimer reveal increased ligand-binding capacity of both components of 5- to 10-fold, resulting in virtually complete functionality. Based on these studies we find that bacterially expressed receptors can exist in one of three distinct states: insoluble, soluble but unable to bind ligand, or soluble with full ligand-binding capacity. These results suggest that coexpression may represent a general strategy for biophysical and structural analysis of receptor complexes.
The lipophilic hormones, including the steroids, retinoids, thyroxin, and vitamin D3, are potent regulators of development, cell differentiation, and organ physiology. The pleiotropic effects of these hormones are mediated through nuclear receptors. Based on their hormone-binding, DNA-binding, and dimerization properties, the nuclear receptors can be grouped into four classes: steroid receptors, homodimeric orphan receptors, monomeric receptors, and obligate heterodimeric receptors with the retinoid-X receptor (RXR) as a partner (1). In contrast to the first three classes, the heterodimeric receptors have the potential to regulate gene expression in response to two different hormones. This introduces increased complexity in the signaling pathways.
Retinoic acid receptors (RARs) and RXRs [each of which has three different mammalian isoforms (2–7)] mediate the effects of metabolites deriving from vitamin A (retinol) (3, 8–10) and are hence involved in a wide spectrum of biological activities, including cell proliferation and differentiation, embryonic development, and spermatogenesis (11–14). Similarly, the insect ecdysone receptor (ECR) has been shown to bind to and activate target genes by forming a heterodimer with either RXR or its Drosophila counterpart ultraspiracle (USP) (15, 16).
Mechanisms of hormone action are unclear but involve allosteric changes between ligand-bound and free receptor complexes. Substantial evidence suggests that the heterodimeric partners communicate with each other, presumably through their dimer interface, as well as potentially through their associated cofactors. To explore the mechanistic basis for these interactions and their functional relevance, we designed a novel double cistronic vector that directs expression of the receptor heterodimer in a single cell. In this paper we describe the expression and characterization of the RAR and RXR ligand-binding domains (LBDs). When expressed alone, the RAR LBD was found to be largely insoluble, and the formation of heterodimeric complexes was inefficient. However, by employing a double cistronic expression system, we were able to express high levels of soluble tightly associated heterodimer. This suggests that the coexpression of RXR improves the solubility and perhaps assists correct folding of RAR, and RXR may thus serve as a molecular chaperone. Furthermore, bacterial cultures expressing heterodimer yield substantially more functional protein than those expressing either RXR or RAR alone. Ligand binding does not affect the stability of the heterodimer, nor does the binding of one ligand (agonist or antagonist) significantly influence the binding of another ligand to the other LBD in the heterodimer. In addition, the ligand-binding capacity of the singly expressed proteins is only 10–20% of that of the complex, which is close to 100% for both components of the heterodimer. The power and generality of this approach is further explored by testing the solubility of the ecdysone receptors. As with the RAR, production of soluble ECR is dramatically improved by coexpression with USP, suggesting that this strategy provides a general approach to the study of the biophysical and structural properties of the nuclear receptor complexes.
MATERIALS AND METHODS
Preparation of Expression Constructs.
The human RXRα, RXRβ, and RARβ LBDs were first cloned into the BamHI site of pGEX-KT (17) and the NdeI site of pET-15b (Novagen) vectors. Following this, DNA sequences encoding the RXR and RAR LBDs along with bacterial ribosome-binding sites were obtained from the pET-15b–receptor constructs using PCR techniques with the following primers: GGA TCC CCG GGA ATT CAG ATC TCG ATC CCG CGA AAT TAA TAC and GTC AGT CAC GAT GAA TTC TTT GTT AGC AGC CGG ATC CTC GAG for the RXR LBD; CAT ATG CTC GAG GAT CCC CTC TAG AAA TAA TTT TGT TTA ACT T and CGG GCT TTG TTA GCA GCC GGA TCC for the RAR LBD. The PCR products were then cloned into the EcoRI site of pGEX-KT–RXR LBD or the BamHI site of pET-15b–RAR LBD using the exonuclease-mediated subcloning technique (18, 19). The ECR and USP LBDs were cloned in the same way using these primers: GGT TCC GCG TGG ATC CCA AGA CTT TGT TAA GAA GGA GAT T and GAA TTC CCG GGG ATC CCT AGG CAT GAA CG CCC AGA TCT CCT C for the ECR LBD; GCG CGG CAG CCA TAT GAC CAA TAG CGT GTC CAG GGA T and GGA TCC TCG AGC ATA TGC TAC TCC AGT TTC ATC GC AGG CC for the USP LBD. All constructs were confirmed by DNA sequencing.
Protein Expression and Purification.
The expression constructs were transformed into either DH5α or BL21(DE3) cells. Cultures were grown at 37°C in Luria–Bertani medium containing ampicillin. These cultures were induced at an optical density (at 595 nm) of 0.5–1.0 using 0.4 mM isopropyl β-d-thiogalactopyranoside and grown for a further 3 h before harvesting by centrifugation. The cell pellet was frozen and then resuspended in HKE buffer (20 mM Hepes, pH 7.5/100 mM KCl/1 mM EDTA) for purifying glutathione S-transferase (GST) fusion proteins or HKI buffer (20 mM Hepes, pH 8.0/100 mM KCl/20 mM imidazole) for purifying histidine-tagged and coexpressed proteins. The cells were lysed by sonication in the buffer containing 0.4 mM freshly added protease inhibitor 4-(2-aminoethyl)benzolsulfonyl fluoride hydrochloride (AEBSF; Boehringer Mannheim). Cleared lysates were prepared by centrifugation and mixed at 4°C with either glutathione agarose or Ni2+ nitrilotriacetate acid (NTA) agarose beads. The agarose beads were washed five times with either HKE or HKI buffer. The proteins were eluted from the beads in HKE buffer containing 15 mM glutathione or in HKI buffer containing 250 mM imidazole.
