Abstract
Crk, which belongs to the adaptor family of proteins composed of Src homology 2 (SH2) and SH3 domains, has a putative role in signaling. However, the downstream events of Crk signaling remain unclear. In this study, we found that Jun kinase (JNK) is moderately activated by v-Crk in both NIH 3T3 cells and chicken embryo fibroblasts. Transient expression of v-Crk, c-Crk-I, or c-Crk-II activated JNK1 in human embryo kidney cells, 293T. Coexpression of a guanine nucleotide exchange protein C3G, which specifically binds to Crk’s SH3 domain, further enhanced the JNK activity as well as growth rate and anchorage-independent growth of v-Crk NIH 3T3 cells. Furthermore, overexpression of a dominant-negative form of C3G lacking the guanine nucleotide exchange domain abolished both the JNK activity and the colony forming potential of v-Crk NIH 3T3 cells. The requirement for JNK activation in v-Crk induced transformation was demonstrated by the suppression of colony forming activity of v-Crk NIH 3T3 cells when a dominant-negative form of JNK kinase, Sek1/MKK4 is expressed in these cells. These data strongly suggest the existence of a novel signaling cascade involving an adaptor protein v-Crk, which transmits signals through C3G toward JNK activation.
The adaptor family of proteins composed primarily of Src homology 2 (SH2) and SH3 domains may mediate signals involved in various cellular responses (1). The identification of the signaling cascade from Grb2 adaptor protein, Grb2/Sos/Ras/mitogen-activating protein (MAP) kinase (MAPK) pathway, has revealed a precise mechanism of various cellular responses triggered by many growth factors (1). Crk also belongs to the adaptor family of proteins, originally reported as an avian retrovirus encoding oncogene product v-Crk (2). The cellular homologs of v-Crk were subsequently identified as c-Crk-I and c-Crk-II, which are alternative spliced forms of single gene (3). In addition, another closely related gene product, CrkL, has been isolated (4). Although Crk has been suggested to be involved in growth factor- or integrin-mediated signaling pathways, a physiological role for Crk is not clearly understood, mainly due to the lack of information about the downstream signaling pathway(s) of Crk (5, 6).
In v-Crk-expressing cells, the level of tyrosine phosphorylation of a limited number of specific cellular proteins is increased, despite the lack of any tyrosine kinase catalytic domain in v-Crk itself (7). Furthermore, this increased level of cellular phosphotyrosine correlates with the transforming activity of v-Crk (8). Among these tyrosine-phosphorylated proteins, components of focal adhesion, p130Cas and paxillin, have been shown to associate with the SH2 domain of v-Crk (6, 9). On the other hand, the SH3 domain of Crk can bind to multiple target molecules, including two guanine nucleotide exchange proteins, C3G (10) and Sos (11), and a tyrosine kinase c-Abl (12), and their binding suggests the possibility that v-Crk activates Ras/MAPK pathway similar to Grb2 .
In this study, to find out whether Crk has an unique signaling cascade or has a function redundant with that of Grb2, we analyzed the downstream events of Crk-mediated signaling pathway in v-Crk-transformed fibroblasts.
MATERIALS AND METHODS
Plasmid DNA Constructions.
pCAGGS-C3GΔ, a mutant C3G lacking CDC25 homology region (amino acids 1–826) was constructed by removing the 3′ region of C3G from the SmaI site (nucleotide position 2479) to the end of the ORF and ligated into pCAGGS expression vector (13), creating a new stop codon, TGA, at the end of truncated C3G sequences.
Jun Kinase (JNK) and MAPK Assays.
JNK and MAPK activities were measured by in vitro kinase assay described elsewhere (14) using glutathione S-transferase (GST)-c-Jun (encompassing amino acids 1–79) and myelin basic protein as substrates, respectively.
Transient Transfection.
A total of 1.5 μg of pcDNA3-HA-JNK1 (HA, hemagglutinin); 5 μg of pMex-v-Crk, pMex-hCrk-I, pMex-hCrk-II; and 3 μg of pCAGGS-C3G, pCAGGS-C3G-F, pEBG-Sek1, pEBG-Sek1-KR, pcDNA3-HA-Sek-AL DNAs were used for transfection of 293T cells cultured in a 60-mm-diameter dish by a modified calcium phosphate transfection system (Stratagene).
Establishment of Stable Cell Lines.
The methods for preparing v-Crk-expressing NIH 3T3 cell line (clone V4) was described (15). Another v-Crk NIH 3T3 cell line (clone 1-1) was established by Chris Marshall in our laboratory. NIH 3T3 cells and v-Crk NIH 3T3 cell lines (V4 and 1-1) were cultured in a 10-cm-diameter dish and 10 μg of each pCAGGS-C3G, pCAGGS-C3G-F, pCAGGS-C3GΔ, pEBG, pEBG-H-Ras, pEBG-Rap-1, pEBG-Sek1, or pEBG-Sek1-KR was cotransfected with 2 μg of pBabe-puro vector DNA, carrying the puromycin-resistant gene by the calcium phosphate method, and 48 h after transfection cells were cultured in DMEM with 10% calf serum containing 2 μg/ml puromycin.
Measurement of Growth Rates.
Cells (1 × 105) of each cell line were initially plated in a 60-mm-diameter culture dish with 5 ml of DMEM supplemented with 5% calf serum, and the medium was refreshed every other day. The numbers of cells were counted using a hemocytometer.
Colony Formation Assay.
Cells (5 × 105) of each cell line were plated in 0.4% noble agar in DMEM with 10% fetal bovine serum as an upper layer. Bactoagar (0.5%) in DMEM with 10% calf serum was used for the bottom layer. Colonies larger than 0.25 mm in diameter evaluated under the microscope were counted as positives 14 days after initial plating. Representative data from three independent experiments are shown as a bar graph.
RESULTS
Activation of JNK by v-Crk in NIH 3T3, Chicken Embryo Fibroblast, and 293T Cells.
