Abstract
The visual transduction processes in rod and cone photoreceptor cells begin with photon absorption by the different types of visual pigments. Cone visual pigments exhibit faster regeneration from 11-cis-retinal and opsin and faster decay of physiologically active intermediate (meta II) than does the rod visual pigment, rhodopsin, as expected, due to the functional difference between rod and cone photoreceptor cells. To identify the amino acid residue(s) responsible for the difference in molecular properties between rod and cone visual pigments, we selected three amino acid positions (64, 122, and 150), where cone visual pigments have amino acid residues electrically different from those of rhodopsin, and prepared mutants of rhodopsin and chicken green-sensitive cone visual pigment. The results showed that the replacement of Glu-122 of rhodopsin by the residue containing green- or red-sensitive cone pigment converted rhodopsin’s rates of regeneration and meta II decay into those of the respective cone pigments, whereas the introduction of Glu-122 into green-sensitive cone visual pigment changed the rates of these processes into rates similar to those of rhodopsin. Furthermore, exchange of the residue at position 122 between rhodopsin and chicken green-sensitive cone pigment interchanges their efficiencies in activating retinal G protein transducin. Thus, the amino acid residue at position 122 is a functional determinant of rod and cone visual pigments.
Keywords: rhodopsin, G protein, retina, vision, molecular evolution
Most vertebrates have two types of photoreceptor cells, rods and cones, which are responsible for twilight (scotopic) and daylight (photopic) vision, respectively. Rods are more sensitive to light than cones, while cones display rapid photoresponse and rapid adaptation compared to rods (1, 2). In contrast to our extensive knowledge of the visual transduction process in rods, little is known about the process in cones. However, recent biochemical and electrophysiological studies have revealed that both types of cells have signal transduction proteins, the functions of which are similar but the molecular properties of which are different (1). Thus, the differences in photoresponse patterns between rods and cones should originate from the different properties of these proteins. Since the signal transduction proteins present in cones have amino acid sequences different from those of their counterparts in rods (1–3), identification of the amino acid residue(s) responsible for the molecular properties is important for furthering our understanding of the molecular mechanisms that underlie physiological differences between rods and cones.
Among the signal transduction proteins, visual pigment receives a light signal from the environment using a light absorbing chromophore, 11-cis-retinal (3, 4) and transfers the signal to the retinal G protein transducin by binding to it and catalyzing the GDP–GTP exchange reaction (1–3). In most vertebrates, different types of visual pigments are present in rod and cone photoreceptor cells. Further, it has been revealed that vertebrate visual pigments are classified into four groups of cone visual pigments and a single group of rod visual pigments, the rhodopsins (5–9). The presence of multiple types of cone visual pigments with different spectral sensitivities is the molecular basis of color discrimination, and these pigments have diverged from an ancestral pigment with replacements of amino acid residues in the course of evolution (5–10). On the other hand, our recent investigations clearly showed that, independent of the different spectral sensitivities among the cone visual pigments, they exhibit faster regeneration from 11-cis-retinal and opsin and faster formation and decay of physiologically active meta II intermediate than does rhodopsin (11). Since the faster regeneration and the faster decay of the active state in cone visual pigments might correlate with the faster recovery after photoresponse (12) and the lesser activation of signal transduction cascade (13, 14), respectively, these properties of cone pigments should give a clue to the elucidation of the molecular mechanism leading to the faster adaptation and less sensitive photoresponse of cones. Thus, it is of interest to investigate which kind of replacement(s) of amino acid residue(s) would discriminate between the molecular properties of rod and cone visual pigments.