Solubility Analysis.
Cell pellets were resuspended in the appropriate buffer, sonicated as above, and then centrifuged at 3,000 × g for 10 min to remove intact cells and cell debris. The supernatants were then further centrifuged at 10,000 × g for 10 min to pellet the insoluble protein. Soluble proteins were purified from the supernatant using the appropriate affinity resin.
Ligand Binding.
All-trans-[3H]retinoic acid (t-[3H]RA) (50 mCi/mmol; 1 Ci = 37 GBq) and cold t-RA were purchased from Dupont/NEN and Sigma, respectively. 9-cis-[3H]RA (58 mCi/mmol), [3H]LG100069 (58 mCi/mmol), 9-cis-RA, LG100069, LG100268, LG100754, LG100550, and LG100629 were supplied by Ligand Pharmaceuticals.
The receptor proteins were purified to greater than 90% homogeneity. The protein was quantitated using the Bio-Rad protein assay as well as Coomassie staining on SDS/PAGE gels. In both cases BSA was used as a standard.
For the saturation binding assays, 50 nM protein (bound to affinity resin) was incubated with radiolabeled ligand in HKBGC buffer (20 mM Hepes, pH 7.5/100 mM KCl/1 mM 2-mercaptoethanol/8% glycerol/10 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) at room temperature for 4 h or overnight with continuous mixing. The resin was then transferred to 4°C and mixed for a further 30 min. The agarose beads were pelleted by centrifugation at 4,000 rpm for 3 min at 4°C and subsequently washed two times using the same buffer. The supernatants, washes, and beads were transferred to scintillation vials containing 5 ml of Ecolume solution (ICN), and tritium counts per minute were measured on a Beckman scintillation counter. For competition binding analysis, 50 nM protein (bound to affinity resin) was incubated with 10 nM radioactive ligand and 1,000 nM unlabeled ligand. The buffer and experimental procedures were the same as described above. To protect the ligands from light-induced isomerization, all operations were carried out under dim yellow light (Sylvania F40G0) or in the dark. The ligands were dissolved in dimethyl sulfoxide (DMSO) and added to protein so that the final concentration of DMSO did not exceed 4%. Nonspecific binding to the affinity resin was assessed using a 100-fold excess of unlabeled ligand.
RESULTS
Double Cistronic Plasmid Construction.
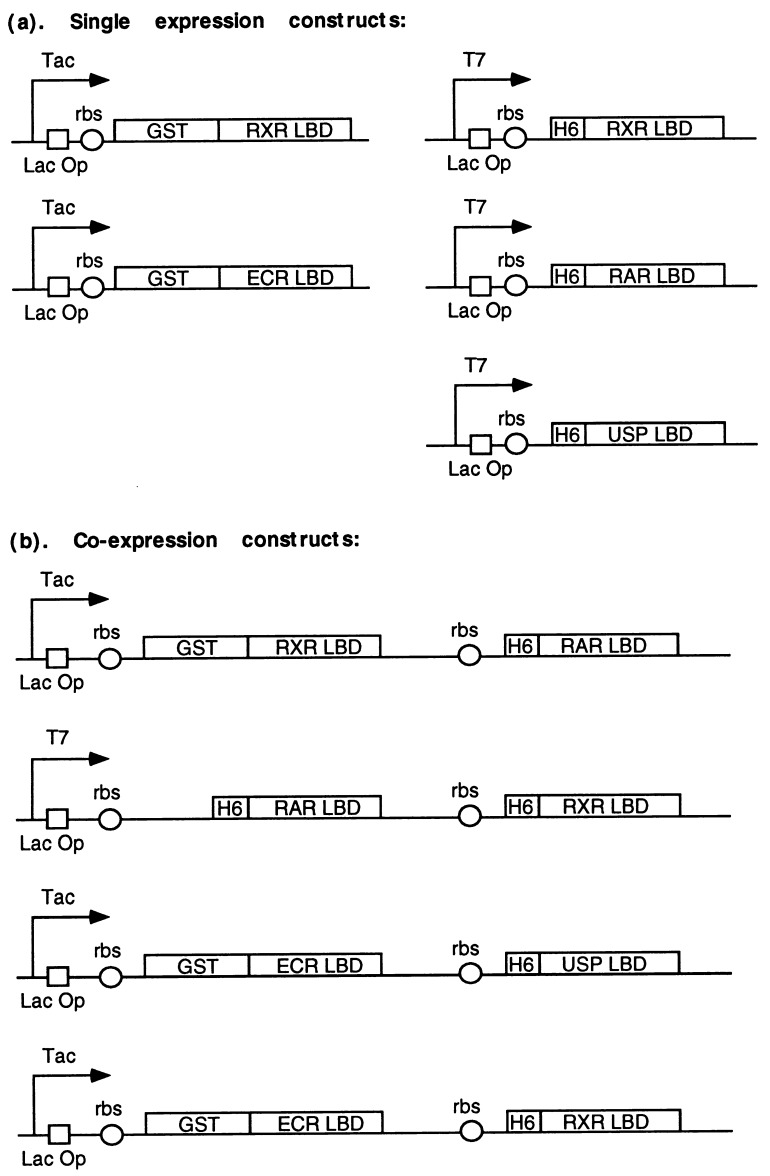
The LBDs of RXR, RAR, and USP were cloned into a pET expression vector to produce receptors tagged N terminally by six histidine residues (H6). The RXR LBD (RXRL) and ECR LBD (ECRL) were cloned into a pGEX expression vector so as to express a GST-tagged protein. The single cistronic expression constructs are shown in Fig. 1a. The double cistronic expression vectors were constructed with the idea that the two cistrons should be driven by a single regulated promoter. A lac operator binding site was placed downstream of the promoter with a ribosome-binding site preceding each cistron, as illustrated in Fig. 1. In the first double cistronic construct, the RXR LBD is fused with GST and the RAR LBD is tagged with six histidine residues. Both of the two cDNAs are driven by a Tac promoter under the control of the lac operator. The GST RXR LBD is upstream of the H6 RAR LBD (Fig. 1b). In the second construct, both RXR and RAR LBDs are tagged with six histidine residues, and they are driven by a T7 promoter under the control of the lac operator. The third and fourth constructs are similar to the first one, but contain ECR USP and ECR RXR, respectively.
