Abstract
Using the cytoplasmic domain of the insulin receptor (IR) in a yeast two-hybrid screen, we identified a cDNA clone encoding the C-terminal 308 amino acids of human Stat5b (Stat5b-Ct). Stat5b-Ct is tyrosine phosphorylated by purified IR kinase domain in vitro. Insulin stimulates tyrosine phosphorylation of overexpressed Stat5b-Ct and endogenous Stat5 in cells overexpressing IR. Stat5 may be a direct target of the IR and, as a member of the Stat family of transcription factors, may play a role in the regulation of gene transcription by insulin. In support of this hypothesis, perfusion of mouse liver with insulin promotes rapid tyrosine phosphorylation of Stat5 and activation of Stat5 DNA binding. Moreover, refeeding of fasted mice leads to rapid tyrosine phosphorylation and stimulation of enhanced DNA-binding activity of Stat5 extracted from liver, skeletal muscle, and adipose tissues. Taken together, our data strongly suggest that IR interacts with and phosphorylates Stat5 in vitro and in tissues physiologically sensitive to insulin.
In addition to playing a vital role in the regulation of glucose homeostasis in mammals, the polypeptide hormone, insulin, exerts a broad spectrum of physiological effects in many different cell types. These effects include stimulation of glucose, ion, and amino acid uptake, modification of the activities of rate-limiting enzymes involved in intermediary metabolism, redistribution of membrane proteins such as the glucose transporter, regulation of gene expression, and promotion of cell growth and differentiation (1). Insulin elicits these pleiotropic effects through the insulin receptor (IR) (2), whose intrinsic protein tyrosine kinase activity is required for its function (3, 4). Extensive research has defined how signaling by the IR is initiated and has identified many of the signal transduction pathways that lead to those biological effects (5). Unlike other receptor protein tyrosine kinases, such as platelet-derived growth factor (PDGF) and epidermal growth factor (EGF) receptors, IR uses its major substrate, insulin receptor substrate-1 (IRS-1) (6) as the main docking protein for recruitment of various SH2 domain-containing signaling proteins, such as PI 3-kinase (7), Syp, a tyrosine phosphatase (8), SH2/SH3-containing adaptor molecules Grb2 (9), and Nck (10). In addition to IRS-1, the SH2/SH3 domain-containing adaptor, Shc, also has been shown to directly interact with IR (11, 12). Binding of Grb2 and Shc to phosphorylated IRS-1 and activated IR, respectively, serves to recruit Sos, a Ras guanine nucleotide exchange factor, resulting in activation of the Ras/Raf/mitogen-activated protein kinase signaling cascade, leading to insulin-activated gene transcription (13, 14).
A potential mitogen-activated protein kinase-independent mechanism for the regulation of gene expression is the JAK-STAT pathway. While this was originally defined through cytokine receptors, more recently it has been shown that the key activating event, STAT tyrosine phosphorylation, is regulated by receptor protein tyrosine kinases (15). For example, Stat1 and Stat3 become tyrosine phosphorylated in response to EGF, PDGF, and CSF-1 in cultured cell lines, and these Stats as well as Stat5 become tyrosine phosphorylated in the livers of mice injected intraperitoneally with EGF (16–20). Unlike the cytokine-mediated activation of Stats, recent studies indicate that tyrosine phosphorylation and activation of Stat1 and Stat3 in response to PDGF or EGF appears to be independent of JAKs, suggesting that Stats may be phosphorylated directly by the receptors (21, 22).
We have been interested in exploring novel substrates of insulin receptor. Using the yeast two-hybrid system (23, 24), we have found that Stat5 is capable of interacting with insulin receptor in vitro and intracellularly. Stat5 is tyrosine phosphorylated in response to insulin in cells overexpressing IR and is specifically activated in mouse liver perfused with insulin. Moreover, refeeding of fasted mice leads to rapid tyrosine phosphorylation and stimulation of DNA-binding activity of Stat5 extracted from liver. Taken together, our results strongly suggest that Stat5 is a physiological substrate and downstream-signaling molecule of IR.
MATERIALS AND METHODS
Cells and Antibodies.
The yeast strains, plasmids, and a HeLa cell-derived cDNA library were provided by Roger Brent (23) (Harvard Medical School, Boston). Chinese hamster ovary (CHO)-IR, a human IR-overexpressing line (2), and COS1 cells were grown in DMEM with 10% calf serum. HIR 3.5 cells (25), an NIH 3T3 line overexpressing human IR, were grown in DMEM with high glucose and glutamine and containing 10% FBS and 1 mM sodium pyruvate. Stat1 polyclonal antibody was a gift from X.-Y. Fu (Yale University). Anti-Stat3 and Stat5 antibodies were purchased from Santa Cruz Biotechnology. Recombinant anti-phosphotyrosine (PTyr) antibody RC20 was from Transduction Laboratories (Lexington, KY). A monoclonal Ab 12CA5 against the influenza hemagglutinin tag and the horseradish peroxidase-coupled 12CA5 (12CA5-P) were purchased from Boehringer Mannheim. The antibodies used for supershifting were as follows: anti-Stat1 monoclonal from Transduction Laboratories, anti-Stat3, anti-Stat5b N-terminal (Stat5b-nt) and C-terminal (Stat5b-Ct), and anti-Stat5a C-terminal (Stat5a-Ct) antibodies from Santa Cruz Biotechnology, and polyclonal serum specific for Stat5a or Stat5b from L. Hennighausen (26).