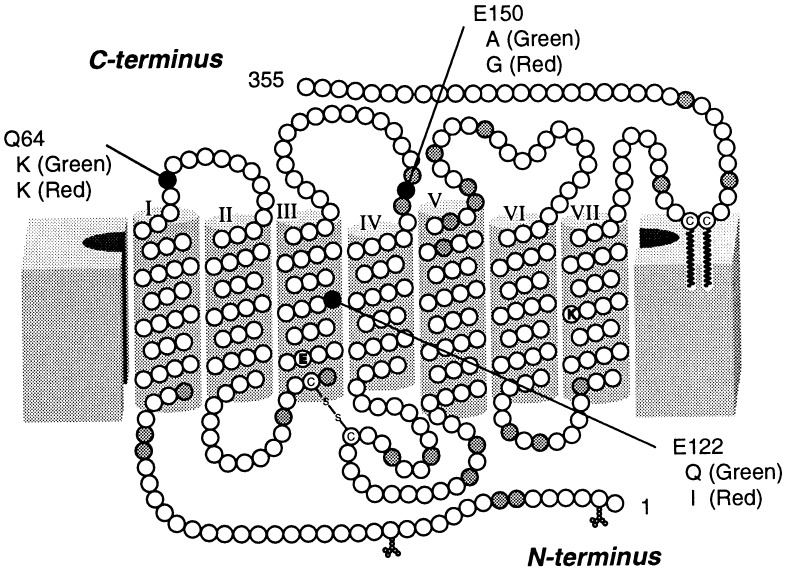
Based on the amino acid sequences of all the visual pigments investigated so far, the common property of cone visual pigments is that they have many basic amino acid residues, whereas rhodopsins have many acidic residues (6). These facts suggested that the differences in rate of regeneration and thermal stability of meta II intermediate between rod and cone visual pigments might be regulated by a dissociative amino acid residue(s) (6, 15). Thus, we selected the amino acid positions where the amino acid residues in cone visual pigments display electric properties different from those of the corresponding residues of rhodopsin (Fig. 1). Whereas each cone visual pigment has several positions where amino acid residues are different from those of rhodopsin, there are three positions (64, 122, and 150) where almost all the cone visual pigments have amino acid residues that are electrically equivalent to each other but are electrically different from those of rhodopsin.† Therefore, we designed and expressed site-directed mutants of rod and cone pigments at these positions. Our current findings clearly show that, among the three positions, only the amino acid at position 122 is responsible for the dramatic changes in the molecular properties of pigments and in their efficiencies in activating transducin. The significance of the replacement at position 122 on the regulation mechanism of signal transduction and its molecular evolution are discussed.
Figure 1.
Amino acid positions in which the electrical properties between the residues chicken rhodopsin and cone visual pigments differ. The transmembrane topography is based on the model of Hargrave et al. (16). Amino acid positions indicated with white circles are those in which rhodopsin and the four types of cone visual pigments have residues similar in electric properties. The gray or black circles indicate the positions at which some (gray) or almost all (black) of the cone pigments have residues electrically different from those of rhodopsin. The residues of rhodopsin replaced in this study are denoted by single-letter codes and numbered using the bovine rhodopsin numbering system. Corresponding residues of chicken green and red are also denoted.
MATERIALS AND METHODS
Preparation of Visual Pigment Mutants.
The mutant visual pigments were expressed as described previously (18). Briefly, the coding regions of chicken rhodopsin and chicken green-sensitive pigment were derived from cDNA clones pZf9 and pZf7, respectively (6). The DNA fragments were attached with a HindIII and EcoRI sites to the 5′ and 3′ ends, respectively, and subcloned into pBluescript II KS+ (Stratagene). Point mutation was introduced by means of phosphorothioate method using Sculptor (Amersham). The entire coding regions were sequenced by the dideoxy termination method with Sequenase (Amersham). Each of the fragments was recloned into the expression vector pUSRα (19) and transfected into 293S cells using calcium phosphate method (20). After harvesting, the cell membrane was isolated by means of sucrose flotation methods (21), and the opsin was extracted with buffer E [0.75% (wt/vol) CHAPS/1 mg/ml l-α-phosphatidylcholine (from egg yolk)/50 mM Hepes/140 mM NaCl/1 mM DTT/1 μg/ml aprotinin/1 μg/ml leupeptin, pH 6.5 at 4°C]. The absorption maxima of the expressed rhodopsins were 507 (wild type), 491 (Q64K/E122Q/E150A), 508 (Q64K), 504 (E150A), 486 (E122Q), 504 (E122I), and 487 (E122D) nm, the latter three of which were in good agreement with those reported previously in bovine rhodopsin system (21–27). The maxima of the expressed chicken green were 511 (wild type) and 525 (cG-Q122E) nm, the former of which is similar to that reported previously (28).
Regeneration Rates of Visual Pigments.
The regeneration rates of visual pigments were measured as described (11), with some modification. The courses of increase of absorbance at 530 nm were recorded with time scan mode of a MPS-2000 spectrophotometer (Shimadzu). The amount of pigments regenerated were estimated by bleaching the fully regenerated pigments with yellow light (>480 nm) at 2°C in the presence of hydroxylamine. The regeneration rate constants of native pigments were reported previously (11).
Decay Rates of Meta II Intermediates.