Figure 1.
Schematic diagram of expression constructs. (a) The single cistronic expression constructs. (b) The double cistronic expression constructs. Bacterial (Tac) or phage (T7) promoters are indicated by arrows. Repressor binding sites are represented by open boxes. E. coli ribosome-binding sites are represented by open circles. In the first construct, RXR and RAR LBDs are fused at their N termini to GST and H6, respectively. In the second construct, both receptor LBDs are fused to H6. The order of transcription of RXR and RAR in the two constructs is reversed. The third and fourth constructs are similar to the first one except that the receptors are ECR/USP and ECR/RXR, respectively.
Protein Expression.
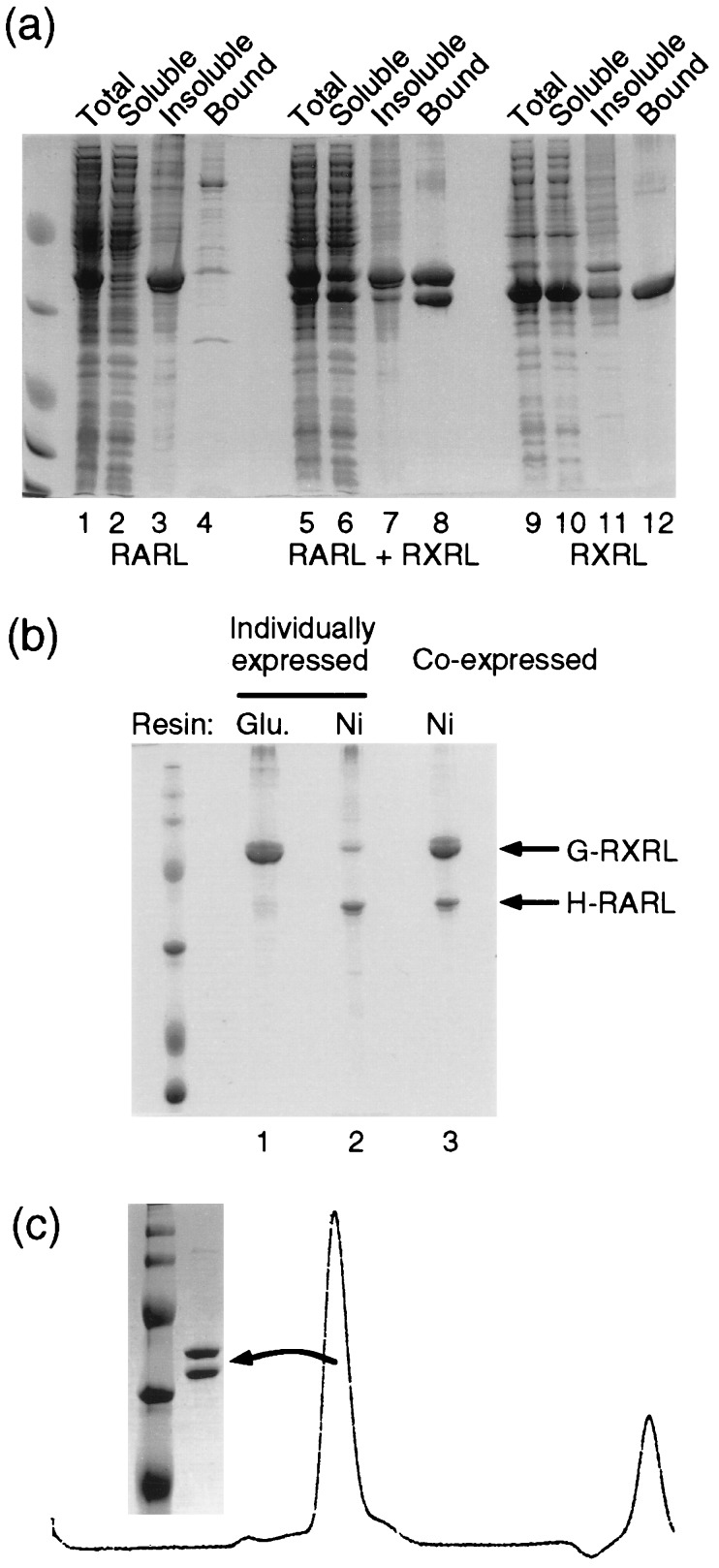
The RXR LBD was found to be soluble and expressed at a high level using both single expression constructs (Fig. 2a, lanes 9–12). In contrast, whereas the H6 RAR LBD is also well expressed, the majority of this protein was found to be insoluble (Fig. 2a, lanes 1–4). Nevertheless, about 500 μg of protein could be purified from the soluble fraction of a 1-liter culture. However, this protein cannot be effectively pulled down by GST RXR LBD on glutathione agarose beads (Fig. 2b, lane 1). Neither could it effectively pull down GST RXR LBD while bound to Ni2+ NTA agarose (Fig. 2b, lane 2).
