Abstract
Thrombopoietin (TPO) acts through its receptor, Mpl, to stimulate the proliferation and maturation of megakaryocytes and their progenitors. The Mpl cytoplasmic domain controls this process through assembly of an active signaling complex using various receptor docking sites. In this report, eight carboxyl truncations of the 121-aa murine Mpl cytoplasmic domain were tested for the ability to support growth of a cytokine-dependent cell line (Ba/F3) and for their capacity to induce TPO-stimulated tyrosine phosphorylation of specific signaling proteins. Point mutations of the five tyrosine residues in the cytoplasmic domain of the receptor were subsequently used to confirm our conclusions. From these studies we demonstrate that: (i) TPO-induced proliferation is moderately reduced by truncation of as many as 53 C-terminal amino acids of Mpl, including the sites of receptor tyrosine phosphorylation; (ii) truncation/mutation of residues 69–83 of the Mpl cytoplasmic domain enhances proliferative signaling, perhaps mediated by a decrease in receptor-driven cellular differentiation; (iii) Mpl can be phosphorylated at either Y112 or Y117 but not at the three proximal cytoplasmic tyrosine residues (Y8, Y29, and Y78); (iv) Y112 of Mpl is necessary for tyrosine phosphorylation of Shc and Shc-associated p145 (SHIP); and (v) unlike STAT3, STAT5 is partially phosphorylated in the absence of any tyrosine residues in the Mpl cytoplasmic domain. These studies identify subdomains of Mpl necessary for activation of several critical signaling pathways and point to two potentially novel mechanisms of TPO-induced signal transduction, an indirect pathway to STAT5 activation and a differentiation domain that acts by limiting proliferation.
Keywords: tyrosine phosphorylation, thrombopoietin, signal transduction, JAK, STAT
The cloning of thrombopoietin (TPO) and its receptor, Mpl, has opened promising avenues toward understanding the molecular basis of megakaryocyte development. In the presence of TPO, multipotential hematopoietic cells undergo proliferation and maturation along the megakaryocyte lineage, characterized by nuclear endoreduplication, cytoplasmic expansion, display of lineage-restricted surface proteins, and platelet production (1, 2). This programmed response is initiated by the interaction of TPO and Mpl and is coordinated by the cytoplasmic domain of the receptor.
Like the receptors for growth hormone, erythropoietin, and granulocyte colony-stimulating factor, Mpl is believed to be activated through homodimerization, independent of the three receptor subunits shared by other cytokines [i.e., βc, gp130, and interleukin (IL)-2 γ chain; refs. 3–5 and unpublished results]. The cytoplasmic domain of murine Mpl contains 121 aa, including two short, membrane-proximal motifs (box1 and box2) that are conserved among most members of the hematopoietic cytokine receptor superfamily and are critical for all aspects of receptor function (6–8). Box1, identified by two spatially conserved proline residues (PXXP), is located between cytoplasmic residues 17 and 20 of Mpl. Box2 is more imprecisely defined by a region with increased serine and glutamic acid content that extends between residues 46 and 64 and by a proposed core motif between residues 52 and 61 (9). Bénit et al. (9) have demonstrated that deletion of either box1 or box2 abolished the transforming potential of v-mpl. Similarly, Gurney et al. (10) have shown that loss of either the Mpl box1 or box2 motifs blocks both activation of the Janus kinases (JAKs) and cellular proliferation.
The Mpl cytoplasmic region does not contain any recognizable enzymatic motif, yet through the recruitment of nonreceptor tyrosine kinases, it promotes rapid tyrosine phosphorylation of numerous intracellular proteins after TPO stimulation. In previous reports, we and others (11–21) have demonstrated that TPO induces rapid tyrosine phosphorylation of Mpl, Shc, SHIP, JAK2, TYK2, STAT3, and STAT5. For the JAKs and STAT family of latent transcription factors, tyrosine phosphorylation is essential for activation. Mpl and Shc acquire phosphotyrosine residues that function as potential binding sites for proteins that contain Src homology 2 (SH2) or phosphotyrosine-binding domains. Differences within the SH2 domains of distinct molecules (e.g., STATs) direct binding to specific phosphotyrosine residues, depending on the adjacent amino acids (22–24). Analysis of the erythropoietin receptor has identified the individual tyrosine residues that recruit STAT5 to the signaling complex (25–27).
The carboxyl portion of the Mpl cytoplasmic domain (residues 70–121) shares little homology with other cytokine receptors, whose length and primary sequence vary tremendously. Several reports have begun to explore the function of this receptor subdomain. Baumann et al. (28) showed that Mpl does not contain a C-terminal box3 motif, which is required for lineage-specific gene expression in the G-CSF, gp130, and leukemia inhibitory factor receptors (28, 29). Gurney et al. (10) demonstrated that truncation of the final 20 aa of the Mpl receptor abrogated Shc phosphorylation and c-fos mRNA induction. Hill et al. (30) showed that a dominant-negative form of the Shc adapter protein blocked TPO-induced myeloid differentiation of 32D-mpl cells. Shc, which contains multiple protein-interactive domains, is believed to be involved in activation of the Ras pathway by simultaneously interacting with several signaling molecules, such as Grb2, SOS, and the recently cloned 145-kDa inositol phosphatase, SHIP (31–33). Finally, Porteu et al. (34) reported that amino acids 71–94 of the Mpl signaling domain, just downstream of box2, were required for megakaryocytic differentiation.
