Abstract
α1C- and α1E-based Ca2+ channels differ in that the former are inhibited by Ca2+ entering through its pore, while the latter are not. It has been proposed on the basis of analysis of α1E/α1C chimeras that the molecular determinants responsible for Ca2+ inhibition involve both a conserved Ca2+-binding motif (EF hand) plus additional sequences located C-terminal to the EF hand. Through construction of similar α1E/α1C chimeras, we transferred Ca2+ inhibition from α1C to α1E by replacing a 134-aa segment of α1E with the homologous 142-aa segment of α1C. This segment is located immediately after the proposed Ca2+-binding EF hand motif. Replacement of the α1C EF hand with the corresponding EF hand of α1E did not interfere with inhibition of α1C by Ca2+, and a triple mutant of α1C, α1C[D1535A,E1537A,D1546A], that disrupts the potential Ca2+-coordinating ability of the EF hand continued to be inhibited by Ca2+. These results indicate that a small portion of the α1C C terminus is essential for inhibition by Ca2+ and place the Ca2+-binding site anywhere in α1C, with the exception of its EF hand-like motif.
Keywords: heart, neurons, EF hands, chimeras
Ca2+ entry into cells through voltage-activated Ca2+ channels is transient as a result of voltage-induced inactivation and/or because of feedback inhibition by Ca2+ itself. We reported previously that, in Xenopus oocytes, concentrations of 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (BAPTA) that are able to block activation of Ca2+-activated Cl− currents by Ca2+ entering through the mouth of the Ca2+ channel do not inhibit Ca2+-dependent inhibition of the channel (1) and concluded that the Ca2+-binding site should be very close to the conduction pathway. But its actual location remained unknown. Biochemical studies and molecular cloning have shown that Ca2+ channels are formed of an α1 subunit and accessory regulatory subunits. Some α1 subunits—e.g., α1C—are feedback-inhibited by Ca2+ (1), while others—e.g., α1E—are feedback-regulated only by voltage (2). Recently, de Leon et al. (3) reported that they were able to confer Ca2+ inhibition to α1E by replacing the entire amino acid C terminus of α1E with a 217-aa truncated form of the α1C C terminus. This segment includes a 29-aa motif with homology to classical Ca2+-binding domains called EF hands (4). This motif thus was proposed by de Leon et al. (3) to be the Ca2+-binding site responsible for channel inhibition. We have also been studying the structural basis for Ca2+ inhibition, and we have also found this general region of α1C to be essential for feedback inhibition by Ca2+. However, as we report below, it excludes both the EF hand and a Ca2+-binding function that may be associated with it.
METHODS
Construction of α1C/α1E Chimeras and Mutants.
To facilitate the engineering of α1C and α1E cDNAs, we subjected both cDNAs to silent mutagenesis to remove undesirable restriction sites and introduce new ones without altering the amino acid sequence encoded in the cDNAs. For α1C, we used the DN60 derivative of clone Va.33 (ref. 5; GenBank accession no. X15539X15539) in which amino acids 1–59 have been removed and eliminated the restriction sites SspI at nucleotide 1970, SacII at nucleotide 4116, and ScaI at nucleotide 4969, and we created sites for BstBI at nucleotide 465, ScaI at nucleotide 1288, HpaI at nucleotide 1745, SalI at nucleotide 2348, SacII at nucleotide 2845, SspI at nucleotide 3569, and BstBI at nucleotide 3752. Va.61 is the engineered α1C cDNA clone that resulted from these manipulations. For α1E, we removed from clone E239 (ref. 2; GenBank accession no. L27745L27745 with the 5′ extension shown below) the restriction sites KpnI at nucleotide 658, BstBI at nucleotide 1231, SspI at nucleotide 1736, and BstBI at nucleotide 6753, and we created sites for HpaI at nucleotide 1510, SalI at nucleotide 2109, SacII at nucleotide 3505, SspI at nucleotide 4247, BstBI at nucleotide 4436, and MluI at nucleotide 5181. E101 is the engineered α1E cDNA clone that resulted from these manipulations. α1E/α1C chimeras (EC chimeras) and mutants/deletions