Figure 2.
Coexpression increases the solubility of RAR LBD and promotes RXR and RAR heterodimer formation. (a) Solubility analysis of individually expressed RAR and RXR LBDs and coexpressed RAR and RXR LBDs. All the RAR and RXR LBDs are tagged with six histidine residues at their N termini. The total cell lysates (Total), the soluble (Soluble) and insoluble (Insoluble) fractions, and the purified protein bound on the Ni2+ NTA beads (Bound) are indicated above each lane. (b) Heterodimer formation of individually or coexpressed RAR and RXR LBDs. Lane 1, the individually expressed H6 RAR LBD purified by interacting with GST RXR LBD on glutathione agarose beads; lane 2, the individually expressed GST RXR LBD was purified by interacting with the H6 RAR LBD on Ni2+ NTA agarose beads; lane 3, the coexpressed GST RXR LBD and H6 RAR LBD were purified by Ni2+ NTA agarose beads. (c) The coexpressed RXR and RAR LBDs are heterodimers. The coexpressed H6 RAR and RXR LBDs were passed through a gel filtration column. A single peak was observed at around the 56-kDa position. The proteins in the peak fraction were run on SDS/PAGE.
In contrast, the double cistronic vectors gave high yields of both soluble proteins (about 10 mg/liter of bacterial culture; Fig. 2a, lanes 5–8). For the mixed tagged construct both proteins could be purified using Ni2+ NTA agarose (Fig. 2b, lane 3). Under the same condition singly expressed GST RXR LBD could not be purified efficiently based on its association with RAR LBD (Fig. 2b, lane 2). In the case of the double H6-tagged construct, following purification using Ni2+ NTA agarose in the protein was shown to elute as a single peak (molecular mass 50–60 kDa) from a gel filtration column at an estimated molecular mass of 56 kDa (Fig. 2c). Furthermore, the soluble fraction consists almost entirely of a 1:1 ratio of RXR to RAR, indicating that coexpression dramatically increases the solubility of the RAR LBD and promotes the dimerization of both RAR and RXR (Fig. 2a, compare lane 6 with lane 2; also Fig. 2 b and c). Remarkably, more than 95% of the expressed protein was in the form of a tight heterodimer (Fig. 2c). Interestingly, this dimer was found to be extremely stable under many different conditions and could only be dissociated by denaturation (data not shown).
Analysis of Ligand Binding.
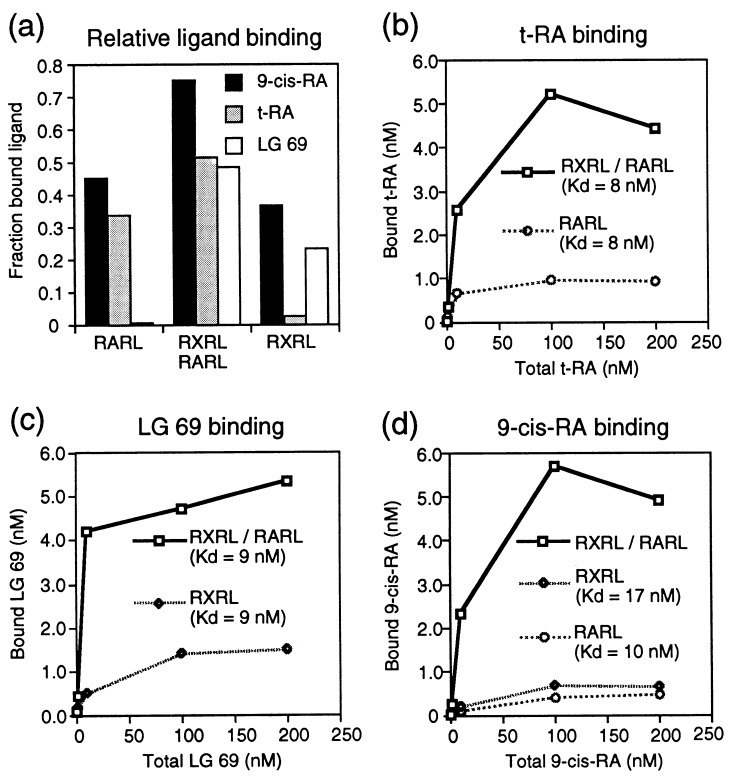
Currently the natural ligands for RAR and RXR are believed to be t-RA and 9-cis-RA. 9-cis-RA activates both receptors (20–22), whereas t-RA is specific for the RAR (22–24). In addition, many synthetic ligands have been shown to have a range of different activities. LG100550 (19) and RO-41253 (20) are specific agonists and antagonists of RAR, respectively. LG100069 and LG100268 (25, 26) are specific agonists of RXR. LG100754 (27) binds specifically to RXR and serves as an antagonist in the absence of RAR. However, in the RXR–RAR heterodimer LG100754 appears to act as an agonist (28).