The decay rates of meta II intermediates were measured as described (11). The pigment was regenerated by the addition of 2.5 nmol of 11-cis-retinal solubilized in ethanol (5 μl) to each of opsin extracts (220 μl of each). Then the pigment sample was subjected to low-temperature time-resolved spectroscopy to investigate formation and decay of meta II (29). The decay rate constants of meta II in native pigments were reported previously (11, 15, 30).
GTPase Activities of Native and Mutant Visual Pigments.
Native rhodopsin and chicken green were purified from chicken retina according to the method previously reported (15, 31). Expressed pigments were regenerated by addition of 11-cis-retinal before solubilization with buffer E. These pigments were incorporated into l-α-phosphatidylcholine liposome by dialysis against buffer Pm (50 mM Hepes/140 mM NaCl/3 mM MgCl2/1 mM DTT/1 μg/ml aprotinin/1 μg/ml leupeptin, pH 6.5 at 4°C). Transducin was prepared from the fresh bovine retina by the method previously reported (32). After the irradiation of the pigment solution with orange light for 30 s and incubation at 2°C (native) or 21°C (expressed pigments), the pigment was added to the reaction solution (0.1 μM pigments/1.5 μM transducin/2.4 μM [γ-32P]GTP). Then the mixture was incubated at 25°C for 2 min for the GTPase reaction, and the reaction was stopped with EDTA solution. The released Pi was collected by addition of activated charcoal and assayed by liquid scintillation counter (model LS6000IC; Beckman). The activity immediately after the irradiation was measured from the sample mixed before the irradiation and was confirmed to be almost identical to that of pigment added to the reaction mixture after the irradiation. The activity of the sample not irradiated before the reaction was measured and calculated as background. The linear relationship between the active pigment concentration and GTPase activity was confirmed using the native rhodopsin. It should be noted that the activities induced by native and expressed rod and cone pigments immediately after the irradiations were within 10% of each other.
RESULTS
Whereas chicken green-sensitive cone pigment (chicken green) has an amino acid sequence more similar to rhodopsins than any other cone visual pigments (6, 28), it exhibits molecular properties similar to the other cone visual pigments but clearly different from those of rhodopsin. Therefore, we prepared site-directed mutants of rhodopsin (Q64K, E122Q, and E150A), each of which had the amino acid residue present at the indicated sites in chicken green.
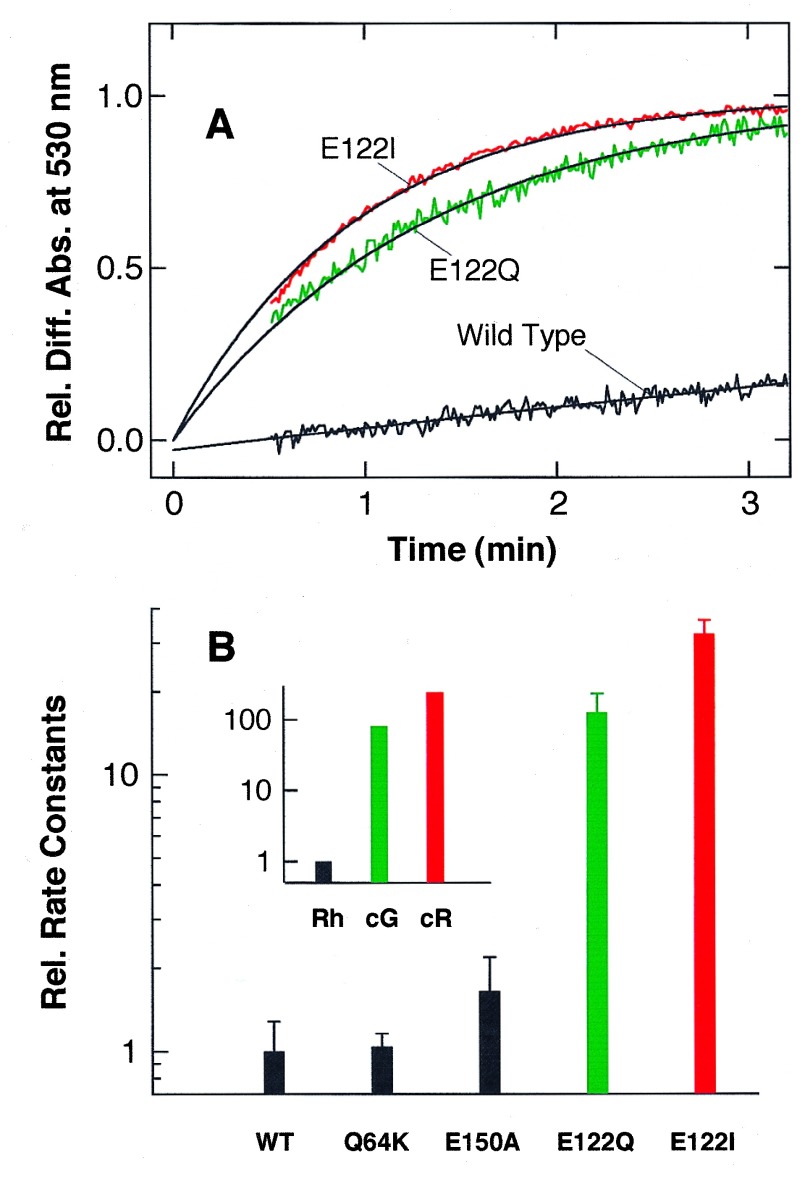