In the present study, eight truncation mutations of murine Mpl were used to identify important regions for TPO-induced proliferation and signaling. Receptor point mutations of specific tyrosine (Y) residues to phenylalanine (F) were then constructed and expressed in Ba/F3 cells to confirm that these residues are critical docking sites for the SH2-containing signaling proteins under study. Our results reveal that Y112 is critical for Shc and SHIP phosphorylation and that Y112 and Y117 are both involved in Mpl and STAT3 phosphorylation. However, STAT5 is partially phosphorylated in response to TPO, even in cells bearing mutant receptors that contain no cytoplasmic tyrosines, suggesting potentially novel mechanisms of receptor-mediated signaling. In addition, deletion or mutation of residues 69–83, a region similar to the differentiation domain recognized by Porteu et al. (34), enhances Mpl-induced proliferation. Together, these experiments further our understanding of the signaling pathways employed by Mpl and pose interesting questions about the molecular mechanisms of TPO-induced megakaryocytopoiesis.
MATERIALS AND METHODS
Antibodies.
Polyclonal Mpl antiserum was raised against the extracytoplasmic domain of the murine Mpl protein and was provided by Zymogenetics. Monoclonal phosphotyrosine antibody (4G10), polyclonal Shc IgG, and JAK2 antiserum were purchased from Upstate Biotechnology (Lake Placid, NY) and were used for immunoprecipitation and Western blotting. STAT3 and TYK2 polyclonal IgG were purchased from Santa Cruz Biotechnology. STAT5B polyclonal antiserum was provided by Chris Saris (Amgen, Thousand Oaks, CA; ref. 16). SHIP was evaluated by coprecipitation with Shc.
Cell Lines.
Parental Ba/F3 cells were provided by Alan D’Andrea (Dana Farber Cancer Institute) and were maintained in Ba/F3 medium [RPMI with 10% heat-treated newborn calf serum (HyClone), l-glutamine (BioWhittaker), penicillin, streptomycin, fungizone (BioWhittaker), and 0.2% conditioned medium containing recombinant murine (m)IL-3]. Transfection with c-mpl cDNA was achieved by electroporation of parental Ba/F3 cells in the presence of 20–30 μg of the appropriate linearized plasmid. After 24 hr, the cells were collected by centrifugation and resuspended in Ba/F3 medium with 1000 μg/ml synthetic neomycin (Geneticin; Life Technologies). Individual clones were isolated through limiting dilution in 96-well plates.
Receptor Constructs.
Native murine c-mpl cDNA was provided by Zymogenetics. The expression vector (pHZ-1) uses a constitutive, truncated metallothionine promoter for Mpl expression and has a neomycin resistance gene, driven by the SV40 promoter. Truncated receptors were generated by PCR mutagenesis, incorporating an XhoI cloning site and termination codon at the desired position. Point mutations were created by two methods: (i) complementary oligonucleotides spanning the site of mutation were annealed and ligated into existing restriction sites of the c-mpl cDNA; or (ii) the high-fidelity Pfu DNA polymerase (Stratagene) was used to introduce new site-directed mutations. Each plasmid was sequenced to confirm PCR fidelity and accurate mutagenesis (Prism sequencing kit; Applied Biosystems).
Proliferation Assay.
Cells in logarithmic-phase growth were washed twice in Ba/F3 medium without exogenous cytokines. Equal numbers of cells from multiple clones bearing the same Mpl construct were combined before each experiment. The cytokine-free cell suspension was divided into 96-well plates, with 15,000–20,000 cells per well. Various concentrations of recombinant murine TPO (Zymogenetics) or mIL-3 (conditioned supernatant or pure protein; Genzyme) were added. After 36–48 hr at 37°C, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma) was added, at 1 mg/ml, and incubated for 5 hr. The formazan granules were solubilized by adding 100 μl of MTT lysis buffer (40% dimethylformamide/2% glacial acetic acid/20% sodium dodecyl sulfate/0.215% concentrated hydrochloric acid) and absorbance (570 nm-630 nm) was determined on an ELISA plate reader. Each experiment was performed in triplicate. Proliferation was expressed as a percentage of maximal mIL-3-induced growth. Statistical significance was tested for the combined results of several experiments using two-factor analysis of variance with replications.
Preparation of Cellular Extracts.
Cells in logarithmic-phase growth were washed and resuspended in serum-free, cytokine-free medium for 16 hr. Samples were either unstimulated or exposed to TPO-containing medium (10–15 ng/ml) for 10 min. Lysis was performed in a 1% Triton X-100 buffer, 100 μl per 107 cells as previously described (11). Final protein concentrations were determined by modified Bradford assay (Bio-Rad).
Immunoprecipitations.
Cellular extracts (750 μg) were incubated with appropriate antiserum (2 μl) or IgG (1 μg) for 2 hr at 4°C. Immune complexes were collected for 1 hr with 20 μl of protein A-agarose (Santa Cruz Biotechnology). The beads were washed three times in lysis buffer and boiled in 30 μl of denaturation buffer containing 2-mercaptoethanol and sodium dodecyl sulfate (11).
Western Blots.
Immunoprecipitated proteins were analyzed on 7.5% polyacrylamide gels with prestained molecular weight markers. Transfer to nitrocellulose, blocking, probing with antibodies, and chemiluminescence were performed as previously described (35).
RESULTS
Design and Expression of Truncated Forms of Mpl.