The ligand-binding activities of both the individually expressed RXR and RAR LBDs and the coexpressed heterodimer were assessed using a pull-down assay with radiolabeled ligands (see Materials and Methods). The expressed proteins demonstrated the expected specificity for the three ligands: 9-cis-retinoic acid is bound by both RAR and RXR; all-trans-retinoic acid is bound only by RAR; and LG69 is specific for RXR (Fig. 3a). The amount of protein and ligand in these assays was very carefully controlled; nevertheless, the heterodimer was able to bind significantly higher amounts of ligand than either individually expressed RXR or RAR LBDs (Fig. 3a). To address the reason for these observations, we performed Scatchard analyses (29) of saturation binding data (Fig. 3 b–d). In each case the heterodimer binds between 5 and 10 times more ligand than the individually expressed LBDs. However, the ligand-binding affinity of the single LBDs is similar to that of the heterodimer, i.e., the Kd for t-[3H]RA of the RAR LBD and heterodimer is the same (8 nM) (Fig. 3b). The Kd of RXR LBD and the heterodimer for [3H]LG69 is 9 nM (Fig. 3c). The Kd values of RXR and RAR LBDs for 9-cis-[3H]RA are 17 and 10 nM, respectively (Fig. 3d). The RXR LBD has a slightly higher maximal binding than the RAR LBD. On the other hand, RAR LBD has higher affinity. Since both RXR and RAR in the heterodimer can bind 9-cis-RA, the radioactivities generated from binding of 9-cis-[3H]RA to RXR, RAR, or both cannot be distinguished in this assay (RXR, RAR, and both receptors of the heterodimer bind ligand). There are potentially four binding equilibria that would display complex kinetics, so a meaningful Kd value cannot be calculated from these data. These results suggest that the majority of the individually expressed LBDs are unable to bind ligand. At the same time this indicates a dramatic benefit of coexpressing heterodimeric partners. It also suggests the existence of two distinct folding states of soluble LBD that correspond to either no or wild-type binding and dimerization.
Figure 3.
Ligand-binding studies of individually and coexpressed RXR and RAR LBDs. (a) Coexpressed RXR–RAR LBDs compared with individually expressed RXR and RAR LBD on binding to RXR-specific/RAR-specific ligand t-RA, ligand LG69, and pan-agonist 9-cis-RA. The concentration of the ligands is 10 nM. (b) Saturation binding analysis of t-RA with coexpressed RXR–RAR LBDs and individually expressed RAR LBD. (c) Saturation binding analysis of LG69 with coexpressed RXR–RAR LBDs and individually expressed RXR LBD. (d) Saturation binding analysis of 9-cis-RA with coexpressed RXR–RAR LBDs and individually expressed RXR and RAR LBDs.
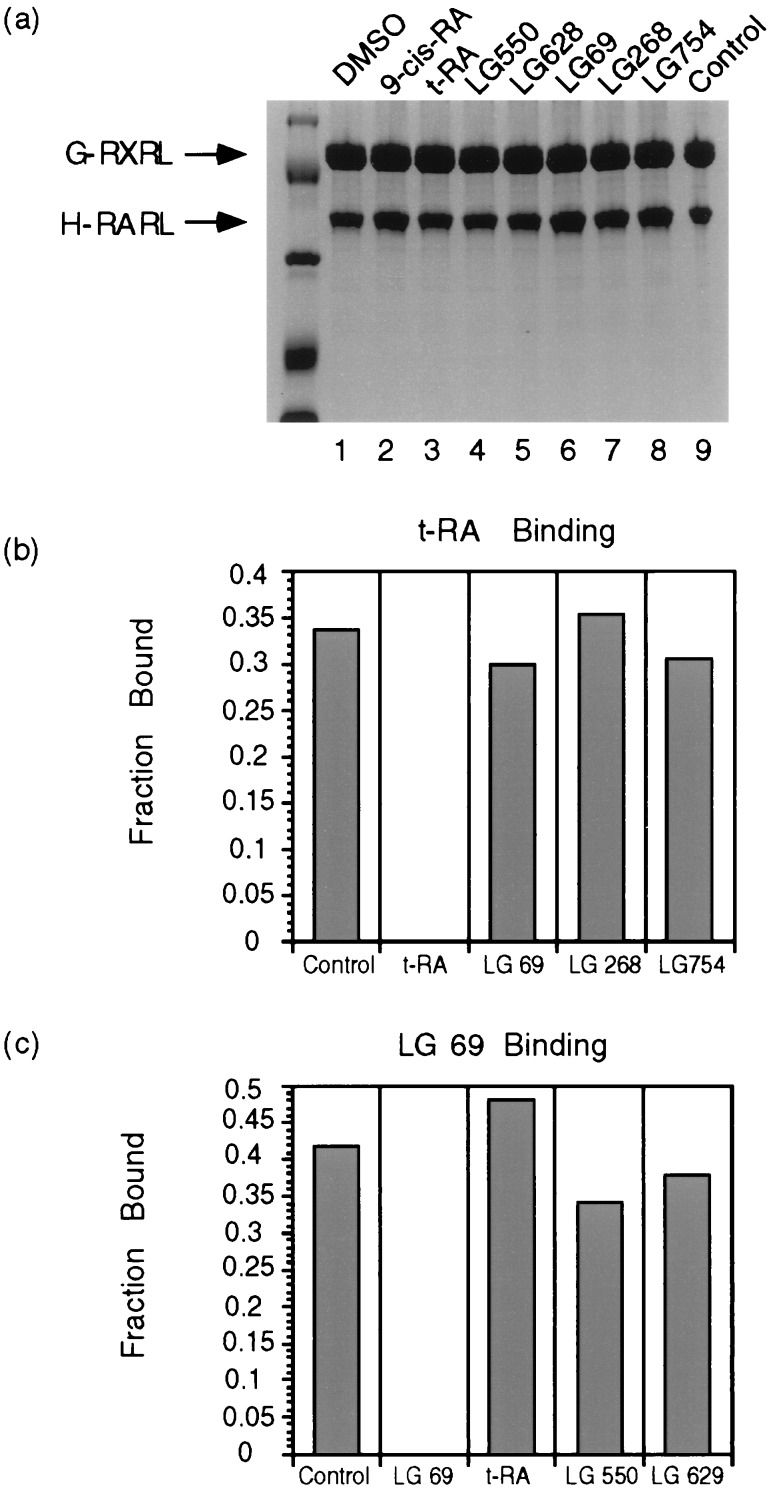