were made by standard two-round PCR or M13-based site-directed mutagenesis (6), using Va.61 and E101 as substrates. The nucleotide sequence of all modified cDNAs was confirmed by the dideoxy chain termination method of Sanger et al. (7) using the double-stranded plasmids as templates. The open reading frames of Va.61 and E101 and their derivatives were placed into a modified pAGA2 vector, which is a pGEM3-based plasmid (5). E101 and its derivatives were placed into pGEM3 downstream of the T7 promoter so that the nucleotide sequence between the T7 promoter and the initiator ATG in L27745 became 5′-GGGAGACCGG AATTGATCCC CGGGTACCAT GGTGTGTCTT CTGTCTGTTT AAACCTCAGG ATG, where the A of ATG corresponds to nucleotide 1 of the open reading frame reported in L27745; the nucleotide sequence between the stop codon and the beginning of the poly(A) tail of pAGA2 is 5′-TAG AGGCTGCTCC CCCCTCCGAT GCATGCTCTT CTCTCACATG GAGAAAACCA AGACAGAATT GGGAAGCCAG TGCGGCCCGG GGGGGAGGAA GAGGAAGAGG GAAAAGTCGT CCTGTTGTAG GCCTCCCCCT AGCATCCTCT TAG, where TAG is the stop codon of the open reading frame. For Va.61 the nucleotide sequence between the T7 promoter and the initiator ATG in X15539X15539 is 5′-TAATACGACT CACTATAGGG AGACCGGAAT TGATCCCCGG GTACC ATG, and the sequence between the stop codon and the beginning of the poly(A) tail of pAGA2 is 5′-TGA GCGCCAGGGC CGGGGGTGCG GGTTTTTTAT TTGTCTCAAT GTTCCTAATG GGTTCGTTTC AGAACGTTTC AGAAGTGCCT CACTGTTCTC GTGACCTGGA GTTAACCGCG GAATTGGGAT CCTCTAGCTA G.
The rat β2a subunit (ref. 8; GenBank accession no. M80545M80545) was cloned as a NcoI/XbaI fragment of clone βb24 into NcoI/XbaI-digested pAGA2 by changing codon 2 from CAG (Q) to GAG (E) and adding a TCTAGA XbaI restriction site immediately after the TGA stop codon.
Synthesis of cRNAs.
HindIII-digested DNA templates (1 μg) were transcribed in a final volume of 20 μl with reagents provided in the mMesSAGE mMACHINE cRNA synthesis kit from Ambion (Austin, TX; catalog no. 1344). After removal of the template DNA by treatment with RNase-free DNase I and precipitation with either LiCl or ammonium acetate, the cRNAs were dissolved in double-distilled diethyl-pyrocarbonate (DEPC)-treated water to a final concentration of 1–2 μg/μl. Wild-type or mutant α1 subunits were expressed in Xenopus oocytes together with the rat β2a subunit. To this end, cRNAs encoding wild-type or mutant α1C, wild-type or mutant α1E, or α1E/α1C chimeras (0.2 μg/μl) were coinjected into oocytes with 0.2 μg/μl of rat β2a cRNA.
Oocyte Preparation.
Frogs (Xenopus laevis) were anesthetized by immersion into 0.15–0.17% tricaine methanesulfonate in water and removed from the tricaine methanesulfonate bath. Ovarian lobes were then exposed through a small incision made into their abdominal wall, removed, and placed into sterilized Ca2+-free OR-2 solution (82.5 mM NaCl/2.5 mM KCl/1 mM MgCl2/5 mM Hepes, pH adjusted to 7.6 with NaOH), and the frogs were returned to tricaine-free water for recovery. The ovarian lobes were then rinsed with sterile water, teased open, and incubated at room temperature in Ca2+-free OR-2 containing 2 mg/ml collagenase (type I; BRL) to cause release and defolliculation of oocytes. After 1 hr on an orbital shaker (≈60 cycles per min), the oocytes were transferred to a Petri dish with OR-2. Dead and too-small oocytes were removed by aspiration, and the selected oocytes were washed several times with collagenase-free and Ca2+-free OR-2 solution, incubated under agitation for an additional 1 hr with solution changes every 7–8 min, and placed into an incubator at 19°C and incubated for an additional 1 hr in a 1:4 mixture of sterile SOS (100 mM NaCl/2 mM KCl/1.8 mM CaCl2/1 mM MgCl2/5 mM Hepes, pH adjusted to 7.6 with NaOH) and Ca2+-free OR-2 solutions and 30 min in 1:3 SOS/Ca2+-free OR-2. The oocytes were then placed into 100% SOS and selected once more, by removing those that are dead or too small and kept at 19°C until injected.
Injected oocytes were kept at 17–19°C, with daily solution changes in sterile SOS containing 50 μg/ml gentamycin, until they were used for electrophysiological testing (4–6 days).