As an initial experiment to examine the downstream signals mediated by v-Crk, we analyzed NIH 3T3 cell lines expressing v-Crk for the association of v-Crk SH3 domain with several target molecules that have been reported such as Sos, C3G, and c-Abl. We found complexes of v-Crk/Sos and v-Crk/C3G (data not shown), but did not observe v-Crk/c-Abl complexes despite the fact that these cell lines have detectable levels of endogenous c-Abl (data not shown). Because Sos bound to v-Crk, we measured MAPK activity to see whether the Ras/MAPK pathway plays a major role in v-Crk signaling in v-Crk NIH 3T3 cells. At the same time, the activity of another member of the MAPK family of proteins, JNK, was also determined.
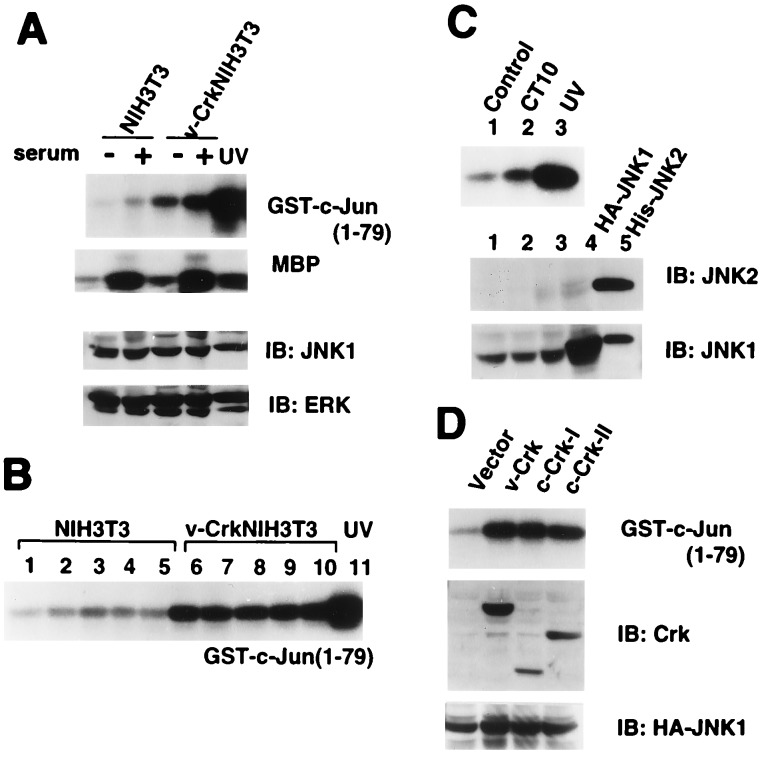
Contrary to our expectation, we found that MAPK activity was increased only 1.3-fold, while JNK was constitutively activated by at least 3-fold in v-Crk NIH 3T3 cells with or without serum stimulation (Fig. 1A). This JNK activation was observed both in exponentially growing cells and in fully confluent cells (Fig. 1B). Furthermore, CT10 virus-infected chicken embryo fibroblasts also had elevated levels of JNK activity (Fig. 1C). In these fibroblasts, the amounts of JNK1, ERK1, and ERK2 were constant while JNK2 was not readily detectable by immunoblotting (Fig. 1 A and C). To verify that the JNK activation in v-Crk NIH 3T3 cells was not due to these specific cell lines, v-Crk, human c-Crk-I, or human c-Crk-II were transiently coexpressed with HA-JNK1 in human embryo kidney cells, 293T. In agreement with the JNK activation seen in the stable cell lines, we also found increased JNK1 activity in the transfected 293T cells (Fig. 1D).
Figure 1.
Activation of JNK by v-Crk. (A) JNK activities were measured by in vitro kinase assay using GST-c-Jun (1–79) as a substrate with lysates from NIH 3T3 cells and v-Crk NIH 3T3 cells after 24 h serum starvation followed by either serum stimulation for 20 min or not. UV-irradiated NIH 3T3 cells were used as a positive control. The activities of MAPK were also measured in vitro using myelin basic protein (MBP) as a substrate. The levels of endogenous JNK1 and ERKs in total cell lysates were examined by immunoblotting (IB) using anti-JNK1 and mixture of anti-ERK1 and ERK2 antibodies (Santa Cruz Biotechnology). (B) JNK activities were measured in NIH 3T3 and v-Crk NIH 3T3 at different growth stages. Cell densities were roughly determined by the percentage of the confluence as 40% (lanes 1 and 6), 60% (lanes 2 and 7), 80% (lanes 3 and 8), 100% (lanes 4 and 9), and 24 h after 100% (lanes 5 and 10). (C Upper) JNK activities were assayed in chicken embryo fibroblasts (CEF) uninfected (lane 1) and infected with avian CT10 virus encoding v-Crk (lane 2). Lane 3, CEF with UV irradiation as a control. (Lower) The endogenous levels of JNKs were determined by immunoblotting using anti-JNK1 and anti-JNK2 antibodies. Lanes: 1, CEF; 2, CEF infected with CT10 virus; 3, CEF with UV irradiation. HA-JNK1 expressed in mammalian cells (lane 4) and bacterially expressed His-JNK2 (lane 5) were used as positive controls. (D) JNK activities in human embryo kidney cells 293T that were transiently coexpressing HA-JNK1 with v-Crk, c-Crk-I, or c-Crk-II.
Enhancement of JNK Activity and Transformation by the Additional Expression of C3G in v-Crk NIH 3T3 Cells.
