Abstract
Destruction of mitotic cyclins by ubiquitin-dependent proteolysis is required for cells to complete mitosis and enter interphase of the next cell cycle. In clam eggs, this process is catalyzed by a cyclin-selective ubiquitin carrier protein, E2-C, and the cyclosome/anaphase promoting complex (APC), a 20S particle containing cyclin-selective ubiquitin ligase activity. Here we report cloning a human homolog of E2-C, UbcH10, which shares 61% amino acid identity with clam E2-C and can substitute for clam E2-C in vitro. Dominant-negative clam E2-C and human UbcH10 proteins, created by altering the catalytic cysteine to serine, inhibit the in vitro ubiquitination and destruction of cyclin B in clam oocyte extracts. When transfected into mammalian cells, mutant UbcH10 inhibits the destruction of both cyclin A and B, arrests cells in M phase, and inhibits the onset of anaphase, presumably by blocking the ubiquitin-dependent proteolysis of proteins responsible for sister chromatid separation. Thus, E2-C/UbcH10-mediated ubiquitination is involved in both cdc2 inactivation and sister chromatid separation, processes that are normally coordinated during exit from mitosis.
The regulated destruction of mitotic cyclins A and B near the end of mitosis is essential for the inactivation of their partner kinase cdc2 and exit from mitosis into G1 of the next cell cycle (reviewed by ref. 1). As with most known cases of regulated proteolysis, cyclins are marked for destruction by the covalent addition of ubiquitin, a modification that targets them for recognition and proteolysis by the proteasome (2, 3). In this pathway, ubiquitin is activated by formation of a thioester with a cysteine residue of the ubiquitin activating enzyme E1. Ubiquitin is then transferred to one of several members of a family of E2 or ubiquitin carrier proteins (UBCs). Transfer of ubiquitin from a specific E2 to a specific target protein usually requires a third activity called E3 or ubiquitin ligase. Recent work using extracts of clam or frog eggs that reproduce the cell cycle stage-specific ubiquitination and destruction of mitotic cyclins has led to the identification of a novel cyclin-selective E2, called E2-C or UBC-x (4–6), and an unusual E3 activity that is part of a 20S particle called the cyclosome or anaphase promoting complex (APC) (7–9). Cyclosome/APC activity is the regulated component of this system, becoming activated by phosphorylation near the end of mitosis and turned off by dephosphorylation (10).
While cyclin destruction is essential for cdc2 inactivation, anaphase onset can proceed in the presence of nondestructible cyclin that lacks an N-terminal destruction box domain (11–15). Addition of this N-terminal domain blocks cyclin destruction and delays anaphase onset, suggesting that “glue” proteins responsible for maintaining sister chromatid cohesion are recognized and degraded by similar mechanisms (11). One such candidate protein, Cut 2, has now been definitively identified in fission yeast (16). Rapid degradation of Cut2 requires an N-terminal domain that can be replaced by that of cyclin B, and nondegradable Cut2 blocks sister chromatid separation, but not cdc2 inactivation or cell division. Furthermore, destruction of full-length Cut2 requires Cut9, a component of the 20S cyclosome/APC. These results suggest that the cell cycle stage-specific degradation of mitotic cyclins and Cut2, which carry out complementary roles in the exit from mitosis, are coordinated as a consequence of their being ubiquitinated by the same machinery at the same time in the cell cycle.
Here we have cloned the human homolog of the cyclin-selective E2, termed UbcH10, which shows 61% amino acid identity with clam E2-C and can substitute for clam E2-C in vitro. Dominant-negative E2-C/UbcH10, created by changing the catalytic cysteine to serine, blocks cyclin ubiquitination and destruction in vitro. When introduced into dividing cells, it inhibits the destruction of mitotic cyclins and causes cells to accumulate in mitosis before anaphase onset. These results show that E2-C/UbcH10 is involved in two normally coordinated processes, inactivation of cdc2 as initiated by destruction of the mitotic cyclins, and anaphase onset, as initiated by destruction of one or more proteins that maintain sister chromatid cohesion.
MATERIALS AND METHODS
Cloning Human E2-C/UbcH10.