First, we investigated the regeneration of pigments from wild-type and mutants opsin with 11-cis-retinal (Fig. 2). Wild-type rhodopsin regenerates with a time constant of 20–30 min, and mutants Q64K and E150A show regeneration rates similar to that of wild-type rhodopsin. Interestingly, mutant E122Q regenerates much faster than the wild-type rhodopsin, suggesting that amino acid residue at position 122 regulates the regeneration rate of visual pigment. To further investigate the role of amino acid residue at position 122, we prepared a rhodopsin mutant (E122I) in which Glu-122 of rhodopsin was replaced with Ile, to mimic chicken red-sensitive cone pigment (chicken red). To our surprise, mutant E122I regenerates with a time constant significantly faster than not only wild-type rhodopsin but also the E122Q mutant, and the relationship correlates well with the rate among native rhodopsin, chicken green, and chicken red (Fig. 2B Inset). Although the relative increase in regeneration rate due to the mutations at position 122 versus wild-type is smaller than the relative difference between native rhodopsin and the cone pigments, these results highly suggest that amino acid residue at position 122 is one of the major determinants in the regulation of regeneration rate of visual pigment.
Figure 2.
Regeneration rates of wild-type and mutant rhodopsins. (A) Regeneration of wild-type, E122Q, and E122I rhodopsins monitored by change of absorbance at 530 nm. 11-cis-Retinal solution (2.5 nmol) in ethanol (5 μl) was added to the respective opsin solution (220 μl) at 2°C, and increase of absorbance at 530 nm due to the regeneration of pigment was recorded. The maximal absorbance due to the full regeneration is normalized. Solid curves are the fitted single exponential curves with time constants of 26 (wild type), 1.3 (E122Q), and 0.94 (E122I) min, respectively. (B) Rate constants of pigment regeneration in wild-type and mutant rhodopsins and native chicken rhodopsin (Rh), chicken green (cG), and chicken red (cR) (Inset). Rate constants of wild-type and native rhodopsins were normalized to 1, and the rate constants of mutant rhodopsins and native cone pigments are represented relative to those of their respective rhodopsins. The standard deviations were estimated from three independent experiments using different preparations.
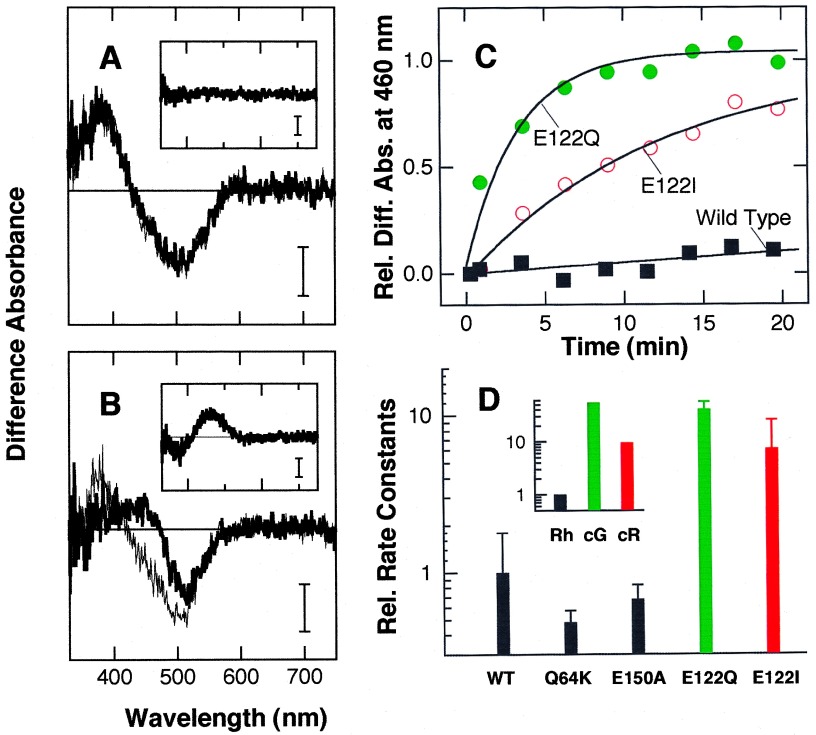
Next, we investigated thermal decay processes of the meta II intermediate of rhodopsin mutants (Fig. 3). Irradiation of the wild-type rhodopsin results in the formation of a meta II with an absorption maximum of ≈380 nm (Fig. 3A). Like meta II produced from native rhodopsin, the meta II from wild-type rhodopsin is stable on the time scale of 10–40 min (Fig. 3A Inset). While mutants Q64K and E150A (Fig. 3D) show meta II stabilities similar to that of wild-type rhodopsin, meta II from mutant E122Q decays to the next meta III intermediate within several minutes (Fig. 3B). Interestingly, the slower decay of chicken red meta II versus chicken green meta II (Fig. 3D Inset) is also reproduced when Glu-122 is replaced by Ile (chicken red) instead of Gln (chicken green).