In Vitro Phosphorylation of His-Stat5b-Ct by IR Cytoplasmic Kinase Domain (CKD).
The 1.0-kb fragment encoding Stat5b-Ct was ligated to expression vector pQE32 (Qiagen, Chatsworth, CA) to obtain pQE32-Stat5b-Ct, which then was transformed into M15 (pREP4) cells for protein expression. Purification of His-Stat5b-Ct using Ni-NTA-resin was according to the conditions recommended by the manufacturer (Qiagen). The CKD of IR (amino acids 957–1355) was expressed in baculovirus and purified (27). The purified His-Stat5b-Ct and CKD were used for preparation of rabbit antisera. In vitro kinase reaction was as described (28). Immunoprecipitation and Western blotting followed the protocol previously described (28).
Insulin-Stimulated Stat5b-Ct Tyrosine Phosphorylation in COS Cells.
Two primers specific to library plasmid pJG4–5 were used to amplify the 1.2-kb Stat5b-Ct and HA tag sequence, which then was cut with HindIII and ligated to pRcCMV (Invitrogen) to obtain the expression vector pRcCMV-Stat5b-Ct. pRcCMV-Stat5b-Ct and pECE-IR encoding the full-length IR were cotransfected into COS1 cells by Lipofectamine following the procedure recommended by the manufacturer (GIBCO/BRL). Western blotting with alkaline phosphatase-coupled RC20 (anti-PTyr) and subsequent color detection followed the conditions recommended by the manufacturer (Boehringer Mannheim).
Analysis of Endogenous Stat5 Activation in Cell Lines Overexpressing the Human IR.
CHO-IR cells (10-cm dishes) were grown to confluence and subjected to insulin stimulation as described in the Fig. 3 legend. The cytoplasmic and high-salt nuclear extracts were prepared as described (16, 17). To assess Stat protein DNA-binding activity, extracts were subjected to nondenaturing gel-mobility shift and supershift assays (16, 17) with 32P-labeled double-stranded oligonucleotide probes containing high-affinity binding sites for Stat1 and Stat3 (m67SIE; refs. 16 and 17) or for Stat5/MGF within the β-casein gene promoter (β-casein PIE; ref. 29). HIR3.5 cells were grown to confluence and then subjected to insulin stimulation as described in the Fig. 3 legend. Whole cell lysates were prepared as described (16).
Figure 3.
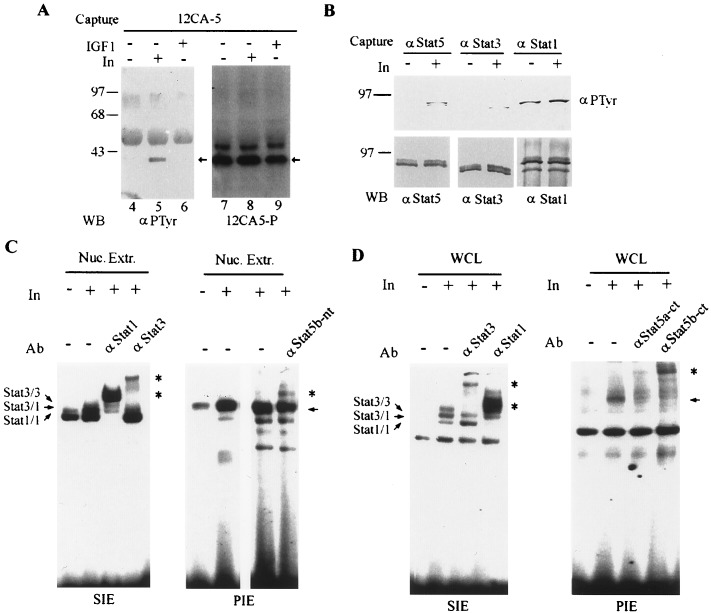
Stat5b-Ct and endogenous Stat5b are tyrosine phosphorylated in response to insulin stimulation in vivo. (A) Tyrosine phosphorylation of Stat5b-Ct in COS cells. Five micrograms each of pRcCMV-Stat5b-Ct and pECE-IR were cotransfected into COS1 cells. After 48 h, the cells were starved for 2 h in serum-free medium and then stimulated with either insulin (50 nM) or IGF1 (20 ng/ml) for 15 min. HA-Stat5b-Ct was immunoprecipitated with 12CA5 antibody, and the immunoprecipitate was divided into two aliquots, one for Western blotting with RC20 (Left) and the other with horseradish peroxidase-12CA5 (Right). Arrows indicate the positions of Stat5b-Ct. (B) Tyrosine phosphorylation of Stat3 and Stat5 in CHO-IR cells. CHO-IR cells were starved in serum-free medium for 4 h in the presence of 200 mM Na3VO4 and then stimulated with insulin (50 nM) for 15 min. One microgram of protein each from the cell lysates was immunoprecipitated with anti-Stat1, Stat3, or Stat5 antibodies. Each immunoprecipitate was divided into two equal parts for detection of Stat tyrosine phosphorylation (Upper) and Stat protein (Lower). (C) Insulin activates Stat3 and Stat5 DNA binding in CHO-IR cells. CHO-IR cells were treated as in B, and nuclear extracts were prepared. Nuclear extracts (20 μg) were subjected to electrophoretic mobility-shift assay (EMSA) analysis with the m67 SIE (Left) or β-casein PIE (Right) probes. Anti-Stat1 and Stat3 antibodies and an antibody against the N-terminal portion of Stat5b (Stat5b-nt) (recognizes both murine Stat5a and Stat5b) were used for the supershift assays. (D) Insulin activates Stat1, Stat3, Stat5a, and Stat5b DNA-binding in HIR3.5 cells. Cells were starved in DMEM with 0.1% BSA for 4 h and then treated with 200 μM vanadate for 40 min, when buffer or insulin (50 nM) was added for the last 15 min. Whole cell lysates were prepared and subjected to gel-mobility shift analysis with the M67 SIE (Left) and β-casein PIE (Right) probes. Antibodies against Stat1, Stat3, Stat5a-Ct, and Stat5b-Ct were used for supershift. Arrows indicate the specific insulin-inducible complexes; asterisks indicate supershifted Stat-containing complexes.