A human HeLa cDNA λ ZAP library (Stratagene) was screened by PCR using a degenerate primer YE2C4 (5′-CARCARGARYTIMGIAC-3′, sense; R is A or G, Y is C or T, M is A or C and I is inosine), which corresponds to amino acids 36–41 (QQELRT) of clam E2-C, and vector primer T7 (5′-TAATACGACTCACTATAGGG-3′, antisense). Reaction products were used in a second nested PCR reaction using the primer YE2C4 and degenerate primer YE2C2 (5′-ATRTCIARRCAIATRTTICC-3′, antisense), which corresponds to amino acids 111–117 (GNICLDI) of clam E2-C. The resulting PCR product of 258 bp was cloned into plasmid vector pCRII (Invitrogen), sequenced, aligned with clam E2-C, and subsequently used to obtain six independently isolated full-length cDNA clones containing human E2-C, termed UbcH10.
In Vitro Mutagenesis.
To subclone UbcH10 into the bacterial expression vector pT7–7 (17), the coding region was amplified by PCR using the primers HSEN (5′-GGAATTCATATGGCTTCCCAAAACCGCG-3′, sense) and HSEC (5′-CCCAAGCTTATCAGGGCTCCTGGCTGGT-3′, antisense). HSEN encodes the first five amino acids of the UbcH10 open reading frame and contains an EcoRI restriction site followed by an NdeI site at the 5′ end. HSEC encodes the last five amino acids of the UbcH10 open reading frame followed by two stop codons, then a HindIII restriction site. The resulting PCR product was digested with NdeI and HindIII, ligated with NdeI/HindIII-cut pT7–7, and transformed into BL-21(DE3) pLysS cells (Novagen).
The UbcH10 C(114)S mutant was generated in two steps by PCR. The amino-terminal portion was amplified from the UbcH10 cDNA clone as above, using the primers HSEN and HSECSR (5′-GATGTCCAGGCTTATGTTACC-3′, antisense). The carboxyl-terminal portion was amplified using primers HSECSF (5′-GGTAACATAAGCCTGGACATC-3′, sense) and HSEC. HSECSR is the antisense sequence of HSECSF and both encode amino acids GNISLDI, which alters residue 114 of UbcH10 from cysteine to serine. To generate a full-length UbcH10 mutant clone the PCR products from the two reactions were mixed, denatured, and allowed to anneal at the GNISLDI overlap, then amplified with primers HSEN and HSEC. The full-length PCR product was digested with NdeI and HindIII and cloned into pT7–7 as described for wild-type UbcH10.
The corresponding clam E2-C mutant was generated by amplification of the amino-terminal portion of E2-C cDNA (5) using the primers CE2FULL (5′-GGGCATATGTCGGGACAAAATATAGATC-3′, sense) and CE2MUTR (5′-CCAGACTTATATTTCCTGACTG-3′, antisense). The carboxyl-terminal portion was amplified using primers CE2MUTF (5′-CAGTCAGGAAATATAAGTCTGG-3′, sense) and CE2REV (5′-GGGAAGCTTCTATTTATCACTCTGAGCCAG-3′, antisense). CE2MUTR has the antisense sequence of CE2MUTF and both encode amino acids ESGNISL, which alters residue 114 of E2-C from cysteine to serine. To generate a full-length E2-C C(114)S the PCR products from the first step were amplified with primers CE2FULL and CE2REV. The second step PCR product was digested with NdeI and HindIII and cloned into pT7–7.
For transfection into human cells, the AU1 epitope (DTYRYI) was added to the C terminus of wild-type UbcH10 and the C114-S mutant by PCR using the primers HSEN and HSEAUC (5′-GGGAAGCTTATCAAATGTACCTGTAGGTGTCGGGCTCCTGGCTGGTGA-3′, antisense). pT7–7 vectors containing the wild-type and mutant genes were used as templates. HSEAUC encodes the last six amino acids of the UbcH10 open reading frame followed by amino acids DTYRYI, two stop codons, then a HindIII restriction site. The resulting PCR product was digested with EcoRI and HindIII and ligated with EcoRI/HindIII-cut pJS55, a derivative of pSG5 (Stratagene) with a modified polylinker (18). All PCR-generated constructs were confirmed by sequencing.
Expression and Purification of Recombinant E2-Cs.