Electrophysiological Recordings of Ca2+ Channel Currents from Oocytes.
The cut-open vaseline gap voltage-clamp method (9) as modified (1, 10) was used throughout. Activation of Cl− current by Ba2+ or Ca2+ influx through the Ca2+ channel was eliminated by injecting 100–150 nl of 50 mM Na4BAPTA before recording (1). The BAPTA solution was adjusted to pH 7.0 with methanesulfonic acid. Ba2+ currents were recorded using an external solution containing 10 mM Ba2+, 96 mM Na+, 0.1 mM ouabain, and 10 mM Hepes, titrated to pH 7.0 with methanesulfonic acid (CH3SO3H). Unless indicated otherwise, Ca2+ currents were recorded with the same external solution but replacing Ba2+ with 10 mM Ca2+. The solution in contact with the oocyte interior was 110 mM K-glutamate/10 mM Hepes, titrated to pH 7.0 with KOH. Low access resistance to the oocyte interior was obtained by permeabilizing the oocyte with 0.1% saponin. The holding potential was −90 mV. The linear components of the currents, corresponding to the scaled currents elicited by small negative control pulses of one-quarter the amplitude of the stimulating pulse, were subtracted online. Data were collected at a sampling frequency of 1 kHz and filtered at 200 Hz.
Presentation of Results.
Inactivation rates (1/τ) in the presence of Ca2+ (irCa) or Ba2+ (irBa) were obtained from least squares fits of the first 350–1000 msec of current inactivation time courses to a first-order decay function (A/Ao = exp(−t/τ) + C). For certain α1E/α1C chimeras, channel inactivation is purely voltage-dependent when measured in external Ba2+, but it is the sum of coexisting voltage-dependent and Ca2+-dependent processes when measured in external Ca2+. Where each of these these effects were very marked—e.g., chimeras EC1 and EC56—we desisted from calculating irCa, as we could not define in the chimera the proportion of inactivation that was due to Ca2+. For these chimeras we thus present only the direct data to indicate whether or not a Ca2+ inactivation component has been introduced into the α1E.
RESULTS AND DISCUSSION
To study Ca2+ channel inhibition by Ca2+, we expressed α1C, α1E, or α1C/α1E chimeras in Xenopus oocytes together with β2a. A β subunit was coexpressed to take advantage of the action of this regulatory subunit to improve the coupling between voltage sensing and pore opening, which lead to an increase in the magnitude of the α1C currents (10). The 2a subtype of β was chosen to take advantage of its effect to delay and reduce voltage-induced inactivation of α1E (11) and thus alleviate the effect of this complicating feature in the analysis of an inactivating effect of Ca2+ in α1E/α1C chimeras. For construction of chimeras, we chose as crossover points sites in the amino acid sequence at which α1C and α1E were identical. The rationale behind this choice was the assumption that the two molecules were not only homologous at the level of linear amino acid sequence composition but also at the level of their three-dimensional structure, so that identical amino acids in the linear sequence could be expected to adopt similar orientations in space. This should then allow the exchange of structures spanned by similarly placed amino acids and minimize folding problems of the chimera. In spite of this, chimera EC58 was not expressed, indicating that additional factors influence the functional expression of a membrane protein.
Assessment of Inhibition by Ca2+.