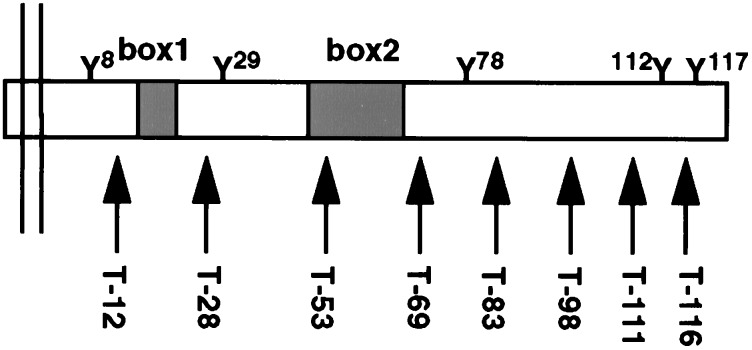
In addition to the putative box1 and box2 motifs, the Mpl cytoplasmic domain includes five tyrosine residues, each of which could potentially be phosphorylated and/or serve as a protein-binding site. Carboxyl truncations of this region were designed to test each of these subdomains for changes in function (i.e., proliferation) or tyrosine phosphorylation. To simplify the nomenclature of Mpl receptor mutants, amino acids are referred to based on their position within the 121-aa cytoplasmic domain, and truncations are named for the number of residues that remain (i.e., T-12 contains only 12 intracellular amino acids; Fig. 1). For each mutant receptor, multiple cellular clones were isolated and tested for Mpl expression by Western blot (data not shown). For all experiments, 3–6 distinct clones were mixed together to minimize the potential for clonal variation among engineered cell lines. However, in preliminary experiments, no significant differences were observed between individual clones and the mixed populations (data not shown).
Figure 1.
Cytoplasmic domain of Mpl is represented with cell membrane on the left and C terminus on the right. The five cytoplasmic tyrosine residues (Y), putative box1 and box2 motifs, and the location of the receptor truncations (arrows) are indicated. The truncations indicate the number of cytoplasmic residues that remain.
Truncation of the Mpl C Terminus and the Proliferative Response.
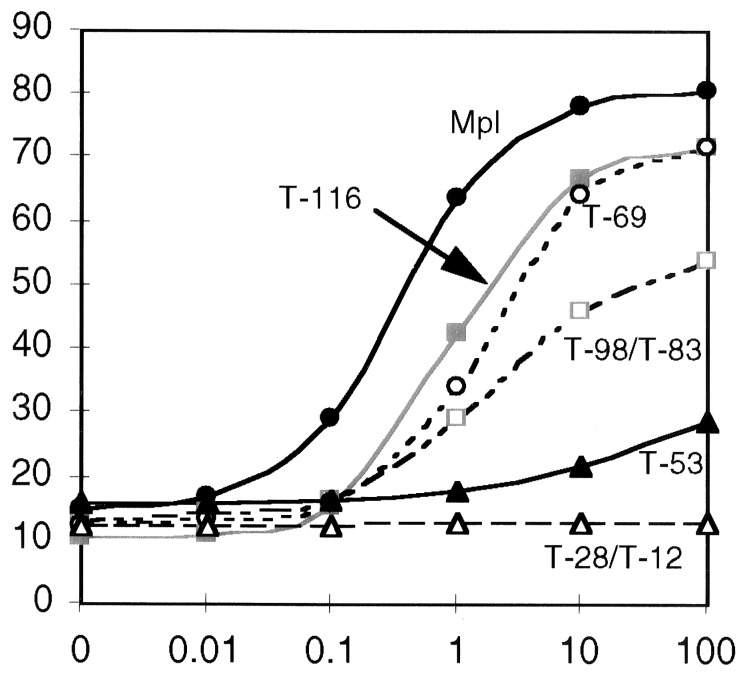
In four separate experiments, cells expressing different receptor constructs were grown in a range of TPO concentrations (0–100 ng/ml) as well as in a concentration of mIL-3 that supports optimal proliferation. Results of MTT proliferation assays are expressed as the mean of triplicate values, corrected for the maximal, mIL-3-induced proliferation (Fig. 2). The full-length Mpl was most sensitive to low cytokine concentrations, with half-maximal proliferation in ≈0.2 ng/ml TPO, and it achieved the highest peak proliferation (≈80% that of mIL-3) at TPO concentrations of 10–100 ng/ml. At higher concentrations (1–10 μg/ml TPO), proliferation was slightly diminished (data not shown), a finding also noted for another homodimeric receptor, growth hormone (36). C-terminal truncation of five residues (T-116) resulted in ≈5-fold decreased sensitivity to TPO (half-maximal proliferation in 1 ng/ml) and a modest decrease in peak proliferation. Both the T-98 and T-83 truncated receptors supported significantly lower proliferation at high TPO concentrations. Surprisingly, the shorter T-69 receptor displayed increased proliferation, comparable to the T-116 truncation. By analysis of variance, the difference between T-69 and T-83 was highly significant (P < 0.001) for TPO concentrations of 1 ng/ml or greater. These data led us to postulate that residues 69–83 might include a subdomain that inhibited TPO-induced proliferation, perhaps similar to the C-terminal inhibitory region of the erythropoietin receptor (37).
Figure 2.
MTT assay of the Mpl truncations. The abcissa shows TPO concentration (0–100 ng/ml), and ordinate indicates percentage of maximum IL-3-induced proliferation. For clarity, curves that are essentially overlapping are represented by a single line. Proliferation curves are shown for Ba/F3 cells bearing the following receptors: full-length Mpl (•), T-116 (▪), T-98 and T-83 (□), T-69 (○), T-53 (▴), and T-28 and T-12 (▵). The data represent the mean values of four separate experiments.
Substitution of Y112 and Y117 Decreases Proliferation.