Liver Perfusion Experiments.
Nine-month-old DBA2 female mice, weighing 22–29 g, were used in most of the perfusion studies. Mice were anethesized by abdominal injection of pentobarbital. Ten milliliters of Hanks solution containing either 1 mM sodium vanadate or 1 mM sodium vanadate plus insulin (50 nM) was used as the perfusate. Perfusion was performed through the portal vein with a 31-gauge needle. After perfusion, the livers were soaked in 5 ml of perfusion solution for an additional 5 min at 37°C. The liver was cut into small pieces and Dounce-homogenized immediately in RIPA buffer (50 mM Tris·HCl, pH 7.2/150 mM NaCl/5 mM EDTA/1% Triton X-100/1% sodium deoxycholate) or frozen on dry ice. Protein concentration in the total cell lysates was quantified by the Bradford method. A similar experiment was performed using 9-month-old C57 black mice with 20 ml of Hanks solution with or without insulin (50 nM) in the absence of sodium vanadate. Liver cells cytoplasmic and nuclear extracts were prepared, and gel-shift analysis was performed as described (16, 17).
Refeeding Experiments.
A pair of 9-week-old mice were starved for 2–3 days. Mouse food was given to one of the starved mice for 80 min, and the mice were then sacrificed. Different tissues were immediately taken and frozen on solid CO2. For protein extraction, the tissues were first disrupted gently using a Polytron homogenizer, followed by Dounce homogenization in RIPA buffer (28). The muscle tissue was homogenized completely with a Polytron homogenizer before RIPA extraction. The gel-shift analysis was performed as described (16, 17).
RESULTS
We have used the yeast two-hybrid system (23, 24) in an attempt to identify potential novel molecules that may mediate IR signaling. Two plasmids, encoding LexA-IR-S and LexA-IR-L, were used as bait in our two-hybrid screening. These constructs and the reporter and library plasmid vectors are depicted in Fig. 1. LexA-IR-S contains nearly all of the cytoplasmic domain of IR (amino acids 992–1355), but lacks 40 amino acids in the juxtamembrane domain. When expressed in yeast, this fusion protein is kinase-inactive (data not shown). An IR-L fragment containing the entire cytoplasmic domain of the IR (amino acids 953–1355) was used to construct the bait plasmid pLexA-IR-L. When expressed in EGY48 cells, this construct yielded a 68-kDa fusion protein with strong in vitro kinase activity and is tyrosine phosphorylated in yeast cells (data not shown). We screened a HeLa cell cDNA library using pLexA-IR-L as the bait construct. Of the 1.5 × 107 colonies screened, 9 were strong positives. These were confirmed by reintroducing each of the purified library plasmids into EGY48 cells containing the bait and reporter plasmids, and then assaying for nutrient requirement and β-galactosidase activity. One of the strong positives encodes the C-terminal 308 amino acids of human Stat5b (30) (Stat5b-Ct), including the SH3-like domain, the SH2 domain, and a potential tyrosine phosphorylation site. Our sequence is identical to that of recently cloned human Stat5b (30), but differs from the sequence published by another group (31) by two amino acids at positions 719 and 721.
Figure 1.
Schematic representation of plasmids used in the two-hybrid system pLexA-IR-S and pLexA-IR-L are the bait fusion plasmids containing the short or long fragment of the insulin receptor β subunit, respectively. pJG4–5-cDNA is the cDNA library derived from HeLa cells mRNAs. pSH18–34 is the reporter plasmid. EC, extracellular; TM, transmembrane domain; PTK, protein tyrosine kinase; MCS, multiple cloning sites; NLS, nuclear localization signal; AD, activation domain; HA, hemagglutinin tag sequence; LexA BS, LexA binding site.