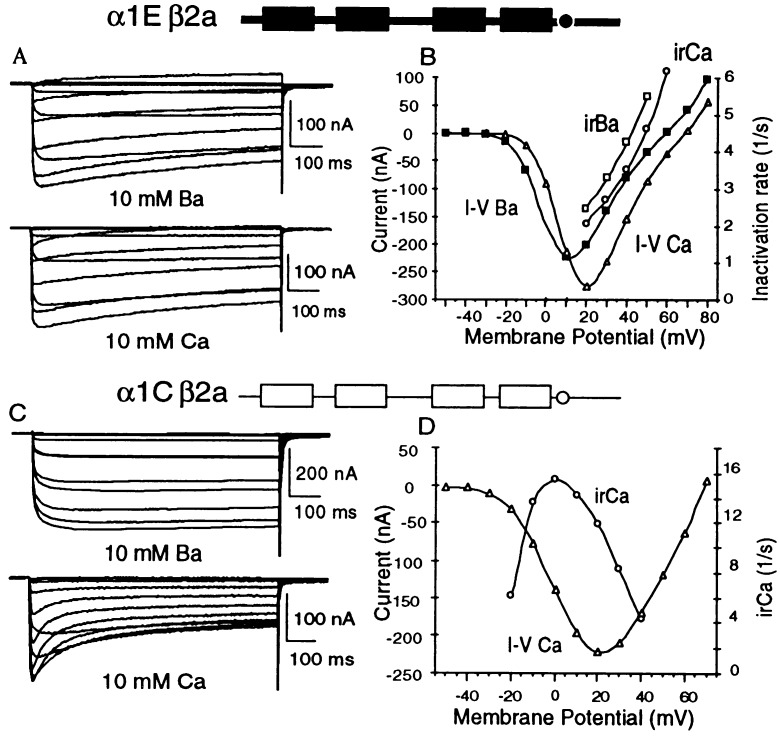
Results reported previously had shown that the site to which Ca2+ binds to inhibit α1C channel activity could be located very close to the inner mouth of the channel, as injection into oocytes of the Ca2+ chelator BAPTA was able to interfere with activation of Cl− currents by Ca2+ entering through the channel’s pore but not with the inactivation of the channel by the incoming Ca2+ (1). As a consequence, α1C inhibition by Ca2+ varies with voltage. It is minimal at low test potentials at which single channel amplitudes are large, mean open times are short, and mean closed times are long. Only minimal Ca2+ influx as a function of time occurs under these conditions. Failure of Ca2+ inhibition at low test potentials could then be due either to failure of Ca2+ binding to the inhibitory site or, because of dissociation during the time the channel is closed and attendant failure in occupying the inhibitory site by Ca2+ for a sufficiently long time to allow for transduction of the binding signal into the conformational cascade responsible for inactivation. Inactivation is also minimal at test potentials close to the reversal potential, at which single channel amplitudes are near zero, even though mean open times are a maximum. Inhibition is maximal at voltages close to those eliciting maximal macroscopic Ca2+ currents, at which influx of Ca2+ per unit time is maximal (or close to it). For reasons that are still under study, a typical characteristic of the rate of inhibition by Ca2+ is that it is maximal at voltages slightly below those at which current–voltage relations show their maxima (Fig. 1).