Figure 3.
Thermal reactions of meta II of wild-type and mutant rhodopsin. (A and B) Formation and decay of meta II monitored by change in the absorption spectrum. Wild-type (A) and E122Q (B) rhodopsins were irradiated with orange light for 30 s at 2°C, followed by continuous recording of the absorption spectra. The thin and bold lines in each panel represent the difference spectra obtained by subtracting the spectra before irradiation from those immediately after and 20 min after irradiation, respectively. The spectra shown in the Insets are the difference spectra between thin and bold lines. (Bars = 0.003 absorbance unit.) (C) Course of conversion from meta II to meta III in wild-type, E122Q, and E122I rhodopsins. Increase in absorbance at 460 nm due to the formation of meta III from meta II is plotted as a function of incubation time after the irradiation. Solid curves are the fitted single exponential curves with the time constants of 190 (wild type), 3.3 (E122Q), and 12 (E122I) min, respectively. (D) Rate constants of meta II decay in wild-type and mutant rhodopsins, and native chicken rhodopsin (Rh), chicken green (cG), and chicken red (cR) (Inset). Rate constants of wild-type and native rhodopsins were normalized to 1, and the rate constants of mutant rhodopsins and native cone pigments are represented relative to those of their respective rhodopsins. The standard deviations were estimated from three independent experiments using different preparations.
Above results highly suggested that the amino acid residue at position 122 regulates the molecular properties of visual pigments. This is also evidenced by the fact that the triple mutant Q64K/E122Q/E150A exhibits regeneration and meta II decay rates similar to those of the single mutant E122Q. To further confirm the effect of the replacement at the position, we have prepared Q122E mutant of chicken green in which Gln-122 is replaced by Glu and investigated its molecular properties. The results showed that the regeneration rate of Q122E mutant is >10 times slower than that of wild-type chicken green, and the meta II decay is >5 times slower than that of wild-type chicken green. However, low expression yields and unstable properties of the expressed cone opsin and its mutant hampered the estimation of the quantitative differences. Nevertheless, our results suggested that replacement of amino acid residue at position 122 in chicken green converts its properties to those of rhodopsin.