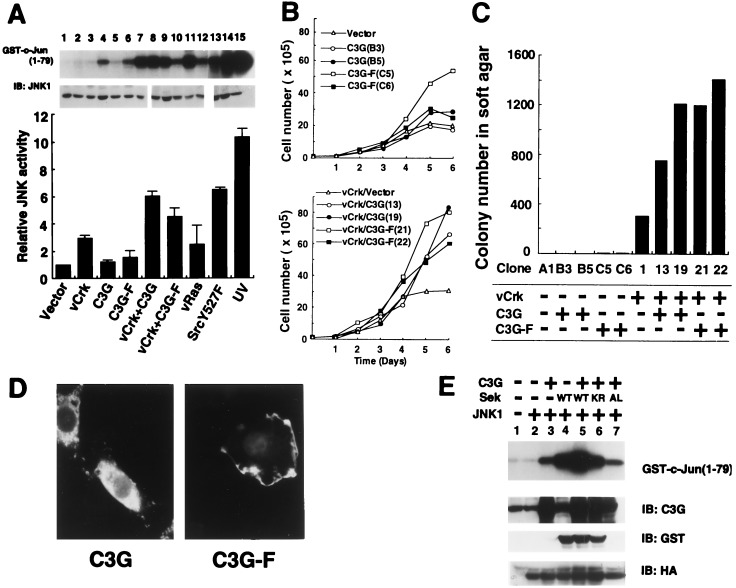
The activation of JNK by v-Crk may involve the guanine nucleotide exchange proteins Sos or C3G, both of which bind to the Crk SH3 domain, because JNK has recently been reported to be regulated by small G proteins, Rac and Cdc42 (14, 43). However, Sos is unlikely to activate JNK without a significant increase in MAPK activity. We therefore asked whether C3G could induce JNK activation, establishing several clonal lines of C3G NIH 3T3 and v-Crk/C3G NIH 3T3 cells by transfecting NIH 3T3 and two independent clones of v-Crk NIH 3T3 cells with an expression plasmid coding for C3G. The level of C3G expression in each cell line was 5 to 10 times higher than that of endogenous C3G (data not shown). Coexpression of C3G and v-Crk enhanced JNK activity by 6-fold in comparison to the 3-fold activation observed in v-Crk NIH 3T3 cells (Fig. 2A). Expression of C3G alone in NIH 3T3 cells did not alter the level of the JNK activity (Fig. 2A). The levels of expression of endogenous JNK1 in these established cell lines were unchanged (Fig. 2A). We also confirmed that stable expression of C3G or v-Crk did not increase the levels of v-Crk or endogenous C3G, respectively, although a possibility exists that the complex formation between two proteins stabilizes them and increases their half-lives (data not shown).
Figure 2.
Enhancement of v-Crk-induced JNK activation and transformation by C3G. (A) JNK activities in NIH 3T3 cell lines stably expressing the following proteins. Lanes: 1, vector control; 2 and 3, C3G (clones B3 and B5); 4 and 5, C3G-F (clones C5 and C6); 6, v-Crk and vector alone (clone 1); 7–10, v-Crk and C3G (clones 13, 19, 3-2, and 3-3); 11 and 12, v-Crk and C3G-F (clones 21 and 22); 13, v-Ha-Ras; 14, c-SrcY527F; and 15, UV-irradiated NIH 3T3 cells. v-Ras and constitutive-activated form of c-Src, Y527F, and UV irradiation were used as a positive controls. Relative activities of JNK are shown as bar graph by assigning the activity of NIH 3T3 alone as 1. The level of endogenous JNK1 in each cell line was determined by immunoblotting. (B) Growth rates of cell lines were determined in culture medium in the presence of 5% calf serum. (Upper) Plot of the number of cells of C3G or C3G-F expressing NIH 3T3 cell lines. (Lower) Plot of the number of cells of v-Crk NIH 3T3 cell lines expressing C3G or C3G-F. The names of cell clones are shown in parenthesis. (C) Anchorage-independent growth of NIH 3T3-derived cell lines expressing either v-Crk or C3G, or both v-Crk and C3G, were measured by soft agar colony forming assay. The cell clones and their expressing proteins are indicated at the bottom. (D) Localization of overexpressed C3G (Left) and C3G-F (Right) in NIH 3T3 cells. Overexpressed C3G and C3G-F were visualized by immunocytochemistry using anti-C3G antibody and fluorescein isothiocyanate-labeled anti-rabbit antibody. (E) Transient expression of C3G-activated JNK and coexpressed Sek1/MKK4 further enhanced its activity in 293T cells. 293T cells were transiently transfected by expression plasmids encoding proteins indicated at the top, and JNK activities were measured by in vitro kinase assay as described in Fig. 1. Wild-type Sek1 was expressed as GST fusion protein (16). Two different dominant-negative forms of Sek, kinase negative GST-Sek1-KR (16) and HA-Sek1-AL (17), in which regulatory tyrosine and threonine residues were substituted by alanine and leucine, were used as control experiment. The expressed protein levels were confirmed by immunoblotting using anti-C3G, anti-GST, and anti-HA antibodies.
In parallel with the increase in the JNK activity, coexpression of C3G and v-Crk considerably augmented the growth rate of NIH 3T3 cells (Fig. 2B Lower), while no significant increase in cell growth was associated with C3G expression alone (Fig. 2B Upper). On average, the saturation density of v-Crk/C3G NIH 3T3 cells was 2.6 times higher than that of v-Crk NIH 3T3 cells (data not shown). Furthermore, the anchorage-independent cell growth was also greatly enhanced in cells coexpressing C3G and v-Crk, in comparison to those expressing v-Crk alone (Fig. 2C). It is noteworthy that no significant increase in the MAPK activity was observed in any of the cell lines expressing v-Crk and C3G (data not shown), and a Mek1 inhibitor, PD98059 (18), did not reduce the number of colonies of v-Crk NIH 3T3 in the soft agar. In v-Crk NIH 3T3 cells, it was found that v-Crk binds equally well to either C3G or Sos (data not shown). However, in v-Crk/C3G NIH 3T3 cells, overexpression of C3G causes the ratio of v-Crk/C3G to v-Crk/Sos complexes to be altered to 10:1 (data not shown). It is conceivable that the increase in the complex formation of v-Crk with C3G rather than Sos is responsible for the activation of JNK and increased cell growth and transformation.
Farnesylation of C3G Alone Is Not Sufficient To Activate JNK in NIH 3T3 Cells.