Figure 1.
Inhibition by Ca2+ of α1C but not α1E and comparison of voltage dependence of irCA to that of the current.
Structural Determinant Conferring Ca2+ Inhibition.
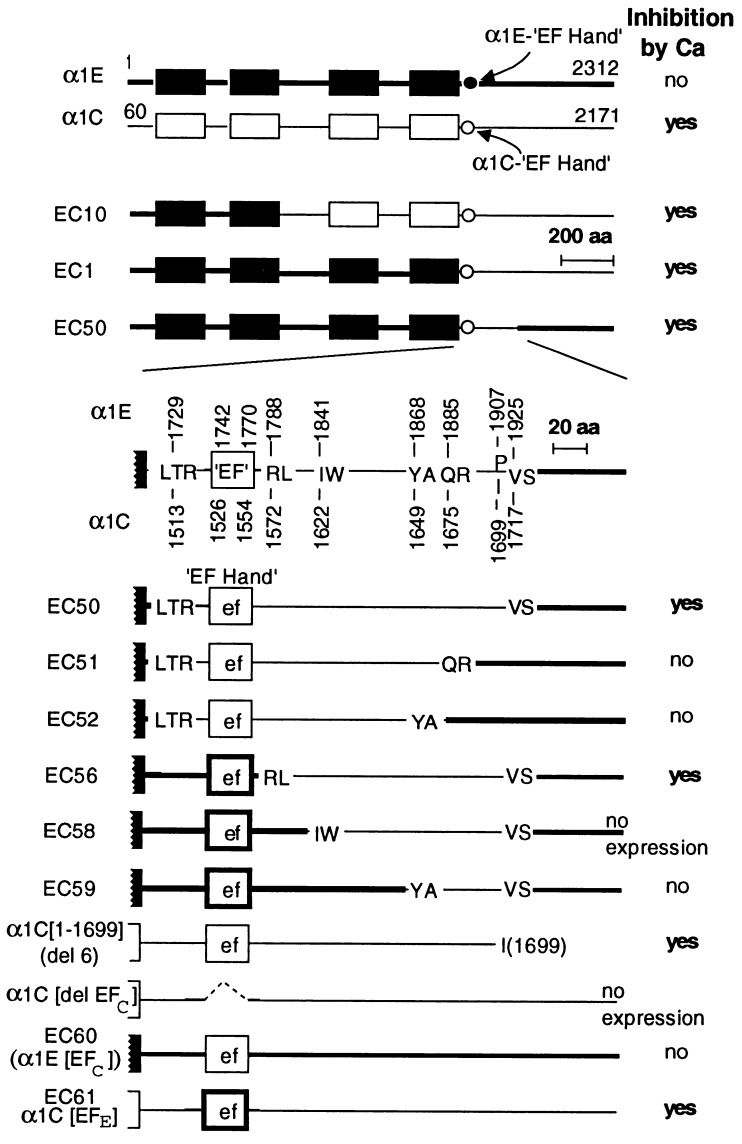
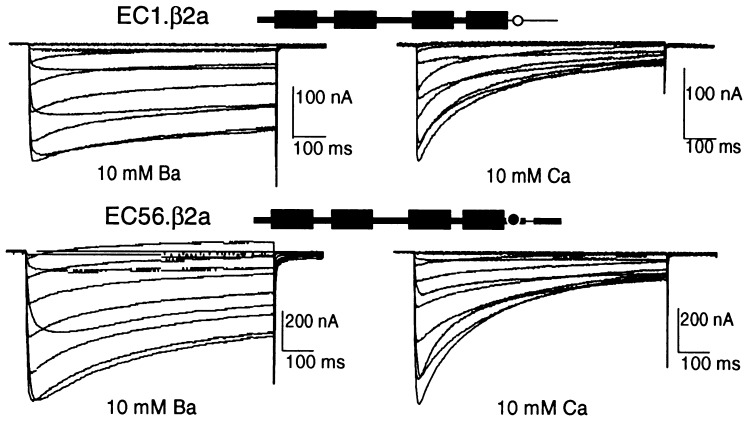
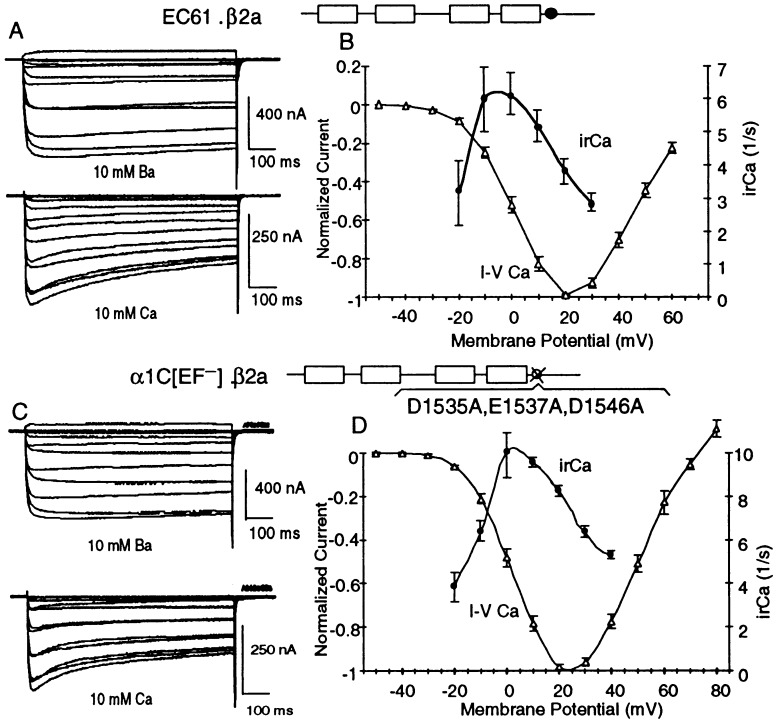
Point mutations and amino acid deletions may have long-range effects on the conformation and function of a protein and therefore do not necessarily pinpoint location of binding and/or regulatory sites. Because of this, we sought to locate the region(s) of cardiac α1C responsible for feedback inhibition by Ca2+ through a gain-of-function approach in which we replaced segments of the Ca2+-unresponsive α1E with the corresponding and structurally homologous segments of the Ca2+-responsive α1C. Fig. 2 depicts the strategy used and a summary of the results obtained with different EC chimeras and α1E and α1C mutants. The patterns of inactivation as a function of test potentials obtained in Ba2+ vs. those obtained in Ca2+ for EC1, α1E with the C terminus of α1C, and EC 56, α1E with the 142-aa RL → VS segment from α1C, are shown in Fig. 3. Those of α1C[EFE], α1C with the EF hand from α1E, are shown in Fig. 4. The results indicate that inhibition by Ca2+ can be conferred to α1E by replacing of a small segment of the C-terminal tail of α1E with the corresponding segment of α1C and that the α1E EF hand can substitute for that of α1C in α1C.