As discussed earlier, since the nuclear receptors act as heterodimers there is a potential for cross talks between different ligand pathways. Indeed, it has been shown that the synthetic ligand LG754 binds to the RXR yet activates through the RAR (30). We wished to determine whether the binding of ligands (agonists or antagonists) to one-half of the dimer influences either the dimerization state or the binding of ligand to the other receptor. Five hundred nanomolar concentrations of seven different ligands were incubated with 50 nM of the GST RXR–H6 RAR LBD heterodimer (bound to Ni2+ NTA agarose) at room temperature overnight. If the heterodimer is dissociated we would expect the GST-tagged receptor to be lost during subsequent wash steps. Fig. 4a indicates that none of the ligands caused heterodimer dissociation. Fig. 4 b and c illustrates the results of competition assays in which we demonstrate that RXR agonists or antagonists do not significantly influence the ability of RAR to bind all-trans-retinoic acid. Similarly, RAR ligands do not significantly affect the ability of RXR to bind its specific ligand LG69.
Figure 4.
Analysis of ligand cross talk. The effects of ligand binding to heterodimer and binding of one ligand to one receptor to the binding of another ligand to another receptor in the coexpressed RXR and RAR heterodimer system. (a) The ligands do not dissociate the RXR–RAR heterodimer. (Lanes 1–8) DMSO, 9-cis-RA, t-RA, LG550, LG628, LG69, LG268, and LG754. (b) Binding of RXR-specific agonists or antagonist to RXR LBD would not significantly affect the binding of RAR-specific ligand t-RA to RAR LBD. (c) Binding of RAR-specific agonists or antagonist would not significantly affect the binding of RXR-specific ligand LG69 to RXR LBD.
Coexpression Increases Solubility and Heterodimer Formation of Other Nuclear Receptors.
To explore the generality of these observations we wished to determine whether coexpression of other receptor combinations would result in more efficient heterodimer formation. Of particular interest was the Drosophila ecdysone receptor LBD, which as seen in Fig. 5a is completely insoluble when expressed in Escherichia coli as a GST fusion protein. Accordingly, two double cistronic vectors were constructed (Fig. 1b) in which the ECR LBD was coexpressed with either ECR’s natural partner USP LBD (H-USPL) or USP’s mammalian homologue RXR LBD (H-RXRL) (Fig. 5 a and b). Remarkably, coexpression with USP led to the first efficient production of soluble ECR LBD (G-ECRL; Fig. 5a, lanes 5–8). Interestingly, this effect is specific for USP since coexpression with RXRL failed to produce significant amounts of soluble ECR LBD (Fig. 5b). In contrast to the singly expressed ECR LBD, which is completely insoluble (Fig. 5a, lane 4), coexpression of ECR LBD with RXR LBD slightly increased the solubility of ECR LBD (Fig. 5b, lane 4). Thus, coexpression seems to be a required feature for ECR LBD production, although not all heterodimer partners are equally efficient in this process.
Figure 5.
Solubility analysis and purification of singly and coexpressed ECRL and USPL (a) and coexpressed ECRL and RXRL (b). ECRL was fused with GST. USPL and RXRL were tagged with histidine. The total cell lysates (Total), the soluble (Soluble) and insoluble (Insoluble) fractions, and the purified protein bound on the Ni2+ NTA beads (Bound) are indicated above each lane.