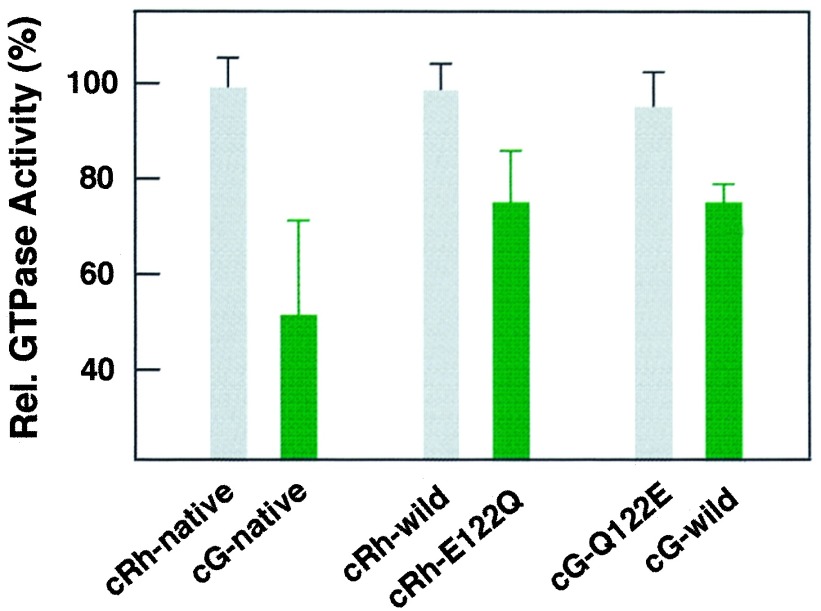
Since it has been well known that meta II of rhodopsin and chicken red activate retinal G protein transducin (30, 33), the thermal stability of the intermediate would influence the efficiency in activating transducin. Thus, to confirm the role of amino acid residue at position 122 on the activation efficiency in transducin activation, we have investigated the efficiency in transducin activation by examining its GTPase activity as a function of incubation time before addition of the irradiated visual pigments into the reaction mixtures containing transducin and GTP. The absolute GTPase activities were similar when both pigments were irradiated in the presence of transducin and GTP, suggesting that catalytic turnover rates in transducin activation were similar in both pigments. However, the transducin activation by the irradiated cone pigments is greatly diminished, as the preincubation time increases, while activation by irradiated rhodopsin is not. In Fig. 4, the relative activities of transducin, which were induced by addition of pigments 6 min after the irradiation, were compared. Whereas the activities of native and wild-type rhodopsin hardly changed, those of native chicken green and its corresponding mutant E122Q were significantly reduced. Furthermore, a reverse mutant Q122E of chicken green rescues a decrease of transducin activity shown by the wild-type chicken green. Therefore, we concluded that amino acid residue at the position 122 changes the visual pigment’s efficiency in transducing signals to transducin as well as the molecular properties of visual pigments.
Figure 4.
Change in the activation of transducin by native, wild-type, and mutant rhodopsin and chicken green. The pigments were added to the reaction mixture containing transducin 6 min after irradiation, and the extents of GTPase activity were measured. The relative activities to those immediately after irradiation were plotted. The standard deviations were estimated from four independent experiments using different preparations.
DISCUSSION
In the present study, we have replaced amino acid residues present in rod and cone visual pigments to identify the amino acid residue(s) responsible for the difference in molecular properties between rod and cone visual pigments. The results showed that, among the three amino acid positions where amino acid residues of rod and cone visual pigments differ in their electrical properties, only the replacement of amino acid residue at position 122 induced dramatic changes of the molecular properties. The replacement changes the different types of molecular properties, namely, regeneration rates of the pigments, the thermal stability of the meta II intermediate, and the efficiency of transducin activation in a time-dependent manner. Thus, it is clear that the amino acid residue at position 122 is one of the major determinants of the molecular properties of visual pigments.
To examine whether it is the loss of Glu or the gain of Gln or Ile that is responsible for the observed changes, we replaced Glu-122 of rhodopsin by Asp. The replacement caused no acceleration of the regeneration rate or of the meta II decay rate, but rather the mutant showed relatively slower (≈2 times slower) rates. These results highly suggest that the presence of carboxyl group at the suitable position is indispensable for the molecular properties of rhodopsin that are different from those of cone visual pigments.
Complete conversions of the molecular properties by the replacement of amino acid residues were not achieved in these experiments. This might be due to the experimental limitations, because we expressed mutant visual pigments in cultured cells where glycosylation different from that in native photoreceptor cells took place (25, 34). However, the possible presence of the other residue(s) that may also regulate the molecular properties of visual pigments is not excluded, although it is hard to pinpoint the residue(s) from the sequence homology between rod and cone visual pigments. The relative increase in regeneration rate due to the mutations at position 122 versus wild type is smaller than the relative difference between the rates of native rhodopsin and the cone pigments. This might be one of the indications that the other residue(s) are also responsible for the fast regeneration of cone visual pigments.