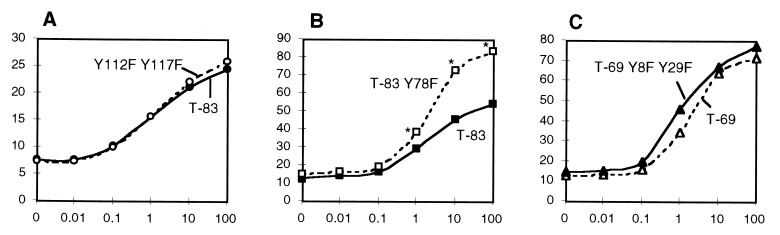
Because TPO-directed proliferation was partially inhibited by C-terminal truncations of Mpl, we hypothesized that the two cytoplasmic tyrosine residues within this region might play an important role in TPO signaling. Thus, we constructed a full-length receptor with each of the distal tyrosine residues replaced by phenylalanine (Y112F Y117F). Cells expressing this altered receptor responded to TPO in a manner similar to T-83 cells (P > 0.05 for all data points), suggesting that loss of these residues is responsible for the observed decrease in proliferation during truncation of the C terminus (Fig. 3A).
Figure 3.
Effect of tyrosine substitution on proliferation. For each assay, the abcissa shows TPO concentration (ng/ml), and the ordinate shows the percentage of maximal IL-3-induced proliferation. (A) MTT assay comparing T-83 (•) and full-length Mpl with two point mutations, Y112F Y117F (○). The curves represent combined results of two experiments. (B) Comparison of T-83 (▪) and T-83 Y78F (□). Combined results of three experiments are depicted; asterisks indicate significant difference between curves (P < 10−6). (C) Comparison of T-69 (▵) and T-69 Y8F Y29F (▴). Combined results of three experiments are shown. (Data in A was obtained in different experiments from those in panels B and C).
Point Mutation of Y78 Reveals a Potential Negative Regulatory Domain.
In view of the increased proliferation of T-69 compared with T-83 (see above; Fig. 2), a point mutation was introduced into Y78 (Y78F), the only tyrosine residue within this region. When compared with T-83 in three separate experiments, the T-83 Y78F receptor substitution resulted in a significant increase in maximal proliferation (Fig. 3B). Analysis of variance confirmed that this difference was significant for TPO concentrations of 1 ng/ml or greater. In contrast, mutation of both of the membrane proximal tyrosine residues (Y8 and Y29) in a T-69 receptor did not alter the proliferative response to TPO (Fig. 3C).
Box2 Is Critical for TPO-Stimulated Proliferation.
Truncation of the Mpl receptor after 53 cytoplasmic residues (T-53) resulted in 100-fold decrease in TPO sensitivity and severely blunted the proliferative response in high cytokine concentrations (Fig. 2). The cytoplasmic amino acids between positions 53 and 69 include much of the core box2 motif (LLEILPK) as defined by Bénit et al. (9). However, the exact size and boundaries of the box2 domain are not known. An Mpl receptor with only 28 cytoplasmic residues (T-28) failed to support any proliferation, despite the presence of a complete box1 motif. Further truncation of the receptor, deleting both box1 and box2 (T-12), also abrogated all TPO-induced proliferation, even at high hormone concentrations.
Tyrosine Phosphorylation of Mpl, Shc, SHIP, and STAT3 Requires the 10 C-Terminal Residues of Mpl.
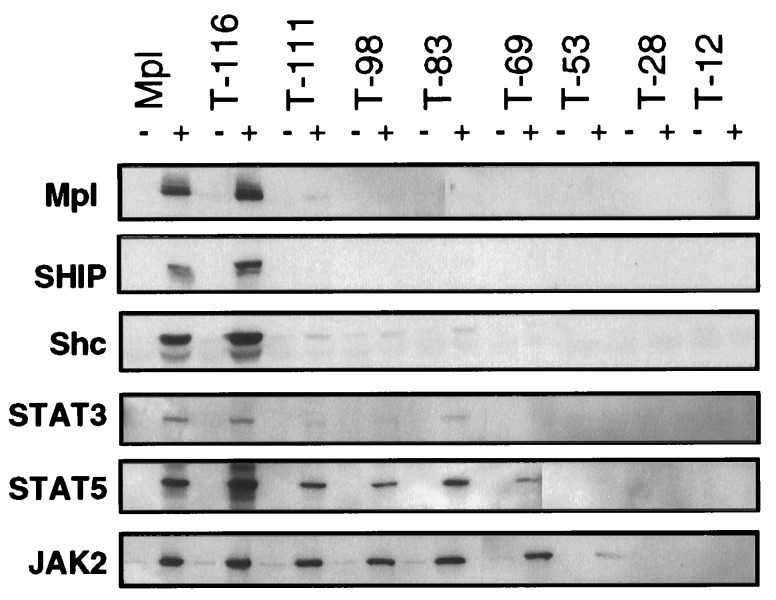
We next determined which regions of the Mpl cytoplasmic domain were necessary for tyrosine phosphorylation of specific intracellular proteins. Protein extracts were generated from Ba/F3 cells expressing each of the Mpl constructs both before and after TPO stimulation. Immunoprecipitations of specific signaling molecules were analyzed by Western blot for TPO-induced changes in phosphotyrosine content (Fig. 4). Both the full-length Mpl and T-116 (lacking five C-terminal residues, including Y117) mediated TPO-dependent phosphorylation of Mpl, Shc, the Shc-associated p145 inositol phosphatase (SHIP), STAT3, STAT5, and JAK2. The T-111 truncation, lacking 10 C-terminal residues including Y112 and Y117, abolished essentially all tyrosine phosphorylation of Mpl, SHIP, Shc, and STAT3 (at long exposures, a small amount of STAT3 phosphorylation could still be induced by T-98 and T-83 receptors). In contrast, JAK2 phosphorylation was unaffected, and STAT5 phosphorylation was only partially reduced.
Figure 4.