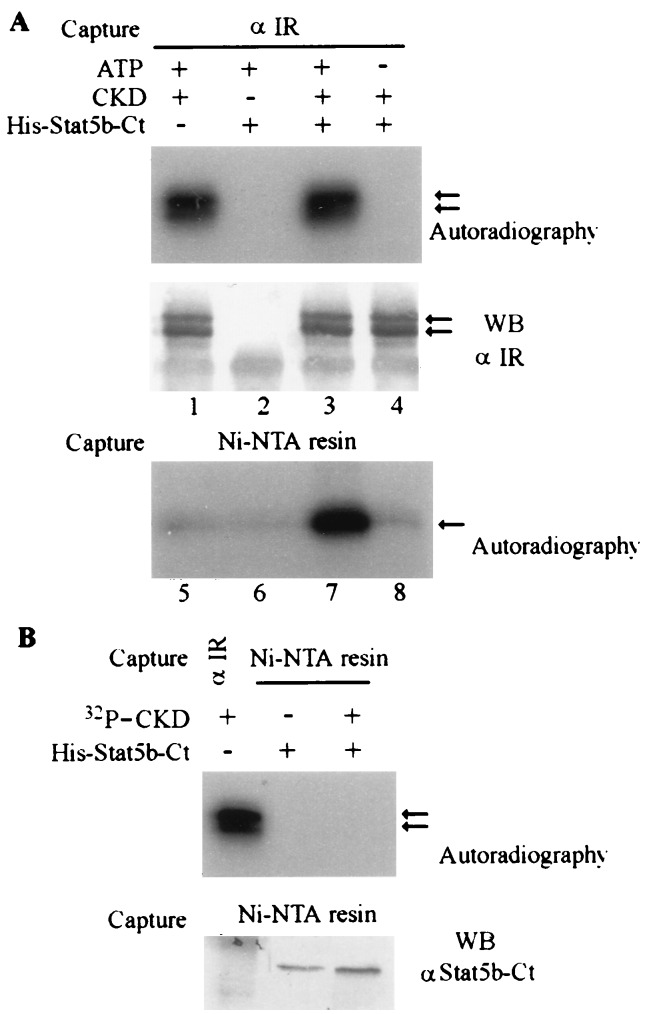
Experiments were performed to begin to define the nature of the interaction between IR and Stat5b-Ct detected in the yeast two-hybrid system. First, the interaction of Stat5b-Ct with the IR in this assay appeared to be specific for the kinase active LexA-IR-L protein, as no β-galactosidase activity was detected when LexA-IR-S (kinase inactive) was used as bait. We then set up an in vitro phosphorylation assay, where purified His-Stat5b-Ct fusion protein was subjected to in vitro phosphorylation by the purified CKD of the human IR (27). The CKD was able to phosphorylate itself (Fig. 2A, lanes 1 and 3, top), as well as His-Stat5b-Ct (Fig. 2A. lane 7). This phosphorylation was dependent on the addition of ATP. Because the CKD and His-Stat5b-Ct migrated to similar positions in SDS gels, we performed an additional experiment to rule out the possibility that the band seen in lane 7 of Fig. 2A actually represented the phosphorylated CKD that had been captured by association with the His-Stat5b-Ct bound to Ni-NTA-resin. In the experiment shown in Fig. 2B, His-Stat5b-Ct and Ni-NTA-resin were added to the in vitro pre-autophosphorylated CKD. Radiolabeled CKD was not captured by the His-Stat5b-Ct plus Ni-NTA-resin (Fig. 2B). We conclude that the band in lane 7 of Fig. 2A indeed represents the Stat5b-Ct phosphorylated by CKD. The phosphorylation of Stat5b-Ct was on tyrosine, because it could be readily detected by Western blotting with an anti-phosphotyrosine antibody (data not shown). We next tested the interaction between IR and Stat5b-Ct in mammalian cells. Plasmids encoding epitope-tagged Stat5b-Ct (pRcCMV-Stat5b-Ct) and the full-length human insulin receptor (pECE-IR) were cotransfected into COS1 cells. Stat5b-Ct became tyrosine phosphorylated in response to insulin, but not to insulin-like growth factor 1 (IGF1), indicating that tyrosine phosphorylation of Stat5b-Ct was the result of IR activation and not the activation of endogenous IGF1 receptor, which is closely related to IR (Fig. 3A).
Figure 2.
In vitro phosphorylation assay of His-Stat5b-Ct. (A) His-Stat5b-Ct can be phosphorylated by CKD in vitro. First 0.3 μg of CKD and 0.75 μg of His-Stat5b-Ct proteins were incubated in 50 μl of kinase buffer (10 mM MnCl2/50 mM Tris·HCl, pH 8.0). [γ-32P]ATP (10 μCi) was added to start the reaction. At the end of reaction, 0.5 ml of RIPA buffer was added, and CKD protein was first immunoprecipitated by anti-IR. (Upper) Kinase assay. (Lower) Amount of CKD protein immunoprecipitated and detected by Western blotting with anti-IR antibody. His-Stat5b-Ct protein subsequently was captured from the supernatant by Ni-NTA-resin, washed with 6 M guanidine hydrochloride, followed by water, and then eluted with Laemmli buffer. The phosphorylated His-Stat5b-Ct was detected by autoradiography. (B) His-Stat5b-Ct does not coprecipitate with CKD in vitro. His-Stat5b-Ct was incubated with preautophosphorylated CKD in RIPA buffer and precipitated with Ni-NTA-resin as above. (Upper) Autoradiography. The left lane is the positive control for 32P-CKD precipitated with anti-IR Ab. (Lower) His-Stat5b-Ct protein captured by Ni-NTA-resin and detected by Western blotting with anti-Stat5b-Ct Ab.