DISCUSSION
We described the development of a novel dual protein expression system that facilitates efficient production of a pure and functional RXR–RAR heterodimer. When expressed singly, the RXR LBD is soluble and the RAR LBD is largely insoluble. Surprisingly, our results show that the singly expressed RAR LBD exists in one of three physical states: insoluble, soluble but unable to bind ligand, and soluble with wild-type ligand-binding capacity. We find no evidence of intermediate states. Furthermore, simple mixing of singly expressed RAR and RXR LBDs is inefficient in promoting heterodimer formation. Our results also show that both of the singly expressed receptors are much less active in ligand binding. In contrast, the bacterially produced heterodimer is highly soluble, not easily dissociated, and fully functional in terms of ligand binding. This seems to suggest that the singly expressed soluble LBDs are in some way defective in proper folding. It is clear that the coexpression with the RXR LBD dramatically increases the solubility of the RAR LBD as well as its appropriate folding. This might be because the RXR masks a hydrophobic patch that otherwise causes the RAR LBD to aggregate and precipitate. However, this would not explain why a portion of the soluble RAR fails to dimerize or bind ligand. It is also possible that the RXR LBD may serve as a form of molecular chaperone assisting the folding of the RAR LBD. All these data imply that the heterodimer forms a true integral unit, whereas single receptors are rather unstable or improperly folded. Interestingly, this situation is also seen for the Drosophila USP–ECR heterodimer. The ECR LBD is totally insoluble in the absence of USP LBD, whereas in the coexpression system substantial amounts of the ECR LBD can be purified in the form of a soluble heterodimer (Fig. 5a). Surprisingly, and for reasons that are not clear, RXR is much less effective than USP in promoting ECR solubility. Taken together, these data strongly suggest that the hormone receptor partners play an important role in the solubility, stability, and function of their partners.
The dissociation constants measured for the bacterially expressed receptors are in the same range of Kd values as reported by others using different assays (20–23, 31, 32). Since our assays involve moving away from equilibrium at the time of measurement (washing unbound ligand away from the receptor), it is likely that the true Kd values are somewhat lower. Equilibrium techniques such as fluorescence quenching are likely to yield more accurate values. Our ligand-binding assays show that the bacterially expressed heterodimeric LBDs bind hormone efficiently and independently. However, it has been reported previously that the RXR–RAR heterodimer binds LG69 much more weakly than RXR alone (33, 34). Other studies suggest that ligand binding by RXR is essentially the same as monomer or dimer, with or without DNA (35, 36). Although the reason for these differences is not clear, our studies suggest that in the heterodimer of LBDs the two partners can bind ligands independently and with similar affinity to their monomeric counterparts. The question remains why LG69 acts as a “silent” ligand in the RXR–RAR heterodimer (26, 33, 34). With the recent studies of nuclear coactivator and corepressor (37–41), it is possible that some ligands (such as LG69) are not efficient in releasing repressor and recruiting activator. Alternatively, based on transcripton-activation studies, a requirement for the activation domain of RAR (42) may make an RXR ligand a poor activator in a heterodimer complex. A key issue remaining is how the formation of the heterodimer leads to the formation of new functional properties of the individual receptors. Based on these studies at least one important role of RXR is to increase the solubility and stability of its partner. Reciprocally, we find that the partner, in this case RAR, dramatically enhances RXR ligand-binding function, indicating a mutually beneficial or cooperative effect from heterodimer formation. Composition analysis of the coexpressed receptor indicates a 1:1 correspondence. The absence of a significant level of free RXR or RAR monomer suggests that the heterodimer probably represents the lowest free energy state, which supports previous conclusions regarding how and why heterodimers might represent the preferential complexes in mammalian cells as well. The most dramatic effects and benefit of coexpression are found in production of the soluble ecdysone receptor LBD, which appears to be almost entirely dependent on the presence of USP. We speculate that RXR serves as a less effective partner probably because of reduced compatibility, even though this heterodimer is functional in mammalian expression systems. Perhaps the presence of heat shock proteins or other chaperones not present in E. coli promotes this process in mammalian cell lines.
In summary, we have successfully coexpressed and purified the RXR–RAR LBD and ECR–USP LBD heterodimers in E. coli. The coexpressed receptors are substantially more soluble than the individually expressed receptors, and thus should be useful for future biological and structural analyses. We suggest that this may represent a general approach to high level expression of nuclear receptor heterodimers and may be useful as well for other classes of heterodimeric protein partners.
Acknowledgments
We are grateful to R. Heyman and M. Boehm (Ligand Pharmaceuticals Inc.) for providing ligands used in this study. We also thank B. Blumberg and H. Chen for helpful discussion, S. Inoue and H. Kao for critical reading of the manuscript, and Elaine Stevens for help in preparing the manuscript. C.L. is a postdoctoral fellow of the Howard Hughes Medical Institute. J.W.R.S. is supported by the Human Frontier Science Program Organization. R.M.E. is an Investigator of the Howard Hughes Medical Institute at The Salk Institute for Biological Studies. This work was supported by the Howard Hughes Medical Institute and by Grants GM 26444 and HD 27183 from the National Institutes of Health (to R.M.E.).