Because the expression of a membrane-targeted form of Sos (farnesylated Sos) has been reported to activate the Ras/MAPK pathway (19), we also tested the effect of a farnesylated form of C3G, C3G-F, on JNK activity and transformation. A farnesylation-acceptable sequence derived from Ki-Ras-2 was attached at the C terminus of C3G. Stable expression of C3G-F in NIH 3T3 cells resulted in weak activation of JNK in only one cell line (Fig. 2A, lane 4) and no transforming activity was detected (Fig. 2C). To verify the effectiveness of our farnesylation sequence, we prepared a Sos-F construction with the same membrane-targeting sequence used for C3G-F and found that it activates MAPK in mouse fibroblast cell lines as previously reported (data not shown). Localization of C3G-F to the plasma membrane of NIH 3T3 cells was confirmed by immunofluorescence microscopy (Fig. 2D). These data suggest that localization of C3G to the plasma membrane is insufficient for the activation of JNK and transformation.
Activation of HA-JNK1 by C3G and Enhancement by Additional Expression of JNK Kinase Sek1/MKK4 in 293T Cells.
To verify that the further enhancement of JNK activation by C3G observed in v-Crk/C3G NIH 3T3 cell lines (Fig. 1A) was not due to these specific stable cell lines, we examined whether C3G was able to activate JNK using the transient protein expression system in 293T cells. We observed that overexpression of C3G activates coexpressed HA-JNK1 in 293T cells (Fig. 2E, lane 3). For further confirmation of the potential of C3G to activate JNK, we tested whether C3G increases the activity of JNK through JNK kinase Sek1/MKK4. Transient expression of JNK kinase Sek1/MKK4 (16, 20) alone increased the activity of HA-JNK1 as expected (Fig. 2E, lane 4), and coexpression of C3G and Sek1/MKK4 cooperatively augmented HA-JNK1 activity (Fig. 2E, lane 5), while coexpression of C3G and a dominant negative form of Sek1/MKK4 did not (Fig. 2E, lanes 6 and 7). These data strongly suggest that C3G has the potential to activate JNK. We should point out here the discrepancy in the results of 293T cells and NIH 3T3 cells: the expression of C3G or C3G-F alone did not significantly activate JNK in NIH 3T3 cells (Fig. 2A, lanes 2–5) but did in 293T cells (Fig. 2E, lane 3). This could be due to the different protein expression levels in these two cell lines. Generally, the levels of transiently expressed protein in a single cell is substantially higher than that seen in stable cell lines. In 293T cells, both C3G and HA-JNK1 were transiently expressed, and we measured the activity of overexpressed HA-JNK1. By contrast, only C3G or C3G-F was stably expressed in NIH 3T3 cells and the activity of endogenous JNK was assayed. We consider that the results in Fig. 2E support the notion that C3G has the potential to activate HA-JNK1 through JNK kinase Sek1/MKK4.
Requirement for the CDC25 Homology Domain of C3G for the Enhancement of v-Crk Induced JNK Activation and Transformation.
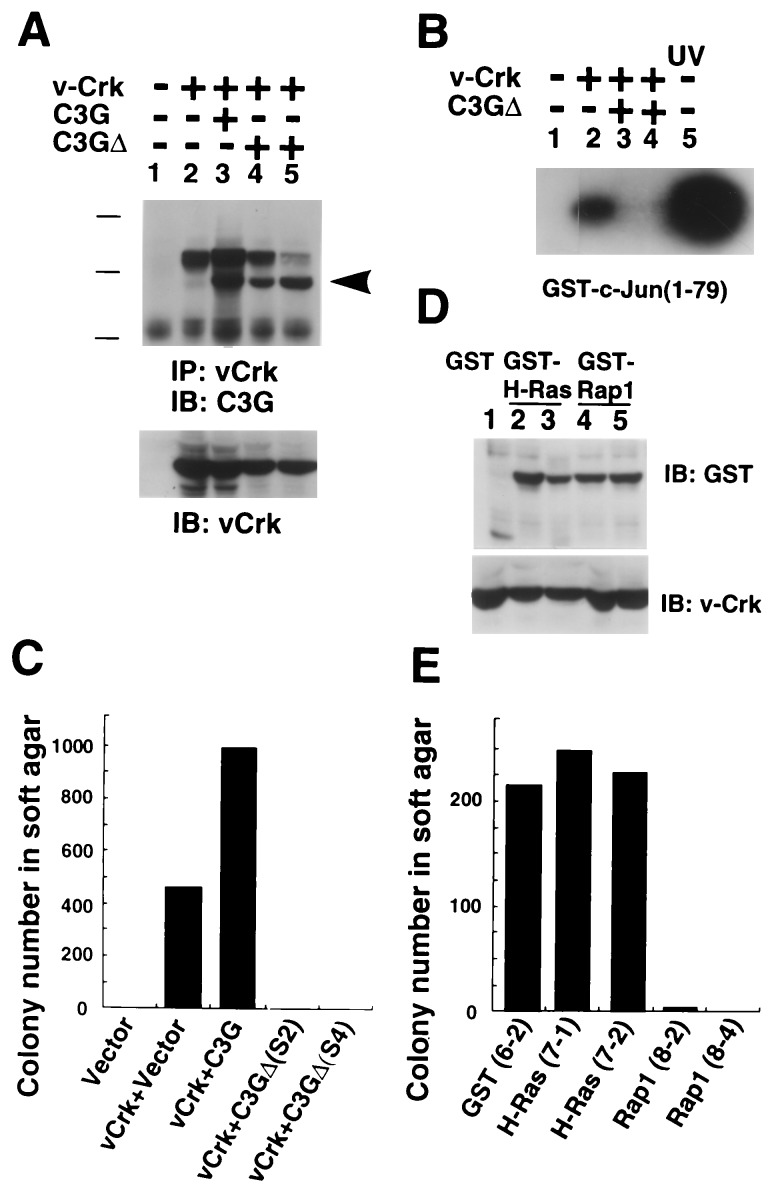
To examine whether the guanine nucleotide exchange domain of C3G is required for JNK activation and transformation, a mutant C3G, C3GΔ, lacking the CDC25 homology region, was stably expressed in v-Crk NIH 3T3 cells. The level of v-Crk was essentially equivalent in each cell line (Fig. 3A Lower), and the association of C3GΔ and v-Crk was detectable by immunoprecipitation with anti-v-Crk antibody (Fig. 3A Upper). We found that the expression of C3GΔ completely abolishes the increased JNK activity found in v-Crk NIH 3T3 cells and suppresses the anchorage-independent cell growth in soft agar (Fig. 3 B and C). These cell lines maintained the same basal level of MAPK activities (data not shown). The dominant negative function of the mutant C3GΔ on JNK activation and anchorage-independent cell growth demonstrates the requirement for the guanine nucleotide exchange domain of C3G in the v-Crk induced JNK activation and transformation.