Figure 2.
Schematic representation of α1E/α1C chimeras and key results obtained. EC1, α1E1728/α1C(1513–2171); EC10, α1E(1–703)/α1C(784–2171); EC50, α1E(1–1728)/α1C(1513–1717)/α1E(1926–2312); EC60, α1E(1–1741)/α1C(1526–1554)/α1E(1771–2312); and EC61, α1C(60–1525)/α1E(1742–1770)/α1C(1555–2171). Amino acid sequences LTR, RL, IW, YA, QR, and VS were common to α1C and α1E and were used as crossover points in the construction of chimeras. Deletion of the EF hand sequence from α1C impeded expression of the protein on the oocyte surface, as neither ionic nor gating currents were detected.
Figure 3.
Chimeras EC1 (α1E with C terminus of α1C) and EC56 (α1E with RL-VS segment of α1C) are inhibited by Ca2+. Ca2+ inhibition of the parental α1C and lack thereof in the parental α1E are shown in Fig. 1.
Figure 4.
EC61, α1C with the EF motif of α1E, and α1C[[D1535A,E1537A,D1546A], an α1C lacking a functional EF motif, are both subject to Ca2+ inhibition.
Lack of a Ca2+-Binding Role for the EF Hand Motif of α1C.
The results on Ca2+ sensitivity of α1C/α1E chimeras shown in Figs. 3 and 4 A and B agree only partially with those reported on similar chimeras by de Leon et al. (3). On one hand, we confirmed that Ca2+ inhibition is encoded in the C terminus of α1C, because replacing both the EF-like motif plus the C-terminal extension of α1E with the corresponding segment of α1C, resulted in appearance of inhibition by Ca2+ in an α1E channel (Fig. 3 Upper). On the other hand, unlike the findings of de Leon et al. (3), our findings showed that an α1C in which its EF-like motif is replaced with the corresponding segment from α1E is still inhibited by Ca2+ (Fig. 4 A and B). Since this α1E segment contains an EF-like motif that is as similar to consensus EF hands as that of the α1C it replaced (Fig. 5), this result can be interpreted in two ways. One assumes that neither the α1C nor the α1E EF hand-like motifs bind Ca2+. The results would then pinpoint the RL → VS segment as the critical determinant required for establishment of Ca2+ inhibition and leave open the question of where Ca2+ binds to exert its action. The second assumes that both EF hand-like motifs have the ability to bind Ca2+ if they are supported by the proper sequence environment and that, in this case, this environment is provided by the RL → VS segment of the α1C. Except for the fact that the α1E EF hand motif was inactive in the context of α1C, this was the general conclusion drawn by de Leon et al. (3).
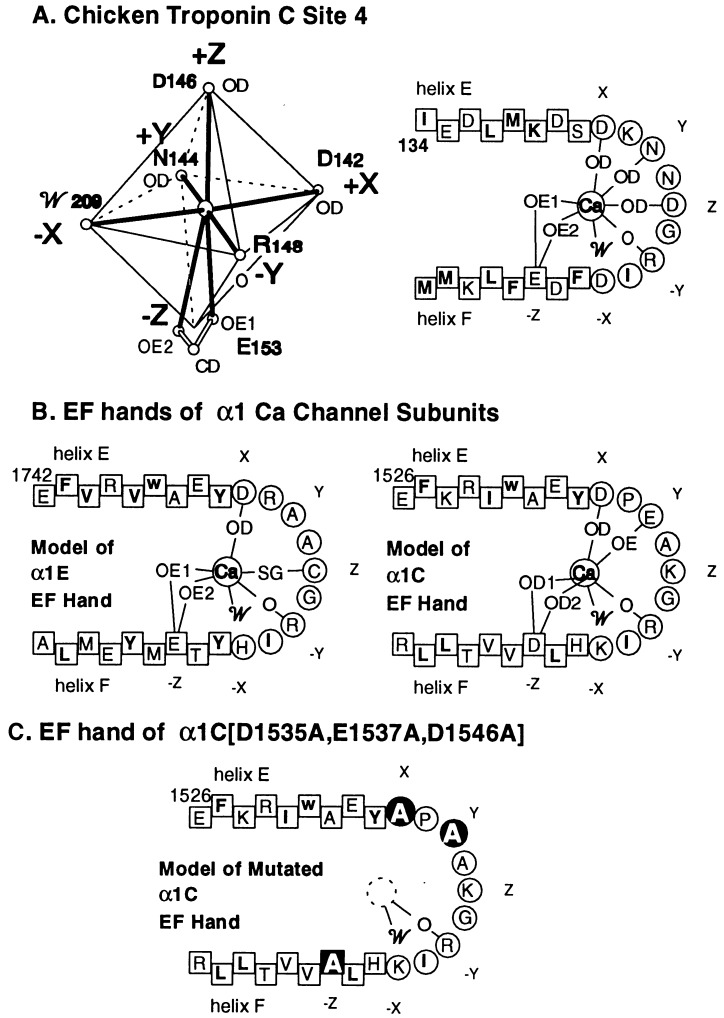
Figure 5.