Four hundred mililiter cultures of bacteria containing expression vectors of the various E2-Cs were grown at 37°C in Luria–Bertani medium containing ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml). At an absorbance of 0.7 at 600 nm, isopropyl β-thiogalactoside (1 mM) was added, and incubation was continued for 3 h. Bacteria were pelleted, washed with PBS, and resuspended in 6 ml of 50 mM Tris·HCl, pH 7.2/1 mM DTT/1 mM EDTA/10 μg/ml leupeptin and chymostatin, and sonicated (four times 30 sec) and centrifuged at 15,000 × g for 10 min. All recombinant E2-Cs were in the supernatant fraction.
For purification, bacterial extracts were diluted with 4 volumes 10 mM potassium phosphate (pH 7.0) and 1 mM DTT, and applied to a column of DE-52 (Whatman) at a ratio of 5 mg of protein per ml of resin. Unadsorbed material was collected and concentrated by centrifuge ultrafiltration (Centriprep-10, Amicon) to 10 mg of protein/ml. This fraction (20–30 mg of protein) was applied to a 120-ml column of Superdex-75 (Pharmacia) equilibrated with 50 mM Tris·HCl, pH 7.4/1 mM EDTA/1 mM DTT. Fractions of 2.5 ml were collected at a flow rate of 1 ml/min. The various E2s eluted in fractions 28–32, well separated from the majority of bacterial proteins. All E2-C preparations were >95% homogenous.
Assay of E2-C Activity.
E2-C activity was assayed as described (7). Briefly, 10 μl reactions contained 40 mM Tris·HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, 0.5 mM ATP, 10 mM creatine phosphate, 50 μg/ml creatine kinase, 1 mg/ml reduced-carboxymethylated BSA, 20 μM ubiquitin, 3 μM ubiquitin-aldehyde, 1 μM okadaic acid, 1 pmol of E1, 1–2 pmol of 125I-cyclin B(13–91) (≈1–2 × 105 cpm), 10 μg of protein of fraction 1A from extracts of clam oocytes, and E2-C as specified. After incubation at 18°C for 60 min, samples were electrophoresed on 12.5% polyacrylamide gels followed by autoradiography and quantitation with a Fuji phosphorimager.
In Vivo Analysis.
COS cells were grown at 37°C under 5% CO2 in DMEM supplemented with 10% fetal bovine serum. For transfection, cells were maintained in log phase, and near-confluent cells were subcultured at a 1:4 dilution the day before transfection. Cells in 100-mm dishes were rinsed twice in serum-free DMEM and incubated for 30 min with 2.5 μg plasmid DNA, 1.5 ml DEAE Dextran (1 mg/ml) in Tris-buffered saline (25 mM Tris·HCl, pH 7.4/140 mM NaCl/5 mM KCl), and 1.5 ml serum-free DMEM. DNA was added to the DMEM first to prevent precipitation. The DNA mixture was removed, and the cells were incubated in DMEM containing 10% fetal bovine serum and 100 μg/ml chloroquine for 3–4 h. At the end of this period the cells were incubated in serum-containing DMEM until fixation or harvesting.
For immunofluorescence, glass coverslips were added to the transfection dishes before subculturing the cells. The coverslips were removed from the dishes at 48 h post-transfection and rinsed briefly with PBS. The cells were fixed for 15 min in 3.7% formaldehyde in PBS, then permeabilized by washing the coverslips four times with 0.1% Triton X-100 in PBS. Coverslips were incubated for 30 min in 1% BSA + 0.1% Triton X-100 in PBS, then incubated for 1 h with AU1 antibody (Babco, Richmond, CA) diluted 1/150 in the same solution. Coverslips were then washed four times with PBS + 0.1% Triton X-100 and incubated for 1 h in the dark with Cy3-conjugated goat anti-mouse antibody (Jackson ImmunoResearch) diluted 1/500 in PBS + 1% BSA + 0.1% Triton X-100. Cells were washed four times with PBS + 0.1% Triton X-100, then incubated for 1 min with 1 μg/ml Hoechst 33342 in PBS + 0.1% Triton X-100. Coverslips were mounted in 70% glycerol containing DABCO (1,4-diazabicyclo[2,2,2]octane, Sigma) as an antifading agent in PBS, sealed with nail polish, and viewed by fluorescence microscopy.
RNA encoding wild-type or mutant E2-C was injected into one cell of two-cell frog embryos as described (19). Embryos were collected at mid-late blastula stage, fixed, stained with Hoechst 33342, squashed, and visualized by fluorescence microscopy.