Figure 3.
Involvement of guanine nucleotide exchange domain of C3G in v-Crk induced JNK activation and transformation. (A) Two v-Crk NIH 3T3-derived cell lines stably expressing C3GΔ, lacking CDC25 homology region, were established (clones S2 and S4). (Upper) The association of C3GΔ and v-Crk was detected by immunoprecipitation. Arrowhead indicates the position of C3GΔ. The proteins expressed in each cell line are indicated at the top. v-Crk was immunoprecipitated from lysate containing 500 μg of total protein by anti-gag antibody (3C2) and resultant blot was probed with anti-C3G antibody. Bars shows the molecular mass 200, 118, and 86 kDa. (Lower) The levels of v-Crk were detected by immunoblotting with anti-gag antibody, using 20 μg of protein in total cell lysate per lane. (B) JNK activity of each cell line was determined by in vitro kinase assay as described in Fig. 1. The proteins expressed in each cell line are indicated at the top. (C) Anchorage-independent growth of NIH 3T3 cell lines expressing v-Crk and C3GΔ. (D) Establishment of v-Crk NIH 3T3 cell line stably expressed GST (lane 1, clone 6-2), GST-H-Ras (lanes 2 and 3, clones 7-1 and 7-2, and GST-Rap-1 (lanes 4 and 5, clone 8-2 and 8-4). Expression levels of GST fusion proteins and v-Crk were confirmed by anti-GST antibody and anti-gag antibody, respectively. (E) Anchorage-independent growth of v-Crk NIH 3T3 cells expressing GST, GST-H-Ras, and GST-Rap-1.
As Rap-1 has been reported to be a substrate of C3G in vitro (21), we prepared stable cell lines expressing both v-Crk and Rap-1 to examine the role of Rap-1 in v-Crk-induced transformation (Fig. 3D). The expression of GST-Rap-1 caused a flat cell morphology (data not shown) and suppressed the anchorage-independent cell growth of v-Crk NIH 3T3 cells, while the expression of GST or GST-H-Ras did not produce these effects (Fig. 3D). Taken together with the data that C3G enhanced the v-Crk-transforming activity (Fig. 2D), we conclude that Rap-1 does not function as a substrate of C3G at least in v-Crk-induced transformation. The different subcellular distribution of Rap-1 and v-Crk may explain the reason why C3G does not activate Rap-1 in vivo. Rap-1 localizes to membranes, especially that of the Golgi apparatus (22) while v-Crk carrying C3G associates with the putative focal adhesion components, p130Cas and paxillin. We are currently exploring potential substrates of C3G involved in JNK activation and transformation.
Requirement for JNK Activity in v-Crk-Induced Transformation of NIH 3T3 Cells.
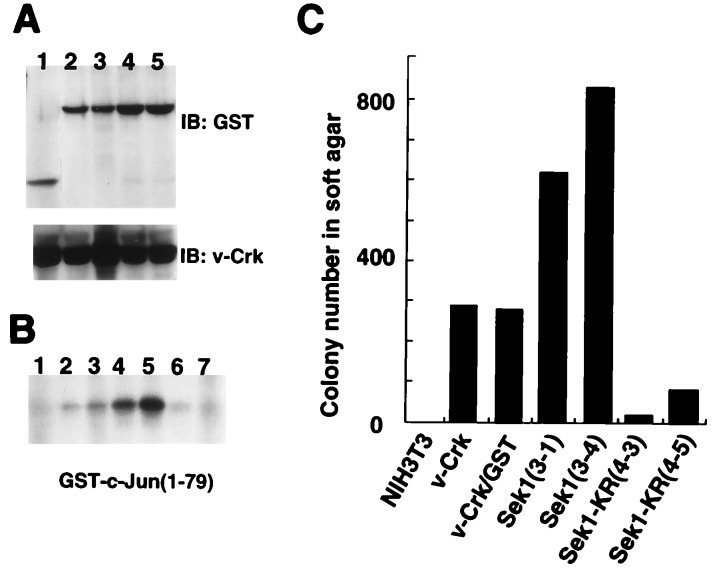
Finally, we examined whether JNK activity is required for v-Crk-induced transformation, because few reports have shown correlation of JNK activation and cell transformation (23). We prepared two clonal cell lines of NIH 3T3 coexpressing both v-Crk and a GST fusion form of wild-type Sek1/MKK4 or both v-Crk and dominant-negative form of Sek1/MKK4, GST-Sek1-KR (18). Immunoblotting using anti-GST antibody showed that the protein expression level of the Sek1/MKK4s were essentially the same in each cell line (Fig. 4A). Comparing with the JNK activity found in v-Crk NIH 3T3 cells expressing GST alone (Fig. 4B, lane 3), coexpression of wild-type Sek1/MKK4 and v-Crk enhanced JNK activity (Fig. 4B, lanes 4 and 5). In parallel with the elevation of JNK activity, growth rate (data not shown) and anchorage-independent cell growth of v-Crk in NIH 3T3 cells were also enhanced by the coexpression of wild-type Sek1/MKK4s (Fig. 4C). On the other hand, we found a profound suppression of JNK activity (Fig. 4B, lanes 6 and 7) and transforming potential of v-Crk NIH 3T3 cells (Fig. 4C) when a dominant-negative form of Sek1/MKK4 was expressed in the same cells. Significant activation of JNK was not detectable even after the serum stimulation in NIH 3T3 cells (Fig. 1A), and the stable expression of dominant-negative form of Sek1/MKK4 caused no suppression of cell growth in NIH 3T3 cells (data not shown). These data clearly demonstrate the requirement of JNK activity for v-Crk induced transformation in NIH 3T3 cells.