TPO-induced tyrosine phosphorylation of Mpl, SHIP, Shc, STAT3, STAT5, and JAK2. The proteins indicated on the left were immunoprecipitated from cellular lysates of Ba/F3 cells bearing either the full-length receptor (Mpl) or one of the truncations. Paired lanes indicate samples that had been unstimulated (−) or TPO-exposed (+). The Western blots were probed with a phosphotyrosine-specific antibody. Blots were stripped and reprobed to ensure equal protein in each lane (data not shown).
Y112 of Mpl Is Essential for Shc and SHIP phosphorylation.
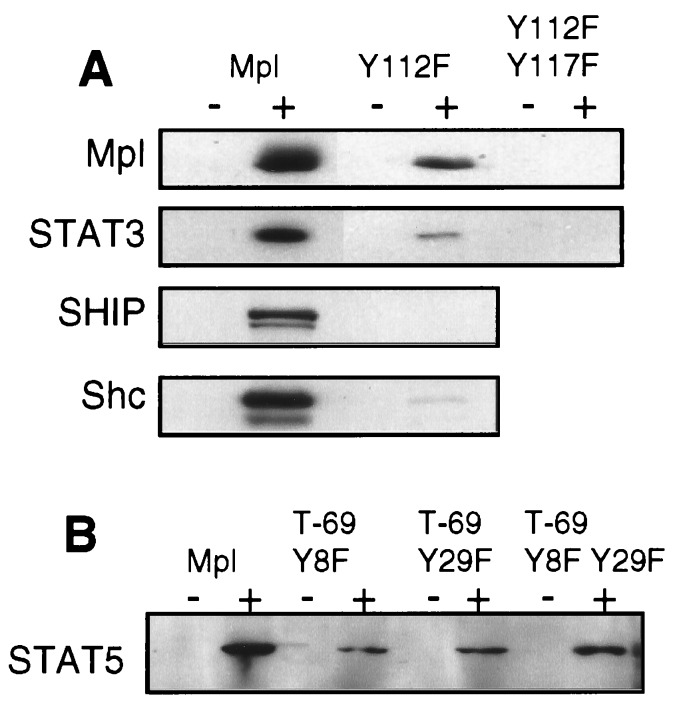
The differences between T-116 and T-111 truncations of Mpl led us to hypothesize that Y112 was a probable site of Mpl phosphorylation and a recruitment site for several proteins. To directly test this premise, a Y112F point mutation was introduced in the full-length c-mpl cDNA and expressed in Ba/F3 cells. This point mutation abolished Shc and coprecipitated SHIP phosphorylation and substantially reduced TPO-dependent tyrosine phosphorylation of Mpl and STAT3 (Fig. 5A). A full-length receptor with mutation of both distal tyrosine residues (Y112F Y117F) eliminated essentially all Mpl and STAT3 phosphorylation, indicating that each of these residues can be phosphorylated and may participate in STAT recruitment (Fig. 5A).
Figure 5.
(A) Effect of Y112F or Y112F Y117F substitution on tyrosine phosphorylation. Lysates from cells expressing either the wild-type receptor (Mpl), the Y112F substitution mutant, or the Y112F Y117F double mutation were used to compare TPO-induced tyrosine phosphorylation of Mpl, STAT3, SHIP, and Shc. (B) TPO-induced STAT5 phosphorylation was compared in cells containing the full-length receptor (Mpl) or a truncated receptor in which one or both of the cytoplasmic tyrosine residues had been replaced by phenylalanine. Paired lanes indicate samples that had been unstimulated (−) or TPO-exposed (+). The Western blot was probed with a phosphotyrosine-specific antibody. Blots were stripped and reprobed to ensure equal protein in each lane (data not shown).
Box2 Is Essential for Tyrosine Phosphorylation of JAK2 and STAT5.
Unlike the molecules discussed above, tyrosine phosphorylation of JAK2 and STAT5 persisted in all truncated receptors until deletion of the putative box2 motif (T-53). In this construct, STAT5 phosphorylation was abolished, and phosphorylation of the JAK2 was greatly reduced but still detectable (Fig. 4). Similar results were obtained for TYK2 phosphorylation (data not shown). Further cytoplasmic truncation (T-28 and T-12) resulted in no detectable TPO-induced tyrosine phosphorylation. By comparing JAK2 phosphorylation with proliferation (Fig. 2), we found these events to be closely related.
STAT5 Is Tyrosine-Phosphorylated in the Absence of Mpl Cytoplasmic Tyrosine Residues.
In the truncated receptors, loss of STAT5 phosphorylation could not be distinguished from loss of JAK2 activation. Because STATs are known to bind receptor phosphotyrosine residues via their SH2 domains, we hypothesized that the two membrane-proximal tyrosines (Y8 and Y29) might recruit STAT5 to the Mpl receptor. Therefore, point mutations of these residues were constructed in the context of the T-69 truncated receptor, which lacks the three distal tyrosine residues. Somewhat surprisingly, whether the tyrosines were mutated singly (T-69 Y8F and T-69 Y29F) or together (T-69 Y8F Y29F), cells expressing these mutant receptors were capable of partial TPO-dependent STAT5 tyrosine phosphorylation (Fig. 5B).
DISCUSSION
In this report, we identify several important functional subdomains of the Mpl receptor, exploring both the capacity of various receptor mutations to support cellular proliferation as well as tyrosine phosphorylation of molecules involved in signaling pathways. Our results indicate that the box2 motif is absolutely required for the phosphorylation of JAKs and for proliferation and that Y78 lies within a subdomain of Mpl that inhibits proliferation. In addition, we found that the C terminus of the cytoplasmic domain includes two sites of receptor phosphorylation (Y112 and Y117), which are necessary for STAT3 and Shc phosphorylation. Finally, TPO-induced STAT5 activation can occur by a mechanism that does not require cytoplasmic tyrosine residues of Mpl.