RESULTS
Cloning the Human Homolog of E2-C, UbcH10.
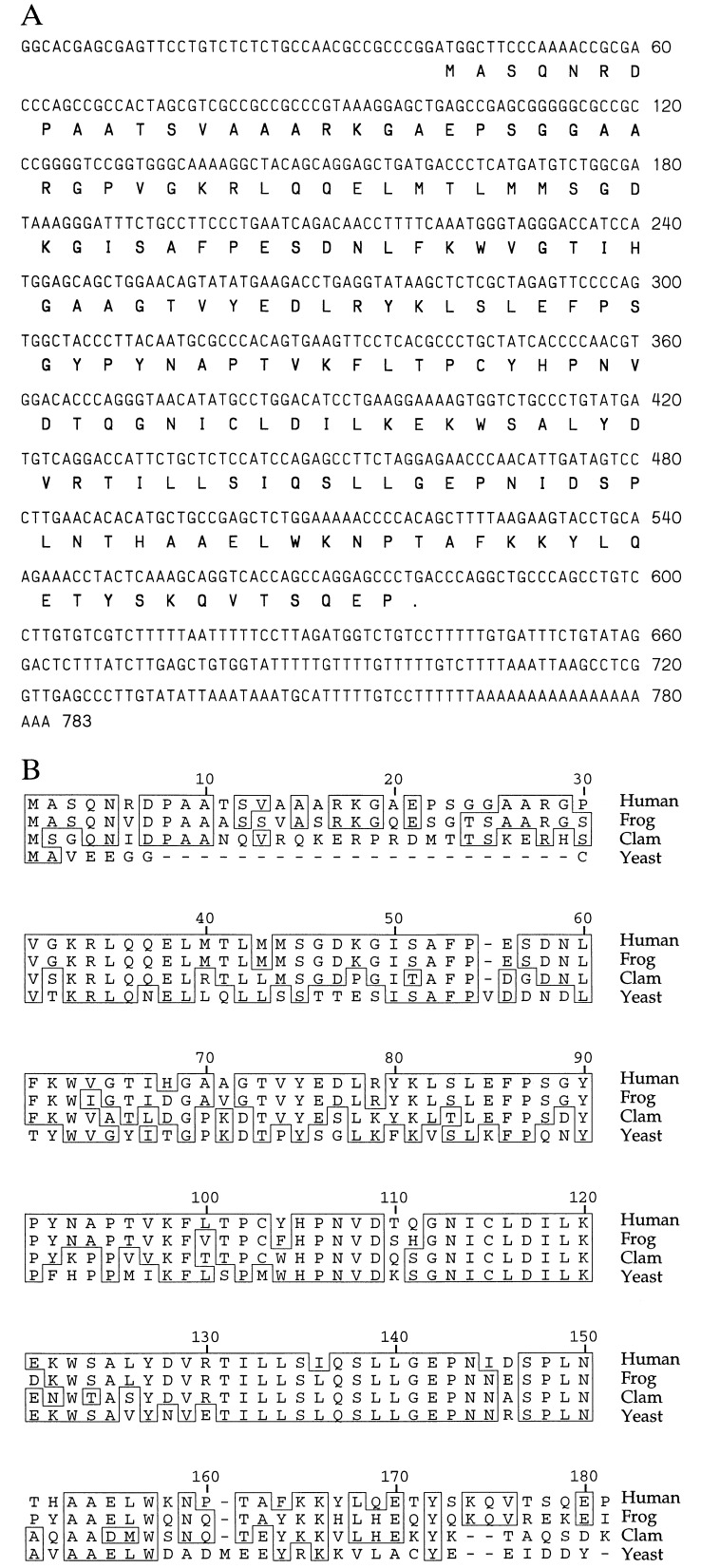
A HeLa cell cDNA library was screened for clones homologous to clam E2-C sequences, as described above. Three candidate clones of identical sequence encoding a 179-amino acid protein (Fig. 1 A) were found. A fourth contained a single nucleotide alteration of G to A at position 114, which alters amino acid 25 from glycine to aspartic acid.
Figure 1.
(A) Nucleotide and amino acid sequence of human UbcH10. (B) Comparison of the amino acid sequences of human UbcH10, clam E2-C, frog UBC-x, and a candidate yeast homolog.