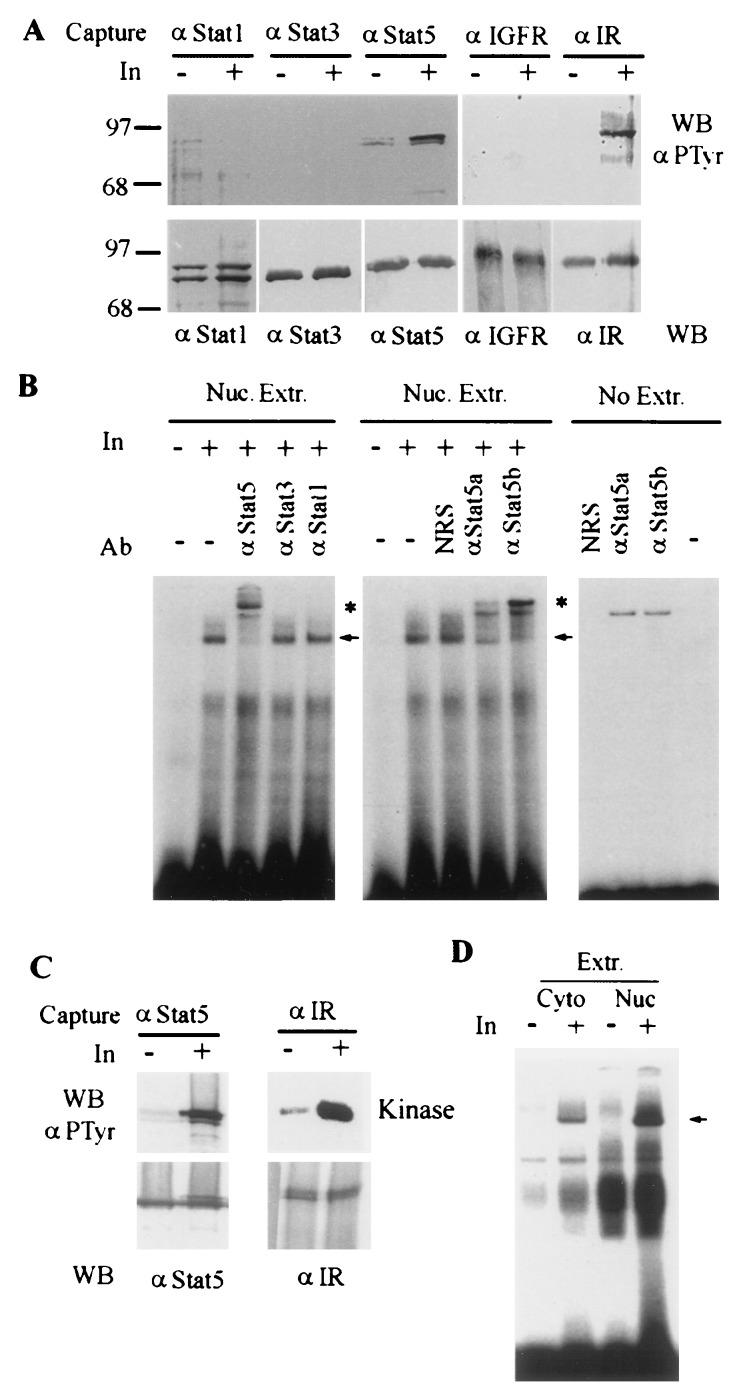
We also examined tyrosine phosphorylation of endogenous Stat5 in response to insulin in a CHO cell line overexpressing the human IR (32). Insulin clearly stimulates tyrosine phosphorylation of endogenous Stat5 in these cells (Fig. 3B). However, we could not detect stable interaction between IR and Stat 5, indicating that the interaction is either transient or with a very low affinity mimicking that between IR and IRS-1 (6). Stat3 also became tyrosine phosphorylated in response to insulin in these cells, whereas Stat1 displayed a high basal level of tyrosine phosphorylation with little or no increase upon insulin stimulation. Moreover, the results of EMSA analysis of nuclear extracts from these cells parallel the tyrosine phosphorylation results. High levels of Stat1 DNA-binding activity was detected in the control cell extract, with obvious insulin-stimulated increase in Stat3 and Stat5 DNA binding (Fig. 3C). The presence of specific Stats in the protein-DNA complex was confirmed by supershift with respective anti-Stat Abs. Because none of the Stat5 antibodies we tested were able to completely supershift/inhibit the insulin-inducible Stat5-PIE complex from CHO cells, we performed a similar experiment with NIH3.5 cells overexpressing the human IR (25). Insulin rapidly stimulates Stat1, Stat3, and Stat5 DNA-binding activity in these cells (Fig. 3D). Maximal activation of Stat1, 3, and 5 in these cells occurs between 10 and 50 nM insulin and by 5 min after insulin addition (data not shown).
The results described so far indicate that Stat5 can be a target for the insulin receptor. In both cases, however, Stat activation was detected in cultured cell lines expressing high levels of the IR. To inquire about the potential for Stat5 activation by the IR in vivo, we performed the liver perfusion experiments. Perfusion of mouse livers with insulin caused increased tyrosine phosphorylation of Stat5, when compared with buffer alone (Fig. 4A). This response is specific to Stat5, as neither Stat1 nor Stat3 becomes tyrosine phosphorylated. However, in the cultured CHO-IR and HIR3.5 cells, we did not observe exclusive activation of Stat5. The increase in Stat5 phosphorylation is accompanied by increased tyrosine phosphorylation (activation) of the IR, but not the IGF1 receptor (Fig. 4A). The perfusion experiments were repeated three times with similar results. To postulate a role for Stat5 in the regulation of gene transcription by insulin, Stat5 tyrosine phosphorylation must be followed by acquisition of Stat5 DNA-binding activity. Therefore, nuclear extracts were prepared from the control and insulin-stimulated liver cells and tested for Stat5 DNA-binding activity by EMSA with a 32P-labeled oligonucleotide probe containing the mammary growth factor (MGF) binding site from the bovine β-casein gene promoter (29). Increased β-casein MGF-site DNA-binding activity was readily detected in nuclear extracts from insulin perfused livers compared with control livers (Fig. 4B). The insulin-induced complexes could be super-shifted/inhibited with an anti-Stat5 specific antibody, but not by anti-Stat3, anti-Stat1, or normal rabbit serum. Moreover, the insulin-induced DNA-binding complexes could be “super-shifted” with antibodies specific to Stat5a or Stat5b, demonstrating that both Stat5 proteins were present in these complexes. A control experiment of antibodies incubated with radiolabeled probe alone demonstrates the nonspecific binding of proteins within the crude sera (Fig. 4B). A similar result, namely, tyrosine phosphorylation and activation of Stat5 in insulin-perfused livers, was observed in the experiment without addition of sodium vanadate in the perfusion buffer, indicating that the Stat5 activation was not due to artificial inhibition of tyrosine phosphatases (Fig. 4 C and D).