ABBREVIATIONS
- RXR
retinoid-X receptor
- RA
retinoic acid
- t-RA
all-trans-retinoic acid
- RAR
retinoic acid receptor
- ECR
ecdysone receptor
- USP
ultraspiracle
- LBD
ligand-binding domain
- H6
six histidine residues
- GST
glutathione S-transferase
- NTA
nitrilotriacetate
- DMSO
dimethyl sulfoxide
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 3.Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, et al. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- 4.Giguere V, Ong E S, Segui P, Evans R M. Nature (London) 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 5.Petkovich M, Brand N J, Krust A, Chambon P. Nature (London) 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 6.Brand N, Petkovich M, Krust A, Chambon P, de The H, Marchio A, Tiollais P, Dejean A. Nature (London) 1988;332:850–853. doi: 10.1038/332850a0. [DOI] [PubMed] [Google Scholar]
- 7.Krust A, Kastner P, Petkovich M, Zelent A, Chambon P. Proc Natl Acad Sci USA. 1989;86:5310–5314. doi: 10.1073/pnas.86.14.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu V C, Delsert C, Andersen B, Holloway J M, Devary O V, Naar A M, Kim S Y, Boutin J M, Glass C K, Rosenfeld M G. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- 9.Kliewer S A, Umesono K, Heyman R A, Mangelsdorf D J, Dyck J A, Evans R M. Proc Natl Acad Sci USA. 1992;89:1448–1452. doi: 10.1073/pnas.89.4.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X K, Hoffmann B, Tran P B, Graupner G, Pfahl M. Nature (London) 1992;355:441–446. doi: 10.1038/355441a0. [DOI] [PubMed] [Google Scholar]
- 11.Roberts A B, Sporn M B. The Retinoids. Vol. 2. New York: Academic; 1984. pp. 209–286. [Google Scholar]
- 12.Thaller C, Eichele G. Nature (London) 1987;327:625–628. doi: 10.1038/327625a0. [DOI] [PubMed] [Google Scholar]
- 13.Thompson J N, Howell J M, Pitt G A J. Proc R Soc London Biol Sci. 1964;159:510–535. doi: 10.1098/rspb.1964.0017. [DOI] [PubMed] [Google Scholar]
- 14.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 15.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 16.No D, Yao T P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakes D J, Dixon J E. Anal Biochem. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- 18.Kaluz S, Kolble K, Reid K B. Nucleic Acids Res. 1992;20:4369–4370. doi: 10.1093/nar/20.16.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao K. Nucleic Acids Res. 1993;21:5528–5529. doi: 10.1093/nar/21.23.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyman R A, Mangelsdorf D J, Dyck J A, Stein R B, Eichele G, Evans R M, Thaller C. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 21.Levin A A, Sturzenbecker L J, Kazmer S, Bosakowski T, Huselton C, et al. Nature (London) 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 22.Allegretto E A, McClurg M R, Lazarchik S B, Clemm D L, Kerner S A, Elgort M G, Boehm M F, White S K, Pike J W, Heyman R A. J Biol Chem. 1993;268:26625–26633. [PubMed] [Google Scholar]
- 23.Yang N, Schule R, Mangelsdorf D J, Evans R M. Proc Natl Acad Sci USA. 1991;88:3559–3563. doi: 10.1073/pnas.88.9.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allenby G, Bocquel M T, Saunders M, Kazmer S, Speck J, et al. Proc Natl Acad Sci USA. 1993;90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boehm M F, Zhang L, Badea B A, White S K, Mais D E, Berger E, Suto C M, Goldman M E, Heyman R A. J Med Chem. 1994;37:2930–2941. doi: 10.1021/jm00044a014. [DOI] [PubMed] [Google Scholar]
- 26.Boehm M F, Zhang L, Zhi L, McClurg M R, Berger E, et al. J Med Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 27.Koch S S C, Dardashti L J, Hebert J J, White S K, Croston G E, Flatten K S, Heyman R A, Nadzan A M. J Med Chem. 1996;39:3229–3234. doi: 10.1021/jm960311d. [DOI] [PubMed] [Google Scholar]
- 28.Lala D S, Mukherjee R, Schulman I G, Koch S S C, Dardashti L J, Nadzan A M, Croston G E, Evans R M, Heyman R A. Nature (London) 1996;383:450–453. doi: 10.1038/383450a0. [DOI] [PubMed] [Google Scholar]
- 29.Scatchard G. Ann NY Acad Sci. 1949;51:660–672. [Google Scholar]
- 30.Schulman, I. G., Li, C., Schwabe, J. W. R. & Evans, R. M. (1997) Genes Dev., in press. [DOI] [PubMed]
- 31.Chen Z P, Shemshedini L, Durand B, Noy N, Chambon P, Gronemeyer H. J Biol Chem. 1994;269:25770–25776. [PubMed] [Google Scholar]
- 32.Cheng L, Norris A W, Tate B F, Rosenberger M, Grippo J F, Li E. J Biol Chem. 1994;269:18662–18667. [PubMed] [Google Scholar]
- 33.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld M G, Heyman R A, Glass C K. Nature (London) 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 34.Forman B M, Umesono K, Chen J, Evans R M. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 35.Apfel C M, Kamber M, Klaus M, Mohr P, Keidel S, LeMotte P K. J Biol Chem. 1995;270:30765–30772. doi: 10.1074/jbc.270.51.30765. [DOI] [PubMed] [Google Scholar]
- 36.Kersten S, Dawson M I, Lewis B A, Noy N. Biochemistry. 1996;35:3816–3824. doi: 10.1021/bi952737k. [DOI] [PubMed] [Google Scholar]
- 37.Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 38.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 39.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 40.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, et al. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 41.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 42.Valcarcel R, Holz H, Jimenez C G, Barettino D, Stunnenberg H G. Genes Dev. 1994;8:3068–3079. doi: 10.1101/gad.8.24.3068. [DOI] [PubMed] [Google Scholar]