Ca2+ coordination octahedron in a typical EF hand (A Left); its two-dimensional projection (A Right), and the predicted coordination of Ca2+ by the EF hands of α1E (B Left), α1C (B Right), and α1C[[D1535A,E1537A,D1546A] (C). Ca2+ binding to site 4 of chicken troponin C (cTnC) is adapted from Satyshur et al. (12). O, carbonyl oxygen of the peptide bond; 0D, δ oxygen of Asp or Asn; OE, ɛ oxygen of Glu; and 𝒲, water. Amino acids in squares are part of either the E or the F helices of the (postulated) EF hand; amino acids in circles form the connecting loop of the helix–loop–helix structure. The amino acids that contribute to the formation of the octahedral Ca2+-coordinating cage are labeled X, Y, Z, −X, −Y, and −Z.
To distinguish between the two possibilities listed in the previous paragraph, we tested validity of the second—i.e., whether the EF-like motif of the Ca2+-sensitive α1C plays a Ca2+-binding role. If so, disruption of the Ca2+-binding ability should interfere with inhibition by Ca2+. We thus introduced “inactivating” mutations into the EF-like motif of α1C.
Ca2+-binding EF hands have been extensively studied by x-ray crystallography and nuclear magnetic resonance (for review see ref. 13). In all cases Ca2+ has been found to be held by seven coordination bonds that define an octahedron with vertices X, Y, Z, −X, −Y, and −Z (Fig. 5A), of which vertex −Z is bidentate. The coordinating oxygens at −Z are most frequently the two ɛ oxygens of a Glu extending from the F helix, but can also be the two δ oxygens of an Asp extending from the same location in helix F. The coordinating atom at −Y is always a carbonyl oxygen of the peptide backbone that forms the loop connecting the E and F helices, making the side chain at this position irrelevant. In most but not all of the EF hands with bound Ca2+ described thus far, the coordinating oxygen at −X is provided by H2O (W in Fig. 5), allowing this residue to be almost any amino acid. The coordinating atoms at positions −X, X, and Z are most frequently a δ oxygen of Asp or Asn or one of the ɛ oxygens of Glu. The coordinating atom at position X has also been found to be the γ oxygen of Ser or Thr or the γ sulfur of Cys (cf. ref. 13). A two-dimensional scheme of a typical EF hand with the corresponding assignments of the coordination bonds is shown for site 4 of chicken troponin C in Fig. 5A Right. The equivalent diagram, predicted by computerized sequence analysis of α1C (4) and α1E (2), shows that the α1E and α1C EF hands are potentially capable of satisfying six of the seven Ca2+ coordination bonds provided by a bona fide Ca2+-binding EF hand (Fig. 5B). It needs to be emphasized, however, that even if all seven coordination bonds can be satisfied by sequence similarity analysis, this by no means proves that the EF hand modeled in this way will indeed bind Ca2+. This would require additional knowledge, including the relative orientations of the E and F helices, and hence the spacial disposition of the residues that provide the atoms that coordinate the Ca2+. Thus, Ca2+ does not bind to site 1 of chicken troponin C and of the four EF hands of recoverin, only one is occupied by Ca2+ (14), even though these sites are predicted to bind Ca2+ by the type of analysis that has predicted the α1C and α1E EF hands.
Fig. 5C illustrates the triple-inactivating mutation introduced into α1C (i.e., the change of Asp, Glu, and Asp at positions X, Y, and −Z of the α1C EF motif to Ala), creating α1C[D1535A,E1537A,D1546A]. This triple change is predicted to render the putative Ca2+-binding motif incapable of binding Ca2+. Involvement of a Ca2+-binding function at the EF motif would then be predicted to render the mutant channel insensitive to Ca2+. We found however, that α1C[D1535A,E1537A,D1546A] is inhibited by Ca2+ (Fig. 4 C and D).