Figure 4.
Requirement of JNK activity for v-Crk-induced transformation. (A) Establishment of v-Crk NIH 3T3 derived cell lines expressing GST alone (lane 1, clone 2-1), wild-type Sek1 (lanes 2 and 3, clones 3-1 and 3-4), and kinase-negative form of Sek1-KR (lanes 4 and 5, clones 4-3 and 4-5). Expression levels of GST fusion Sek1 proteins were examined by anti-GST antibody. (B) JNK activity of Sek1 or Sek1-KR expressing v-Crk NIH 3T3 cells. Lanes: 1, NIH 3T3; 2, v-Crk NIH 3T3 (clone V4); 3, v-Crk/GST NIH 3T3 (clone 2-1); 4 and 5, v-Crk/GST-Sek1 NIH 3T3 (clones 3-1 and 3-4); 6 and 7, v-Crk/GST-Sek1-KR NIH 3T3 (clones 4-3 and 4-5). (C) Anchorage-independent growth of v-Crk NIH 3T3 cells expressing GST, GST-Sek1, and GST-Sek1-KR.
DISCUSSION
We have analyzed downstream signaling pathways of the adaptor protein v-Crk and found that v-Crk activates JNK through C3G (Fig. 5). The identification of an unique signaling pathway of v-Crk, activation of JNK rather than MAPK, solves the initial question as to whether the Crk pathway is redundant with the Grb2-mediated Sos/Ras/MAPK pathway. Several proteins may participate in the C3G-dependent JNK activation because C3G does not have any serine/threonine kinase domain found in JNK activators. The ability of C3G to activate overexpressed JNK1, in concert with JNK kinase, Sek1/MKK4, in 293T cells (Fig. 2E) suggests the involvement of Sek1/MKK4 or related proteins in C3G-dependent JNK activation. A small G protein cascade could be implicated in the pathway from C3G to JNK, since C3G-induced enhancement of JNK activity was blocked by the deletion of the CDC25 homology region (Fig. 3B). Although Rac and Cdc42 have been shown to activate JNK (14, 43), it is unlikely that C3G works as a direct activator for Rac or Cdc42, because C3G shares little homology with the CDC25 domain of Dbl or Ost which are known guanine nucleotide exchange proteins for Rac and Cdc42 (24), while it contains a CDC25 domain highly homologous to that of Sos or Ras-GRF, the representative activator for Ras family of proteins. The small G protein that is a direct substrate of C3G for the activation of JNK remains to be identified.
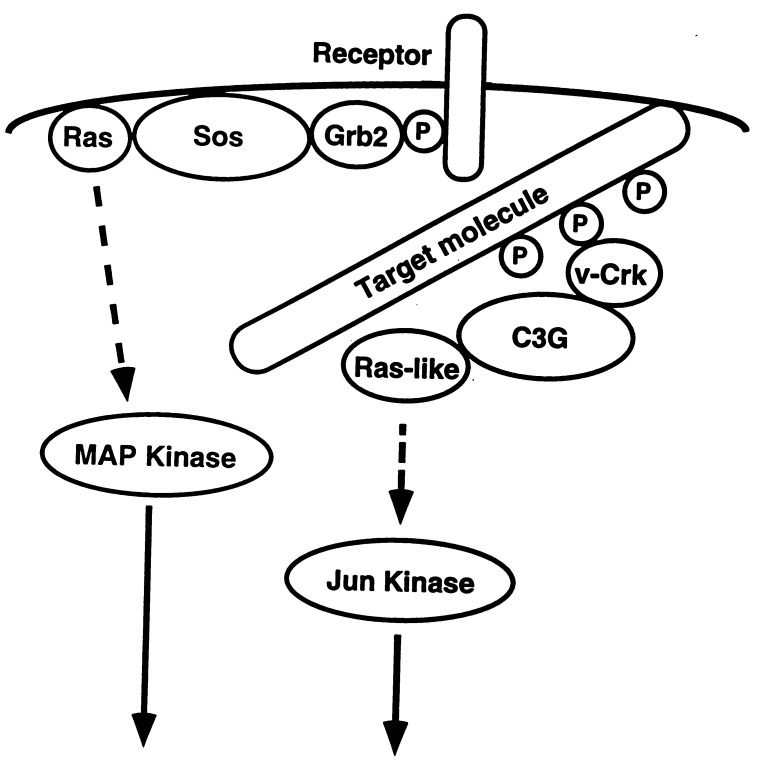
Figure 5.
A model for the v-Crk-mediated signaling pathway. v-Crk binds through its SH2 domain to multiple tyrosine-phosphorylated target molecules such as p130Cas or paxillin that supposedly localize to the focal adhesion. A guanine nucleotide exchange protein C3G, which associates with SH3 domain of v-Crk, can presumably activate Ras-like small G protein and generate signal for the activation of Jun kinase. The well established Grb2-mediated signaling pathway from tyrosine-phosphorylated receptor localized in the plasma membrane through Sos to the activation of MAPK is also described as a comparison of different signaling pathways mediated by two adaptor molecules.