Our results showed that the amino acid residue responsible for the differences in molecular properties between rod and cone visual pigments is situated in the transmembrane region of the proteins. Since position 122 is in between the retinylidene chromophore and the site for transducin activation (cytoplasmic surface), it is reasonable to speculate that the amino acid residue at this position plays the role of regulating the intramolecular signal transduction in visual pigments. In fact, site-directed mutagenesis experiments on bovine rhodopsin showed that the replacement of Glu-122 causes a shift in meta I–meta II equilibrium (25, 27). In addition, Fourier transform infrared spectroscopy of rhodopsin mutants suggested environmental changes of this residue occurred upon formation of metarhodopsin II, although its protonation state was unchanged (26, 27). These results also suggest that the carboxyl group of the Glu-122 forms a hydrogen-bonding network with nearby amino acid residue(s) and/or the peptide backbone. Thus, the absence of Glu-122 in cone visual pigments causes the formation of a hydrogen-bonding network system different from that in rhodopsin and results in faster regeneration and faster decay of meta II intermediates.
In addition to Glu-122, His-211 is also present in all the rhodopsins and its functional role has been highlighted for many years (25, 35, 36). However, chicken green has a histidine residue at the position equivalent to 211 in rhodopsin, but it exhibits molecular properties clearly different from those of rhodopsin. Furthermore, our recent investigation showed that the replacement of His-211 in rhodopsin by cysteine caused no acceleration of regeneration and meta II decay (H.I., N. Tamai, and Y.S., unpublished work). These results suggest that the histidine residue might not act by itself in discriminating the molecular properties between rod and cone visual pigments. On the other hand, site-directed mutagenesis experiments of rhodopsin clearly showed the importance of the histidine on the energetics of meta I-to-meta II transition (25, 35), although it was reported that the replacement of histidine caused little effect on the transducin activation (36). Thus, the role of histidine might be limited in rhodopsin, or it may act in an indirect manner through a hydrogen-bonding network system in visual pigments. If one can assume that the histidine residue might interact with Glu-122 and might support the role of Glu-122, it could explain why rhodopsins have diverged from the group that includes chicken green (see below).
As already described, the role of the amino acid residue at position 122 is to regulate the intramolecular signal transduction in visual pigments. Its role is in marked contrast to the roles of Lys-296 and Glu-113, which are essential for the basic function (photoreception) of the vertebrate visual pigments. Specifically, these residues are responsible for chromophore binding (16, 37, 38) and its stabilization as a counterion (22, 23, 39), respectively. The role of the amino acid residue at position 122 is relatively similar to the roles of the amino acid residues that tune the spectral sensitivity of visual pigments and thus form a molecular basis of color vision (10). From a phylogenetic tree based on the amino acid sequences, it has been suggested that rhodopsin has evolved from a cone visual pigment (6). Therefore, acquirement of the Glu-122 might have caused the molecular evolution of cone visual pigments into rhodopsin, one of the key steps in the divergence into both daylight and twilight vision (visual duplicity).
Acknowledgments
We thank Prof. J. Nathans for his gift of 293S cell line and Prof. F. Tokunaga for providing a pUSRα expression vector and experimental guidance. We also thank Profs. T. Yoshizawa, A. Maeda, and Y. Fukada and Drs. H. Kandori, Y. Imamoto, and T. Okano for their valuable discussions. This work was supported in part by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Science, Sports and Culture, a grant from Takeda Science Foundation, and Japan Society for the Promotion of Science Research Fellowships for Young Scientists.
Footnotes
References
- 1.Yau K-W. Invest Opthal Visual Sci. 1994;35:9–32. [PubMed] [Google Scholar]
- 2.Baylor D. Proc Natl Acad Sci USA. 1996;93:560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hargrave P A, McDowell J H. FASEB J. 1992;6:2323–2331. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]