Figure 4.
Insulin stimulates Stat5 tyrosine phosphorylation and DNA binding activity in perfused liver. (A) Stat5 is tyrosine phosphorylated in mouse liver perfused with insulin. The livers were perfused with 10 ml Hanks solution containing either 1 mM Na3VO4 or 1 mM Na3VO4 plus insulin (50 nM). After 10 min, the liver lysates were prepared. Three milligrams of protein of each lysate was immunoprecipitated with antibodies against Stat1, Stat3, Stat5, IR, or IGF1 receptor and then divided into two parts, one part for Western blotting to detect tyrosine phosphorylation (Upper) and the other for assessing protein amount (Lower). (B) Insulin stimulates Stat5 DNA-binding activity. Nuclear proteins were extracted from the perfused mouse liver and subjected to EMSA using a 32P-labeled double-stranded oligonucleotide probe that contained a MGF/Stat5 binding site from the β-casein promoter (PIE) (29). The Stat-specific antibodies were used for supershift assay. The nonspecific supershift was determined in the absence of nuclear extract. (C) Stat5 is tyrosine phosphorylated in mouse liver perfused with insulin in the absence of sodium vanadate. Conditions were the same as in A except sodium vanadate was not added in the perfusion buffer, and 6 mg of total protein of each lysate was used for analysis. (D) Insulin stimulates Stat5 DNA binding in the absence of sodium vanadate. Frozen pieces of the mouse livers perfused as described in C were thawed and cytoplasmic and nuclear extracts were prepared. Stat5 DNA-binding activity in these extracts (30 μg) was assessed similarly using PIE probe. Arrows indicate the specific insulin-inducible complexes; asterisks indicate supershifted Stat5-containing complexes.
Members of the JAK family of nonreceptor-type tyrosine kinases are critical for phosphorylation of Stat proteins in response to cytokine stimulation. We therefore tested whether JAK kinases were tyrosine phosphorylated (activated) after perfusion of mouse liver with insulin. We could not detect any insulin-stimulated increases in tyrosine phosphorylation of the JAK kinases in liver tissue (data not shown). Therefore, it is likely that the insulin receptor is capable of phosphorylating Stat5, independent of the activation of JAKs.
To determine whether insulin “physiologically” stimulate Stat5 activation in target tissues, we took advantage of a classical fasting-refeeding paradigm, namely postprandial increase in circulating insulin levels. We could detect significant increase in the tyrosine phosphorylation of Stat5 in several tissues after refeeding, including muscle, fat, and liver (Fig. 5A). In contrast, no increase in Stat5 tyrosine phosphorylation was detected in brain, and only a very slight increase of Stat5 tyrosine phosphorylation was detected in heart. Although we cannot rule out potential effects due to other hormones or factors that might be released after refeeding, the simplest interpretation of these results is that postprandial secretion of insulin is responsible for Stat5 activation in those target tissues. The activation of Stat5 after refeeding by growth hormone seems to be very unlikely, because ample evidence indicates that feeding does not increase plasma growth hormone level in adult animals (33, 34). To demonstrate that refeeding of fasted mice also leads to activation of Stat5 DNA-binding activity in the liver, we prepared cytoplasmic and nuclear extracts from the livers of fasted vs. refed mice and performed EMSA analysis with the β-casein PIE probe (Fig. 5B). Extracts from the liver of a refed mouse clearly contain more Stat5 DNA-binding activity than extracts from the liver of a fasted mouse. The fasting and refeeding experiment was repeated several times with similar results.
Figure 5.
Refeeding of the fasted mice leads to Stat5 tyrosine phosphorylation and activation in insulin target tissues. (A) Tyrosine phosphorylation of Stat5 in liver, muscle, and adipose tissues of refed mice. Mice were starved for 3 days, allowed to feed for 80 min, and then sacrificed. Whole cell lysates were prepared from the indicated tissues. Three milligrams each of the total protein lysates was used for immunoprecipitation with anti-Stat5. A second immunoprecipitation from the supernatant was then performed with anti-IR antibody. Anti-Stat5 immunoprecipitates were divided into two parts: one part for PTyr Western blotting (Top; faint signal of Stat5 tyrosine phosphorylation in the heart of refed mice in the original filter did not reproduce well), and the other part for anti-Stat5 blotting (Upper Middle). The complexes captured by anti-IR was assayed in vitro for kinase activity (Lower Middle). The same filter was used for detecting the IR amount by Western blotting with anti-IR antibody (Bottom). (B) Stat5 DNA binding activity is induced by refeeding. Cytoplasmic and nuclear extracts from the livers of fasted and refed mice were subjected to EMSA analysis with the PIE probe (29). Arrow indicates the refeeding-induced complex.