In the Grb2/Sos/Ras/MAPK pathway, translocation of Sos by Grb2 to the plasma membrane, where its substrate Ras is located, is an essential event. In fact, this function of Grb2 can be artificially substituted by the attachment of membrane targeting peptides to Sos (19). A similar effect following farnesylation of C3G was expected; however, no significant activation of JNK was observed in C3G-F expressing cells. This suggests that the target of C3G may not be localized to the plasma membrane. v-Crk/C3G complexes may be primarily associated with p130Cas and paxillin, components of focal adhesion plaques (Fig. 5). This may also explain why MAPK activity is not significantly elevated in v-Crk NIH 3T3 cells despite the presence of v-Crk/Sos complexes in these cells: the majority of v-Crk/Sos complexes localizes to the focal adhesion plaques by the SH2 domain of v-Crk, not to the plasma membrane. These arguments are consistent with the fact that most of the guanine nucleotide exchange proteins including Sos, Ras-GRF, Dbl, Ost, and Vav possess a pleckstrin homology domain (24) one of whose functions is thought to bind to inositol triphosphates (25) resulting in the association with plasma membranes in vivo, while the consensus sequences for pleckstrin homology domain are not found in C3G.
Rap-1 was originally reported as a tumor suppressor protein against v-Ki-Ras in NIH 3T3 cells (26). It is argued that Rap-1 may inhibit Ras-induced colony formation by competing the effector proteins with Ras. As Rap-1 was recently found to be a substrate of C3G (21), we first hypothesized that the overexpression of C3G might suppress the v-Crk induced transformation. However, stable expression of C3G enhanced cell growth and the transforming activity of v-Crk in NIH 3T3 cells. The direct analysis of the effect of Rap-1 on v-Crk-induced transformation showed that Rap-1 suppressed colony forming activity of v-Crk NIH 3T3 cells (Fig. 3E). Taken together with the recent report showing that Rap-1 activates MAPK pathway via B-Raf in vitro (27), we conclude that Rap-1 is not a substrate of C3G, at least for JNK activation and transformation in v-Crk-expressing NIH 3T3 cells. The binding of the v-Crk/C3G complex to p130Cas and paxillin may not activate Rap-1, which primarily localizes in the membrane. However, we cannot completely exclude the possibility of Rap-1 functioning as a substrate of C3G and transducing a signal to MAPK in vivo because c-Crk or CrkL may bring C3G to the plasma membrane or Golgi apparatus under certain conditions.
In this study, we found a requirement for the activity of JNK for v-Crk induced transformation of NIH 3T3 cells (Fig. 4). Although stress-induced JNK activation has been well characterized (28), induction of apoptosis is not only the effect of JNK. Recently it was shown that tumor necrosis factor α activates JNK, but this activity is not necessary for the induction of apoptosis (29). JNK isoform-dependent differential activation of several transcription factors such as c-Jun, Elk-1, or ATF2 and its related families of proteins may contribute to the various effects of JNK (30). We are now investigating the responsible transcription factor(s) involving in the v-Crk-induced transformation. Although we showed a requirement for JNK activation in v-Crk-induced transformation, the elevated activity of JNK alone may not be sufficient for cellular transformation in general. In v-Crk-expressing cells, the increased amount of tyrosine-phosphorylated proteins suggests the activation of tyrosine kinase(s) or inactivation of tyrosine phosphatase(s) in v-Crk-expressing cells. Together with the activation of JNK by v-Crk, these increased levels of tyrosine phosphorylated proteins may generate signals and contribute to the elevation of transforming potential of v-Crk. Furthermore, the dominant-negative form of Ras has been shown to suppress v-Crk-induced transformation of NIH 3T3 cells (15). v-Crk moderately activated MAPK in chicken embryonal fibroblasts while no significant increase of MAPK activity was found in v-Crk NIH 3T3 cells (15). These data suggest that the contribution of Ras effector(s) other than MAPK may play a role in the v-Crk-induced transformation of NIH 3T3 cells.
While we have elucidated a C3G-dependent signaling pathway mainly based on an analysis of v-Crk function, this pathway may also be involved in c-Crk signaling, because c-Crk-I and c-Crk-II could activate JNK1 (Fig. 1D), and c-Crk and CrkL have been reported to bind to C3G in several cell lines (31, 32). Biochemical analysis has also supported the importance of the complex between c-Crk and C3G by showing that the c-Crk N-terminal SH3 domain, SH3(N), has 12-fold higher binding affinity for C3G than the v-Crk SH3 domain (33). In addition, c-Crk SH3(N) has a 5-fold higher affinity for C3G than Sos (33). Since Crk has been reported to be involved in signaling in many different types of cells (34–36), the signaling pathway, Crk/C3G/Ras-like G protein/JNK, proposed in our study may function in various cellular responses. For example, integrin engagement was found to increase the tyrosine phosphorylation of p130Cas and its association with c-Crk-II (37, 38), or to activate JNK by a kinetics different from that of integrin-induced MAPK activation in the same cells (39). Therefore, Crk-induced JNK activation may play an important role in the integrin-mediated signal transduction.
The significance of Crk-mediated JNK activation in human disease is also suggested from the analysis of Philadelphia chromosome-positive leukemia expressing Bcr-Abl chimeric protein. CrkL has been reported to be tyrosine phosphorylated and to bring Bcr-Abl oncoprotein to the focal adhesion in these leukemic cells (40, 41). Taken together with the recent evidence showing activation of JNK by Bcr-Abl and the requirement for c-Jun in Bcr-Abl-induced transformation (42), signaling by the Crk/C3G/Ras-like G protein/JNK pathway may play an important role in the oncogenesis of a certain type of cancer.
Acknowledgments
We thank Michiyuki Matsuda (National Institute of Health, Japan) for pCAGGS-C3G, pCAGGS-C3G-F, pEBG-H-Ras, pEBG-Rap-1, pMex-Crk-I, pMex-Crk-II, and anti-Crk-SH3 antiserum; Bruce Mayer (Harvard Medical School) for pEBG-Sek1 and pEBG-Sek1-KR; J. Silvio Gutkind (National Institutes of Health) for pcDNA3-HA-JNK1; Brent Zanke (Ontario Cancer Institute, Canada) for pcDNA3-HA-Sek-AL; Robert Glassmann for anti-GST antiserum; Junichi Miyazaki (Touhoku University, Japan) for pCAGGS vector; Rosemary Williams for technical assistance; Raymond B. Birge for discussion; and Avery August for critical reading of the manuscript. S.T. was supported by the Sasagawa Medical Research Foundation and by the Charles H. Revson Foundation, and is on leave from Department of Pathology, Hokkaido University School of Medicine, Japan. This work is supported in part by Grant CA44356 from the National Institutes of Health.