Taken together, these results indicate that neither α1E nor α1C has an operational Ca2+-binding EF hand and that Ca2+ inhibition can be conferred to an α1 by introduction of a segment of the α1C C terminus. A likely interpretation of the data is therefore that inhibition by Ca2+ is due to binding to an as yet undefined site on α1. Based on our previous finding that Ca2+ inhibition of α1C is not interfered with by injection of up to 100 nl of 100 mM BAPTA, which is sufficient to suppress Ca2+-activated Cl− currents (1), the inhibitory site for Ca2+ should be very close to the internal mouth of the channel. Results from Eckert and Tillotson (15) and Imredy and Yue (16), showing blockade of inhibition by Ca2+ by chelation of Ca2+ in cells that naturally express Ca2+ channels, place the Ca2+-binding site outside the conduction path proper. In as yet unreported experiments, instead of injecting BAPTA, we perfused oocytes expressing α1Cβ2a with 500 μM BAPTA and observed first unmasking of the Ca2+ inhibition, as Cl− currents were suppressed, and then its progressive suppression, indicating that also for the Ca2+ channel complex expressed in the oocyte, the Ca2+-binding site is outside of the conduction pathway (F.N., R.O., and E.S., unpublished work). Further studies are needed to determine the exact location of the Ca2+-binding site and the mechanism by which Ca2+ inactivates the channel in the manner it does.
Acknowledgments
This work was supported in part by National Institutes of Health Grants AR43411 and AR38970 (L.B.), by a National Institutes of Health Research Service Award (N.Q.), and by American Heart Association grants (R.O. and N.Q.).
ABBREVIATION
- BAPTA
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate
References
- 1.Neely A, Olcese R, Wei X, Birnbaumer L, Stefani E. Biophys J. 1994;66:1895–1903. doi: 10.1016/S0006-3495(94)80983-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider T, We X, Olcese R, Costantin J, Neely A, Palade P, Perez-Reyes E, Qin N, Zhou J, Crawford G D, Smith G R, Appel S H, Stefani E, Birnbaumer L. Recept Channels. 1994;2:255–270. [PubMed] [Google Scholar]
- 3.de Leon M, Wang Y, Jones L, Perez-Reyes E, Wei X, Soong T W, Snutch T P, Yue D T. Science. 1995;270:1502–1506. doi: 10.1126/science.270.5241.1502. [DOI] [PubMed] [Google Scholar]
- 4.Babitch J. Nature (London) 1990;346:321–322. doi: 10.1038/346321b0. [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Perez-Reyes E, Lacerda A E, Schuster G, Birnbaumer L, Brown A M. J Biol Chem. 1991;266:21943–21947. [PubMed] [Google Scholar]
- 6.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 7.Sanger F, Nicklen S, Coulson A B. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Reyes E, Castellano A, Kim H S, Bertrand P, Baggstrom E, Lacerda A E, Wei X, Birnbaumer L. J Biol Chem. 1992;267:1792–1797. [PubMed] [Google Scholar]
- 9.Taglialatela M, Stefani E. Proc Natl Acad Sci USA. 1993;90:4758–4762. doi: 10.1073/pnas.90.10.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neely A, Wei X, Olcese R, Birnbaumer L, Stefani E. Science. 1993;262:575–578. doi: 10.1126/science.8211185. [DOI] [PubMed] [Google Scholar]
- 11.Olcese R, Qin N, Neely A, Stefani E, Birnbaumer L. Neuron. 1994;13:1433–1438. doi: 10.1016/0896-6273(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 12.Satyshur K A, Rao S T, Pyzalska D, Drendel W, Greaser M, Sundaralingam M. J Biol Chem. 1988;263:1628–1647. [PubMed] [Google Scholar]
- 13.Nakayama S, Moncrief N D, Kretsinger R H. J Mol Biol. 1992;34:416–448. doi: 10.1007/BF00162998. [DOI] [PubMed] [Google Scholar]
- 14.Flaherty K M, Zozulya S, Stryer L, McKay D B. Cell. 1993;75:709–716. doi: 10.1016/0092-8674(93)90491-8. [DOI] [PubMed] [Google Scholar]
- 15.Eckert R, Tillotson D L. J Physiol (London) 1981;314:265–280. doi: 10.1113/jphysiol.1981.sp013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imredy J P, Yue D T. Neuron. 1992;9:197–207. doi: 10.1016/0896-6273(92)90159-b. [DOI] [PubMed] [Google Scholar]