Within the cytokine receptor superfamily, the membrane-proximal box1 and box2 elements are usually required for both proliferation and JAK activation. Two reports have already demonstrated that point mutations or deletions of box1 abrogate the oncogenic potential of v-mpl and prevent JAK activation by Mpl (9, 10). However, the exact position and role of the Mpl box2 motif has been less clearly defined. From our experiments, we conclude that box2 is absolutely essential for Mpl-induced proliferation because a receptor that contains all of box1 but none of box2 (T-28) is completely inactive. Functionally, box2 does not extend distal to cytoplasmic residue 69, because the T-69 receptor supports full JAK2 phosphorylation, comparable to full-length Mpl. In addition, box2 must extend proximal to cytoplasmic residue 53 because the T-53 receptor retains measurable TPO-dependent proliferation and JAK phosphorylation. This implies that residues 29 to 53 of the Mpl cytoplasmic domain include a crucial part of box2, sufficient for minimal JAK activation.
Although the membrane-proximal portion of receptor cytoplasmic domains is required for proliferation, subdomains have been recognized in the carboxyl portion of several receptors that modify the proliferative response. In the case of the erythropoietin receptor, a tyrosine phosphatase, SHPTP1, binds to the receptor at phosphotyrosine Y453 and dephosphorylates the erythropoietin signaling complex, limiting the stimulatory response (38). Mutations that affect the SHPTP1 binding site result in an erythropoietin receptor with increased proliferative potential (39). After TPO stimulation, Mu et al. (16) showed that SHPTP1 was not substantially phosphorylated. In the present report, we define a region between residues 69 and 83 of the Mpl cytoplasmic domain that partially inhibits proliferation, and, furthermore, we show that a point mutation of Y78 (the only tyrosine within this region) results in substantially increased proliferation of the T-83 truncated receptor. A similar region (residues 71–94) was recently shown to be necessary for TPO-induced megakaryocytic differentiation and inhibited proliferation of UT7-mpl cells (34). Differentiation and proliferation are often inversely related, but our results indicate that the inhibitory effect of this subdomain occurs even in cells (Ba/F3) that do not significantly differentiate. Although Y78 appears to be important for the inhibitory response, it is not a site of measurable Mpl phosphorylation, nor does it contain the SHPTP1 consensus binding motif (pY-Hy-X-Hy, where pY = phosphotyrosine and Hy = hydrophobic; ref. 23). Therefore, other, potentially novel signaling mechanism(s) must be invoked to explain these observations.
A central feature of cytokine signal transduction is tyrosine phosphorylation of the receptor itself, providing potential binding sites for SH2 domain- and phosphotyrosine-binding domain-containing proteins. Our results indicate that the Y112 and Y117 of the Mpl cytoplasmic domain, but none of the three proximal tyrosine residues, are detectably phosphorylated in response to TPO stimulation. Furthermore, we show by truncation and substitution of these residues that Y112 and Y117 contribute to but are not essential for proliferation. Thus, TPO-dependent proliferation occurs in the absence of receptor tyrosine phosphorylation. We demonstrate that Y117 is capable of partial STAT3 recruitment and phosphorylation, which is consistent with the fact that it is part of a STAT3 consensus binding sequence (YXXQ) as determined by Stahl et al. (24). The list of additional SH2 domain-containing proteins that could potentially interact with Y117 is quite extensive, and it is possible that future studies will identify additional molecules that bind at this site.
Tyr-112 of the Mpl cytoplasmic domain is both a site of receptor phosphorylation and a critical residue for the TPO-dependent phosphorylation of Shc, SHIP, and STAT3. This residue likely acts as the Shc recruitment site, because the surrounding sequence (NHSY) matches the Shc phosphotyrosine-binding binding motif (NXXY; refs. 40 and 41). SHIP could either interact directly with the receptor at Y112 via an intrinsic phosphotyrosine-binding domain, or it may be recruited to the signaling complex indirectly, through association with Shc. The role of Shc and SHIP in megakaryocytopoiesis is not yet clear. As for the insulin and epidermal growth factor receptors, Shc may provide a link between the Mpl receptor and Ras activation (31). Gurney et al. (10) demonstrated that truncation of the Mpl C terminus abrogated both Shc phosphorylation and c-fos induction, an event downstream of the Ras signaling pathway. Hill et al. (30) recently reported that a dominant-negative form of Shc blocked TPO-directed differentiation. However, the cell line used for these experiments (32D-Mpl) was of myeloid origin, and the observed differentiation was along the myelomonocytic pathway. Our results suggest that Shc and SHIP may contribute to, but are not essential for, TPO-induced proliferation.
Tyr-112 also contributes to STAT3 tyrosine phosphorylation and plays a partial role in STAT5 phosphorylation. Thus, two signaling pathways (Shc-RAS and JAK-STAT) appear to use the same residue of Mpl. Because Ba/F3-Mpl cells cannot be induced toward megakaryocytic maturation, we are unable to assess whether or not Y112F substitution affects differentiation. Further investigation of the roles played by Shc and STAT activation in megakaryocyte development await studies in knock-out mice, in which expression of these proteins is eliminated, or studies in a more physiologically relevant in vitro setting.
Originally identified for its role in prolactin-stimulated milk production, STAT5 has recently been implicated in signal transduction by multiple cytokines (42). Whether STAT5 is predominantly involved in proliferative or differentiative signaling remains controversial. Recently, altered forms of the erythropoietin and IL-2β receptors were described, which proliferate normally but fail to induce STAT5 phosphorylation or activation (25, 43). Other reports indicate that erythropoietin and granulocyte colony-stimulating factor receptors that lack cytoplasmic tyrosine residues can induce STAT5 phosphorylation, although higher ligand concentrations are required than for the native receptors (26, 27, 44).