ABBREVIATIONS
- JNK
Jun kinase
- MAP
mitogen-activating protein
- MAPK
MAP kinase
- SH2 and SH3
Src homology 2 and 3
- GST
glutathione S-transferase
- HA
hemagglutinin
References
- 1.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 2.Mayer B J, Hamaguchi M, Hanafusa H. Nature (London) 1988;332:272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda M, Tanaka S, Nagata S, Kojima A, Kurata T, Shibuya M. Mol Cell Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ten Hoeve J, Morris C, Heisterkamp N, Groffen J. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- 5.Birge R B, Knudsen B S, Besser D, Hanafusa H. Gene Cells. 1996;1:595–613. doi: 10.1046/j.1365-2443.1996.00258.x. [DOI] [PubMed] [Google Scholar]
- 6.Clark E A, Brugge J S. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda M, Mayer B J, Fukui Y, Hanafusa H. Science. 1990;248:1537–1539. doi: 10.1126/science.1694307. [DOI] [PubMed] [Google Scholar]
- 8.Mayer B J, Hanafusa H. J Virol. 1990;64:3581–3589. doi: 10.1128/jvi.64.8.3581-3589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, Matsuda M. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda M, Hashimoto Y, Muroya K, Hasegawa H, Kurata T, Tanaka S, Nakamura S, Hattori S. Mol Cell Biol. 1994;14:5495–5500. doi: 10.1128/mcb.14.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feller S M, Knudsen B, Hanafusa H. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 14.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 15.Greulich H, Hanafusa H. Cell Growth Differ. 1996;7:1443–1451. [PubMed] [Google Scholar]
- 16.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Nature (London) 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 17.Zanke B W, Boudreau K, Rubie E, Winnett E, Tibbles L A, Zon L, Kyriakis J, Liu F F, Woodgett J R. Curr Biol. 1996;6:606–613. doi: 10.1016/s0960-9822(02)00547-x. [DOI] [PubMed] [Google Scholar]
- 18.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. Proc Natl Acad Sci USA. 1996;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronheim A, Engelberg D, Li N, Al-Alawi N, Schlessinger J, Karin M. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 20.Derijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 21.Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, Kurata T, Matsuda M. Mol Cell Biol. 1996;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wienecke R, Maize J C, Shoarinejad F, Vass W C, Reed J, Bonifacino J S, Resau J H, Gunzburg J D, Yeung R S, DeClue J, E. Oncogene. 1996;13:913–923. [PubMed] [Google Scholar]
- 23.Xu X, Heidenreich O, Kitajima I, McGuire K, Li Q, Su B, Nerenberg M. Oncogene. 1996;13:135–142. [PubMed] [Google Scholar]
- 24.Cerione R A, Zheng Y. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson K M, Lemmon M A, Schlessinger J, Sigler P B. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 26.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 27.Ohtsuka T, Shimizu K, Yamamori B, Kuroda S, Takai Y. J Biol Chem. 1996;271:1258–1261. doi: 10.1074/jbc.271.3.1258. [DOI] [PubMed] [Google Scholar]
- 28.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1996;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 30.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Derijard B, Davis R J. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 31.Reedquist K A, Fukazawa T, Panchamoorthy G, Langdon W Y, Shoelson S E, Druker B J, Band H. J Biol Chem. 1996;271:8435–8442. doi: 10.1074/jbc.271.14.8435. [DOI] [PubMed] [Google Scholar]
- 32.Smit L, Horst G V D, Borst J. J Biol Chem. 1996;271:8564–8569. doi: 10.1074/jbc.271.15.8564. [DOI] [PubMed] [Google Scholar]
- 33.Knudsen B S, Zheng J, Feller S M, Mayer J P, Burrell S K, Cowburn D, Hanafusa H. EMBO J. 1995;14:2191–2198. doi: 10.1002/j.1460-2075.1995.tb07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka S, Hattori S, Kurata T, Nagashima K, Fukui Y, Nakamura S, Matsuda M. Mol Cell Biol. 1993;13:4409–4415. doi: 10.1128/mcb.13.7.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribon V, Saltiel A R. J Biol Chem. 1996;271:7375–7380. doi: 10.1074/jbc.271.13.7375. [DOI] [PubMed] [Google Scholar]
- 36.Khwaja A, Hallberg B, Warne P H, Downward J. Oncogene. 1996;12:2491–2498. [PubMed] [Google Scholar]
- 37.Petruzzelli L, Takami M, Herrera R. J Biol Chem. 1996;271:7796–7801. doi: 10.1074/jbc.271.13.7796. [DOI] [PubMed] [Google Scholar]
- 38.Nojima Y, Morino N, Mimura T, Hamasaki K, Furuya H, Sakai R, Sato T, Tachibana K, Morimoto C, Yazaki Y, Hirai H. J Biol Chem. 1996;270:15398–15402. doi: 10.1074/jbc.270.25.15398. [DOI] [PubMed] [Google Scholar]
- 39.Miyamoto S, Teramoto H, Coso O A, Gutkind J S, Burbelo P D, Akiyama S K, Yamada K M. J Cell Biol. 1996;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ten Hoeve J, Kaartinen V, Fioretos T, Haataja L, Voncken J W, Heisterkamp N, Groffen J. Cancer Res. 1994;54:2563–2567. [PubMed] [Google Scholar]
- 41.Salgia R, Uemura N, Okuda K, Li J L, Pisick E, Sattler M, de Jong R, Druker B, Heisterkamp N, Chen L B. J Biol Chem. 1995;270:29145–29150. doi: 10.1074/jbc.270.49.29145. [DOI] [PubMed] [Google Scholar]
- 42.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minden A, Lin A, Claret F-X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]