DISCUSSION
We have obtained evidence to suggest that tyrosine phosphorylation and activation of the Stat5 transcription factor is part of the response to insulin in target tissues sensitive to physiological stimulation by insulin. In support of this model, we have shown that (i) Stat5b-Ct is tyrosine phosphorylated by IR CKD in vitro; (ii) Stat5b-Ct is tyrosine phosphorylated in vivo in response to insulin stimulation when Stat5b-Ct and IR are coexpressed in COS cells; (iii) Endogenous Stat5 is tyrosine phosphorylated and activated in response to insulin in CHO-IR and HIR3.5 cells; (iv) Stat5 is tyrosine phosphorylated and activated in response to insulin in insulin perfused livers; and (v) Stat5 is tyrosine phosphorylated and activated in refed mice.
Our yeast two-hybrid system results show that the insulin receptor is able to interact directly with a fragment of Stat5b. While this interaction appears to require receptor kinase activity, we do not know the minimal requirements for this interaction. The two-hybrid interaction could reflect kinase-substrate interaction, receptor phosphotyrosine-Stat5 SH2 domain interaction, or both. Mutagenesis of both receptor and Stat5 will be required to fully determine the nature of these interactions. The interaction of Stat5 and IR is not restricted to the yeast two-hybrid system, as we have shown that Stat5b-Ct is phosphorylated in vitro by IR CKD and in COS cells overexpressing IR upon insulin stimulation. Moreover, endogenous Stat5 is rapidly tyrosine phosphorylated when IR-expressing CHO and NIH 3T3 cells are treated with insulin or when mouse livers are perfused with insulin. Furthermore, we have shown that insulin-stimulated tyrosine phosphorylation of Stat5 in the IR-expressing cells and in mouse livers is accompanied by increased Stat5 DNA-binding activity, in the absence of detectable JAK tyrosine phosphorylation. Therefore, we suggest that IR-mediated phosphorylation and activation of Stat5 do not require activation of JAK kinases. IR-mediated activation of Stat5 then appears to be similar to the activation of Stat1 and Stat3 by PDGF and EGF receptors (21, 22) and is distinct from cytokine-mediated activation of JAKs and Stats. Giorgetti-Peraldi et al. (35) reported a very weak tyrosine phosphorylation of JAK1, but not JAK2 upon insulin stimulation in NIH 3T3 cells overexpressing IR. Saad et al. (36) reported rapid JAK2 tyrosine phosphorylation upon injection of insulin into rat liver by portal vein. Our failure to detect significant phosphorylation of JAKs in perfused mouse liver could be due to a very rapid and transient phosphorylation of JAKs such that we have missed the peak of phosphorylation after 15 min of perfusion. Alternatively, the stoichiometry of insulin-induced JAK phosphorylation in mouse liver could be very low such that it was below the sensitivity of our detection. At the present, we cannot totally rule out the possibility of JAK activation in the insulin-perfused mouse liver or after refeeding.
The known substrates of insulin receptor to date include IRS-1, IRS-2 (also called 4PS), and Shc (6, 37–39). Our finding of the interaction and activation of Stat5 adds to the list of IR substrates, in this case, a nuclear effector. This may provide an additional pathway through which the IR signals to the nucleus, one that is independent of the ability of IR to activate gene transcription through the RAS/RAF/MEK/MAPK cascade. We speculate that insulin-induced activation of Stat5 represents an IRS-1 and MAP kinase-independent pathway of insulin-mediated gene regulation. It is interesting to note that potential Stat5 binding sites are present in the liver-specific promoter regions of the glucokinase and albumin genes (H.B.S., unpublished observations). The transcriptional activation of these genes could be regulated by Stat5. As the importance of MAP kinase in mitogenic signaling is well established, we further propose that these two independent pathways may lead to differential activation of genes required for cell growth versus those involved in metabolic activity and differentiation of cells. This may in part explain the pleiotropic effects of insulin.
Acknowledgments
We thank R. Brent for the gift of reagents for the two-hybrid system; R. O’Neill for help in the initial experiments of the two-hybrid system; C. S. Zong for help in liver perfusion and preparation of some of the figures; L. Ellis for the IR expressing CHO cells; X. Y. Fu for anti-Stat1 serum; L. Hennighausen for antiserum specific to Stat5a and Stat5b; J. Baxbaum and M.-H. Teng for the gift of DBA2 and C57 black mice; J. Xu for excellent technical assistance; and J. Chan for assistance in preparing some of the figures. This work was supported by National Institutes of Health Grant CA 55054 (L.-H.W.) and DK 50074 (R.A.K.).