Our results demonstrate that TPO-dependent STAT5 phosphorylation is partially mediated by the C terminus of Mpl but also occurs when no cytoplasmic tyrosines are present. Y112 is a likely STAT5 recruitment site because it has a similar motif (pYLPL) to Y343 of the erythropoietin receptor (pYLVL) and the recently described STAT5 consensus binding site (pYLXL; ref. 45). However, it is tempting to speculate that STAT5 may be activated indirectly through interreceptor association when Mpl does not contain intracellular tyrosine residues. Such “cross-talk” was recently demonstrated between the OKit and erythropoietin receptors, although under somewhat different circumstances (46). In support of this hypothesis, we have recently shown that TPO-stimulated mature megakaryocytes (i.e., those undergoing differentiation only) have markedly diminished STAT5 phosphorylation and activation (35). Thus, a molecule that is part of the Mpl signaling complex in proliferating Ba/F3-Mpl cells, but not in terminally differentiated megakaryocytes, may mediate STAT5 recruitment. These data emphasize the importance of studying signaling events in physiologically relevant cells and also underscore the potential for receptor cross-talk in cytokine signaling.
Acknowledgments
We appreciate the contributions of Kelly Millett (proliferation assays), Tor Bieker (mutagenesis), Todd Berard (sequencing), and Aly Karsan (critical review) at the University of Washington. Several essential reagents were provided by Charlie Hart (Mpl antiserum), Don Foster (purified murine TPO), Si Lok (pHZ-1 expression vector and mMpl cDNA), and Erica Vanaya and Will Lint (oligonucleotides), at Zymogenetics, Inc., and by Chris Saris (STAT5B antibody) at Amgen Corp. This work was supported by the National Institutes of Health Grant R01DK49855. J.G.D. received postdoctoral support from National Institutes of Health Grant K08 HL03498-01 and the American Society of Hematology Fellow Scholar Award.
ABBREVIATIONS
- TPO
thrombopoietin
- IL
interleukin
- SH2
Src homology 2
- m
murine
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
References
- 1.Kaushansky K, Lok S, Holly R D, Broudy V C, Lin N, et al. Nature (London) 1994;369:568–571. doi: 10.1038/369568a0. [DOI] [PubMed] [Google Scholar]
- 2.Kaushansky K, Broudy V, Lin N, Jorgensen M, McCarty J, Fox N, Zucker-Franklin D, Lofton-Day C. Proc Natl Acad Sci USA. 1995;92:3234–3238. doi: 10.1073/pnas.92.8.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander W S, Metcalf D, Dunn A R. EMBO J. 1995;14:5569–5578. doi: 10.1002/j.1460-2075.1995.tb00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broudy V C, Lin N L, Fox N, Taga T, Saito M, Kaushansky K. Blood. 1996;88:2026–2032. [PubMed] [Google Scholar]
- 5.Morella K K, Bruno E, Kumaki S, Lai C F, Fu J, Wang H M, Murray L, Hoffman R, Timour M, Bénit L, Gisselbrecht S, Zhvans H, Wojchowski D M, Bauman H, Gearing D P. Blood. 1995;86:557–571. [PubMed] [Google Scholar]
- 6.Vigon I, Florindo C, Fichelson S, Guenet J L, Mattei M G, Souyri M, Cosman D, Gisselbrecht S. Oncogene. 1993;8:2607–2615. [PubMed] [Google Scholar]
- 7.Skoda R C, Seldin D C, Chiang M K, Peichel C L, Vogt T F, Leder P. EMBO J. 1993;12:2645–2653. doi: 10.1002/j.1460-2075.1993.tb05925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T. Proc Natl Acad Sci USA. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bénit L, Courtois G, Charon M, Varlet P, Dusanter-Fourt I, Gisselbrecht S. J Virol. 1994;68:5270–5274. doi: 10.1128/jvi.68.8.5270-5274.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurney A L, Wong S C, Henzel W J, De Sauvage F J. Proc Natl Acad Sci USA. 1995;92:5292–5296. doi: 10.1073/pnas.92.12.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drachman J G, Griffin J D, Kaushansky K. J Biol Chem. 1995;270:4979–4982. doi: 10.1074/jbc.270.10.4979. [DOI] [PubMed] [Google Scholar]
- 12.Dorsch M, Fan P D, Bogenberger J, Goff S P. Biochem Biophys Res Commun. 1995;214:424–431. doi: 10.1006/bbrc.1995.2304. [DOI] [PubMed] [Google Scholar]
- 13.Pallard C, Gouilleux F, B’enit L, Cocault L, Souyri M, Levy D, Groner B, Gisselbrecht S, Dusanter-Fourt I. EMBO J. 1995;14:2847–2856. doi: 10.1002/j.1460-2075.1995.tb07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sattler M, Durstin M A, Frank D A, Okuda K, Kaushansky K, Salgia R, Griffin J D. Exp Hematol. 1995;23:1040–1048. [PubMed] [Google Scholar]
- 15.Ezumi Y, Takayama H, Okuma M. FEBS Lett. 1995;374:48–52. doi: 10.1016/0014-5793(95)01072-m. [DOI] [PubMed] [Google Scholar]
- 16.Mu S X, Xia M, Elliot G, Bogenberger J, Swift S, Bennett L, Lappinga D L, Hecht R, Lee R, Saris C J M. Blood. 1995;86:4532–4543. [PubMed] [Google Scholar]