The amino acid sequence of human E2-C was aligned with that of clam E2-C and a frog homolog, UBC-x (Fig. 1B). Human E2-C shares 80.4% overall amino acid sequence identity with UBC-x and 61% identity with clam E2-C, and contains the 30-amino acid N-terminal extension that distinguishes clam E2-C from other ubiquitin-conjugating enzymes (5). The human E2-C cDNA sequence is identical to that cited in ref. 6, and we refer to this enzyme as UbcH10, as it is the 10th human UBC to be identified. The most closely related sequence in the yeast genome is a novel UBC family member located on contig X95720, GenInfo ID: 1199839, c3063..2593. It shows 53% overall amino acid identity with human UbcH10, but lacks the distinctive N-terminal extension found in other E2-Cs (Fig. 1B).
UbcH10 Can Substitute for Clam E2-C in the Ubiquitination of Cyclin B in Vitro.
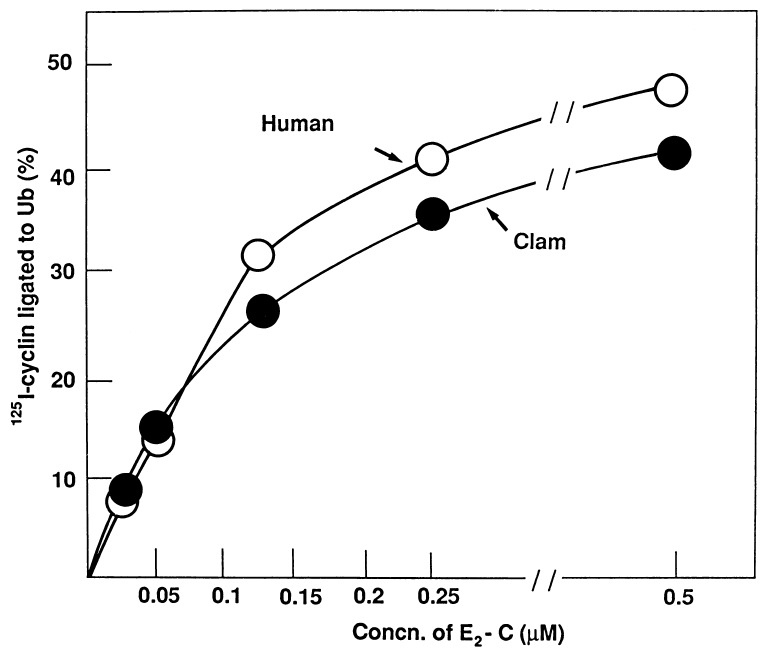
Recombinant UbcH10 was tested for its ability to support cyclin ubiquitination in clam cell free extracts in vitro (see Materials and Methods). Increasing concentrations of UbcH10 or clam E2-C were added to a reaction mixture containing ubiquitin, E1, the 20S E3-containing particle from M-phase clam oocytes, and a radiolabeled cyclin fragment. Recombinant human UbcH10 is as effective as its clam counterpart: half-maximal rates were attained at around 0.15 μM in both cases (Fig. 2). This result indicates that recombinant human UbcH10 is highly active, and that E2-C function is conserved in evolution.
Figure 2.
Stimulation of cyclin-ubiquitin ligation by recombinant human E2-C, UbcH10. Cyclin-ubiquitin ligation was assayed as described in the text. Recombinant, purified human UbcH10 (○) or clam E2-C (•) was added at the concentrations indicated.
The UbcH10 C(114)S Mutant Is a Dominant-Negative Inhibitor of Cyclin Ubiquitination in Vitro.