- 4.Wald G. Science. 1968;162:230–239. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 5.Nathans J, Thomas D, Hogness D S. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 6.Okano T, Kojima D, Fukada Y, Shichida Y, Yoshizawa T. Proc Natl Acad Sci USA. 1992;89:5932–5936. doi: 10.1073/pnas.89.13.5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizawa T. Photochem Photobiol. 1992;56:859–867. doi: 10.1111/j.1751-1097.1992.tb09707.x. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R L, Grant K B, Zankel T C, Boehm M F, Merbs S L, Nathans J, Nakanishi K. Biochemistry. 1993;32:208–214. doi: 10.1021/bi00052a027. [DOI] [PubMed] [Google Scholar]
- 9.Hisatomi O, Kayada S, Aoki Y, Iwasa T, Tokunaga F. Vision Res. 1994;34:3097–3102. doi: 10.1016/0042-6989(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs G H. Proc Natl Acad Sci USA. 1996;93:577–581. doi: 10.1073/pnas.93.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shichida Y, Imai H, Imamoto Y, Fukada Y, Yoshizawa T. Biochemistry. 1994;33:9040–9044. doi: 10.1021/bi00197a002. [DOI] [PubMed] [Google Scholar]
- 12.Wald G, Brown P K, Smith P H. J Gen Physiol. 1955;38:623–681. doi: 10.1085/jgp.38.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilden U, Hall S W, Kühn H. Proc Natl Acad Sci USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langlois G, Chen C-K, Palczewski K, Hurley J, Vuong T M. Proc Natl Acad Sci USA. 1996;93:4677–4682. doi: 10.1073/pnas.93.10.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai H, Imamoto Y, Yoshizawa T, Shichida Y. Biochemistry. 1995;34:10525–10531. doi: 10.1021/bi00033a026. [DOI] [PubMed] [Google Scholar]
- 16.Hargrave P A, McDowell J H, Curtis D R, Wang J K, Juszczak E, Fong S-L, Mohanna Rao J K, Argos P. Biophys Struct Mech. 1983;9:235–244. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]
- 17.Hisatomi O, Iwasa T, Tokunaga F, Yasui A. Biochem Biophys Res Commun. 1991;174:1125–1132. doi: 10.1016/0006-291x(91)91537-m. [DOI] [PubMed] [Google Scholar]
- 18.Kojima D, Oura T, Hisatomi O, Tokunaga F, Fukada Y, Yoshizawa T, Shichida Y. Biochemistry. 1996;35:2625–2629. doi: 10.1021/bi9511548. [DOI] [PubMed] [Google Scholar]
- 19.Kayada S, Hisatomi O, Tokunaga F. Comp Biochem Physiol. 1995;110:599–604. doi: 10.1016/0305-0491(94)00179-x. [DOI] [PubMed] [Google Scholar]
- 20.Gorman C M, Gies D R, McCray G. DNA Protein Eng Tech. 1990;2:3–10. [Google Scholar]
- 21.Nathans J. Biochemistry. 1990;29:937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- 22.Zhukovsky E A, Oprian D D. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 23.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama T A, Khorana H G. J Biol Chem. 1991;266:4269–4275. [PubMed] [Google Scholar]
- 25.Weitz C J, Nathans J. Biochemistry. 1993;32:14176–14182. doi: 10.1021/bi00214a016. [DOI] [PubMed] [Google Scholar]
- 26.Fahmy K, Jager F, Beck M, Zvyaga T A, Sakmar T P, Siebert F. Proc Natl Acad Sci USA. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeCaluwe G L J, Bovee-Geurts P H M, Rath P, Rothschild K J, DeGrip W J. Biophys Chem. 1995;56:79–87. doi: 10.1016/0301-4622(95)00018-s. [DOI] [PubMed] [Google Scholar]
- 28.Wang S-Z, Adler R, Nathans J. Biochemistry. 1992;31:3309–3315. doi: 10.1021/bi00128a002. [DOI] [PubMed] [Google Scholar]
- 29.Imai H, Mizukami T, Imamoto Y, Shichida Y. Biochemistry. 1994;33:14351–14358. doi: 10.1021/bi00251a049. [DOI] [PubMed] [Google Scholar]
- 30.Okada T, Matsuda T, Kandori H, Fukada Y, Yoshizawa T, Shichida Y. Biochemistry. 1994;33:4940–4946. doi: 10.1021/bi00182a024. [DOI] [PubMed] [Google Scholar]
- 31.Okano T, Fukada Y, Artamonov I D, Yoshizawa T. Biochemistry. 1989;28:8848–8856. doi: 10.1021/bi00448a025. [DOI] [PubMed] [Google Scholar]
- 32.Fukada Y, Ohguro H, Saito T, Yoshizawa T, Akimo T. J Biol Chem. 1989;264:5937–5943. [PubMed] [Google Scholar]
- 33.Stryer L, Hurley J B, Fung B K-K. Curr Top Membr Transp. 1981;15:93–108. [Google Scholar]
- 34.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weitz C J, Nathans J. Neuron. 1993;8:465–472. doi: 10.1016/0896-6273(92)90274-h. [DOI] [PubMed] [Google Scholar]
- 36.Cohen G B, Oprian D D, Robinson P R. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 37.Ovchinnikov Y A, Abdulaev N G, Feigina M Y, Artamonov I D, Bogachuk A S, Eganyan E R, Kostetskii P V. Bioorg Khim. 1983;9:1331–1340. [PubMed] [Google Scholar]
- 38.Nathans J, Hogness T. Cell. 1983;34:807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- 39.Nathans J. Biochemistry. 1990;29:9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]