ABBREVIATIONS
- IR
insulin receptor
- IRS-1
insulin receptor substrate 1
- CKD
cytoplasmic kinase domain
- EGF
epidermal growth factor
- PDGF
platelet-derived growth factor
- IGF1
insulin-like growth factor 1
- EMSA
electrophoretic mobility-shift assay
- CHO
Chinese hamster ovary
References
- 1.Draznin B, Melmed S, Leroith D, editors. Insulin Action, Molecular and Cellular Biology of Diabetes Mellitus. II. New York: Liss; 1989. [Google Scholar]
- 2.Ebina Y, Ellis L, Jarnagin K, Edery M, Graf L, Clauster E, Ou J H, Masiarz F, Kan Y W, Goldfine I D, Roth R A, Rutter W J. Cell. 1989;40:747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- 3.Chou C K, Dull T J, Russel D S, Gherzi R, Lebwohl D, Ullrich A, Rosen O M. J Biol Chem. 1987;262:1842–1847. [PubMed] [Google Scholar]
- 4.Ebina Y, Araki E, Taira M, Shimada F, Mori M, Craik C S, Siddle K, Pierce S B, Roth R A, Rutter W J. Proc Natl Acad Sci USA. 1987;84:704–708. doi: 10.1073/pnas.84.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheatham B, Kahn C R. Endocrine Review. 1995;16:117–142. doi: 10.1210/edrv-16-2-117. [DOI] [PubMed] [Google Scholar]
- 6.Sun X J, Rothenberg P, Kahn C R, Backer J M, Araki E, Wilden P A, Cahill D A, Goldstein B J, White M F. Nature (London) 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- 7.Backer J M, Schroeder G G, Kahn C R, Meyers M G, Jr, Wilden P A, Cahill D A, White M F. J Biol Chem. 1992;267:1367–1374. [PubMed] [Google Scholar]
- 8.Kuhue M R, Pawson T, Leinhard G E, Feng G S. J Biol Chem. 1993;16:11479–11481. [PubMed] [Google Scholar]
- 9.Skolnik E Y, Batzer A, Li N, Lee C-H, Lowenstein E, Mohammadi M, Margolis M, Schlessinger J. Science. 1993;260:1953–1955. doi: 10.1126/science.8316835. [DOI] [PubMed] [Google Scholar]
- 10.Lee C-H, Li N, Nishimura R, Zhou M, Batzer A G, Myers M G, Jr, White M F, Schlessinger J, Skolnik E Y. Proc Natl Acad Sci USA. 1993;90:11713–11717. doi: 10.1073/pnas.90.24.11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craparo A, O’Neill T J, Gustafson T A. J Biol Chem. 1995;270:15639–15643. doi: 10.1074/jbc.270.26.15639. [DOI] [PubMed] [Google Scholar]
- 12.Gustafson T A, He W, Craparo A, Schaub C D, O’Neill T J. Mol Cell Biol. 1995;15:2500–2508. doi: 10.1128/mcb.15.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blenis J. Proc Natl Acad Sci USA. 1993;90:5889–5892. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis R J. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 15.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 16.Sadowski H B, Gilman M Z. Nature (London) 1993;362:79–83. doi: 10.1038/362079a0. [DOI] [PubMed] [Google Scholar]
- 17.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 18.Fu X Y, Zhang J J. Cell. 1993;74:1135–1145. doi: 10.1016/0092-8674(93)90734-8. [DOI] [PubMed] [Google Scholar]
- 19.Ruff-Jamison S, Zhong Z, Wen Z, Chen Z, Darnell J E, Jr, Cohen S. J Biol Chem. 1994;269:21933–21935. [PubMed] [Google Scholar]
- 20.Ruff-Jamison S, Chen K, Cohen S. Proc Natl Acad Sci USA. 1995;92:4215–4218. doi: 10.1073/pnas.92.10.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaman D W, Pisharody S, Fickinger T W, Commane M A, Schlessinger J, Kerr I M, Levy D E, Stark G R. Mol Cell Biol. 1996;16:369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignais M L, Sadowski H B, Watling D, Rogers N C, Gilman M. Mol Cell Biol. 1996;1:1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zervous A S, Gyuris J, Brent B. Cell. 1993;72:222–232. [Google Scholar]
- 24.Field S, Song O-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 25.Whittaker J, Okamoto A K, Thys R, Bell G I, Steiner D F, Hofmann C A. Proc Natl Acad Sci USA. 1987;84:5237–5241. doi: 10.1073/pnas.84.15.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Robinson G W, Gouilleux F, Groner B, Hennighausen L. Proc Natl Acad Sci USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohanski R A. Biochemistry. 1993;32:5773–5780. doi: 10.1021/bi00073a008. [DOI] [PubMed] [Google Scholar]
- 28.Jong S M, Wang L-H. J Virol. 1990;64:5997–6009. doi: 10.1128/jvi.64.12.5997-6009.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mui A L-F, Wakao H, O’Farrell A M, Harada N, Miyajima A. EMBO J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J X, Mietz J, Modi W S, John S, Leonard W J. J Biol Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- 31.Silva C M, Lu H, Day R N. Mol Endocri. 1996;10:508–518. doi: 10.1210/mend.10.5.8732682. [DOI] [PubMed] [Google Scholar]
- 32.Ebina Y, Edery M, Ellis L, Standring D, Beaudoin J, Roth R A, Rutter W J. Proc Natl Acad Sci USA. 1985;82:8014–8018. doi: 10.1073/pnas.82.23.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassett J M. Aust J Biol Chem. 1972;25:1277–1287. doi: 10.1071/bi9721277. [DOI] [PubMed] [Google Scholar]
- 34.Bassett J M, Weston R H, Hogan J P. Aust J Biol Chem. 1971;24:321–330. doi: 10.1071/bi9710321. [DOI] [PubMed] [Google Scholar]
- 35.Giorgetti-Peraldi S, Peyrade F, Baron V, Van Obberghen E. Eur J Biochem. 1995;234:650–660. doi: 10.1111/j.1432-1033.1995.656_b.x. [DOI] [PubMed] [Google Scholar]
- 36.Saad M, Carvalho C, Thirone A, Velloso L. J Biol Chem. 1996;271:22100–22104. doi: 10.1074/jbc.271.36.22100. [DOI] [PubMed] [Google Scholar]
- 37.Araki E, Lipes M A, Patti M E, Burning J C, Haag B, III, Johnson R S, Kahn C R. Nature (London) 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 38.Sun X J, Wang L M, Zhang Y, Yenush L, Meyers M G, Glasheen E, Lane W S, Pierce J H, White M F. Nature (London) 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 39.Pellicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci P. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]