- 17.Miyakawa Y, Oda A, Druker B J, Miyazaki H, Handa M, Ohashi H, Ikeda Y. Blood. 1996;87:439–447. [PubMed] [Google Scholar]
- 18.Miyakawa Y, Oda A, Druker B J, Kato T, Miyazaki H, Handa M, Ikeda Y. Blood. 1995;86:23–27. [PubMed] [Google Scholar]
- 19.Bacon C M, Tortolani P J, Shimosaka A, Rees R C, Longo D L, O’Shea J J. FEBS Lett. 1995;370:63–68. doi: 10.1016/0014-5793(95)00796-c. [DOI] [PubMed] [Google Scholar]
- 20.Tortolani P J, Johnston J A, Bacon C M, McVicar D W, Shimosaka A, Linnekin D, Longo D L, O’Shea J J. Blood. 1995;85:3444–3451. [PubMed] [Google Scholar]
- 21.Sasaki K, Odai H, Hanazono Y, Ueno H, Ogawa S, Langdon W Y, Tanaka T, Miyagawa K, Mitani K, Yazaki Y, Hirai H. Biochem Biophys Res Commun. 1995;216:338–347. doi: 10.1006/bbrc.1995.2629. [DOI] [PubMed] [Google Scholar]
- 22.Heim M H, Kerr I M, Stark G R, Darnell J E., Jr Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 23.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T, Baltimore D, Ratinofsky S, Feldman R A, Contley L C. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl N, Farruggella T J, Boulton T G, Zhong Z, Darnell J E, Jr, Yancopoulos G D. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 25.Quelle F W, Wang D, Nosaka T, Thierfelder W E, Stravopodis D, Weinstein Y, Ihle J N. Mol Cell Biol. 1996;16:1622–1631. doi: 10.1128/mcb.16.4.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damen J E, Wakao H, Miyajima A, Krosl J, Humphries R K, Cutler R L, Krystal G. EMBO J. 1995;14:5557–5568. doi: 10.1002/j.1460-2075.1995.tb00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin H, Nakamura N, Kamiyama R, Miyasaka N, Ihle J N, Miura O. Blood. 1996;88:4415–4425. [PubMed] [Google Scholar]
- 28.Baumann H, Gearing D, Ziegler S F. J Biol Chem. 1994;269:16297–16304. [PubMed] [Google Scholar]
- 29.Fukunaga R, Ishizaka I E, Nagata S. Cell. 1993;74:1079–1087. doi: 10.1016/0092-8674(93)90729-a. [DOI] [PubMed] [Google Scholar]
- 30.Hill R J, Zozulya S, Lu Y-L, Hollenbach P W, Joyce-Shaikh B, Bogenberger J, Gishizky M L. Cell Growth Differ. 1996;7:1125–1134. [PubMed] [Google Scholar]
- 31.Pronk G J, deVries-Smits A M, Buday L, Downward J, Maassen J A, Medema R H, Bos J L. Mol Cell Biol. 1994;14:1575–1581. doi: 10.1128/mcb.14.3.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold A, Rohrschneider L R. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 33.Damen J E, Liu L, Rosten P, Humphries R K, Jefferson A B, Majerus P W, Krystal G. Proc Natl Acad Sci USA. 1996;93:1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porteu F, Rouyez M C, Cocault L, Bénit L, Charon M, Picard F, Gisselbrecht S, Souyri M, Dusanter Fourt I. Mol Cell Biol. 1996;16:2473–2482. doi: 10.1128/mcb.16.5.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drachman J G, Sabath D F, Fox N E, Kaushansky K. Blood. 1997;89:483–492. [PubMed] [Google Scholar]
- 36.Fuh G, Cunningham B C, Fukunaga R, Nagata S, Goeddel D V, Wells J A. Science. 1992;256:1677–1680. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- 37.D’Andrea A D, Yoshimura A, Youssoufian H, Zon L I, Koo J W, Lodish H F. Mol Cell Biol. 1991;11:1980–1987. doi: 10.1128/mcb.11.4.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klingmuller U, Lorenz U, Cantley L C, Neel B G, Lodish H F. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 39.de la Chapelle A, Traskelin A L, Juvonen E. Proc Natl Acad Sci USA. 1993;90:4495–4499. doi: 10.1073/pnas.90.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laminet A A, Apell G, Conroy L, Kavanaugh W M. J Biol Chem. 1996;271:264–269. doi: 10.1074/jbc.271.1.264. [DOI] [PubMed] [Google Scholar]
- 41.van der Geer P, Wiley S, Gish G D, Lai V K, Stephens R, White M F, Kaplan D, Pawson T. Proc Natl Acad Sci USA. 1996;93:963–968. doi: 10.1073/pnas.93.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pallard C, Gouilleux F, Charon M, Groner B, Gisselbrecht S, Dusanter-Fourt I. J Biol Chem. 1995;270:15942–15945. doi: 10.1074/jbc.270.27.15942. [DOI] [PubMed] [Google Scholar]
- 43.Fujii H, Nakagawa Y, Schindler U, Kawahara A, Mori H, Gouilleux F, Groner B, Ihle J N, Minami Y, Miyazaki T, Taniguchi T. Proc Natl Acad Sci USA. 1995;92:5482–5486. doi: 10.1073/pnas.92.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian S-S, Tapley P, Sincich C, Stein R B, Rosen J, Lamb P. Blood. 1996;88:4435–4444. [PubMed] [Google Scholar]
- 45.May P, Gerhartz C, Heesel B, Welte T, Doppler W, Graeve L, Horn F, Heinrich P C. FEBS Lett. 1996;394:221–226. doi: 10.1016/0014-5793(96)00955-6. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, Klingmuller U, Besmer P, Lodish H F. Nature (London) 1995;377:242–246. doi: 10.1038/377242a0. [DOI] [PubMed] [Google Scholar]