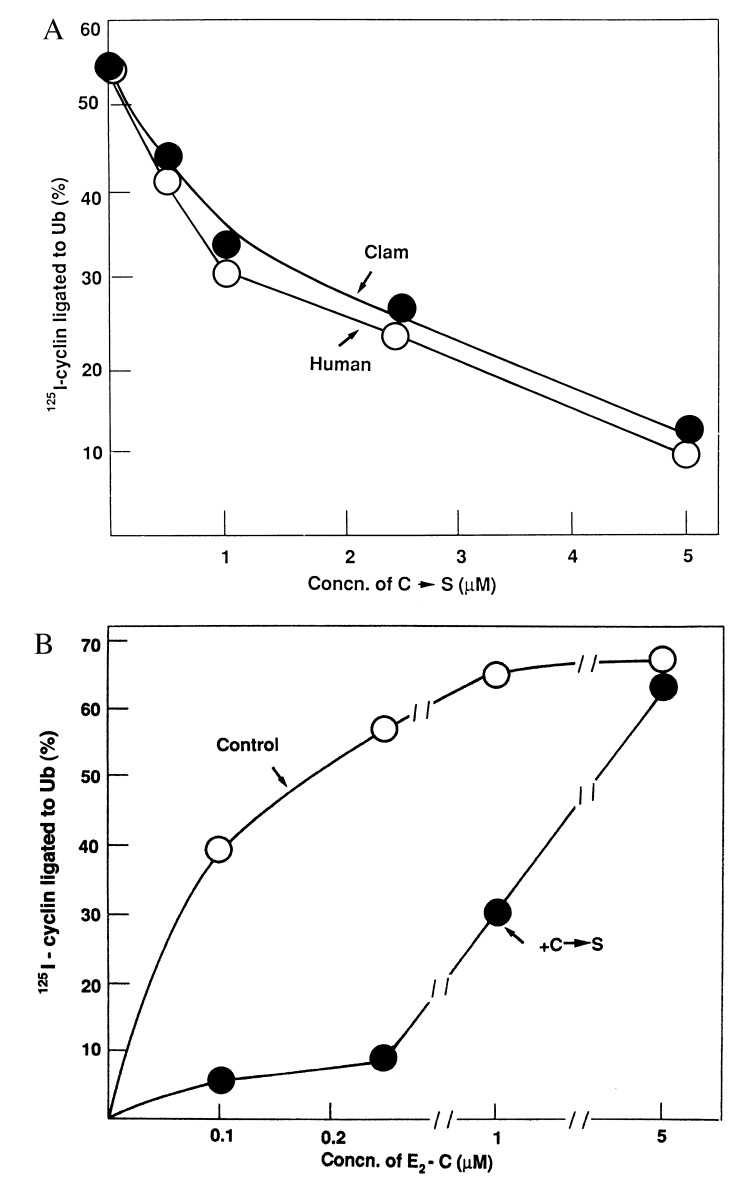
All E2 enzymes contain an active site cysteine residue near the middle of the coding sequence that is used for ubiquitin-thiolester formation. Alteration to alanine results in the loss of E2 activity (20, 21). In UBC2 (RAD6) alteration to serine allows formation of a ubiquitin ester that cannot be transferred to target proteins (22). In the case of CDC34, a UBC required for G1 progression in yeast, simultaneous replacement of cysteine and a nearby leucine residue with serine created a dominant-negative mutant allele that arrested the cell cycle (23). The effect of the single cysteine → serine (C→S) mutation in both clam E2-C and human UbcH10 on cyclin ubiquitination was assayed. In Fig. 3A, all incubations contained a constant concentration (0.25 μM) of wild-type clam E2-C, and the concentrations of human or clam C→S constructs were varied. Mutant enzymes from both species inhibited this system with equal effectiveness. Nearly complete inhibition required about 20-fold excess of the C→S mutants in both cases.
Figure 3.
Inhibition of cyclin-ubiquitin ligation by C→S mutants of human UbcH10 and clam E2-C. (A) All incubations contained 0.25 μM wild-type clam E2-C. Where indicated, increasing concentrations of recombinant, purified C→S mutants of human UbcH10 (○) or clam E2-C (•) were added. Cyclin-ubiquitin ligation was assayed as described in the text. (B) Recombinant, purified human UbcH10 was added at the concentrations indicated in the absence (○, Control) or presence (•) of human UbcH10 C→S mutant (1 μM).
To test if the mutants act as competitive or noncompetitive inhibitors of cyclin-ubiquitin ligation, a constant concentration (1 μM) of the human C→S mutant was examined at increasing levels of wild-type human UbcH10. As shown in Fig. 3B, 1 μM C→S mutant strongly inhibited cyclin-ubiquitin ligation at low concentrations of wild-type UbcH10, but inhibition was overcome by high concentrations of wild-type UbcH10. This indicates a competition between wild-type and mutant UbcH10 on a common target.
The UbcH10 C(114)S Mutant Inhibits the Degradation of Native, Full-Length Cyclin B in Clam Extracts.
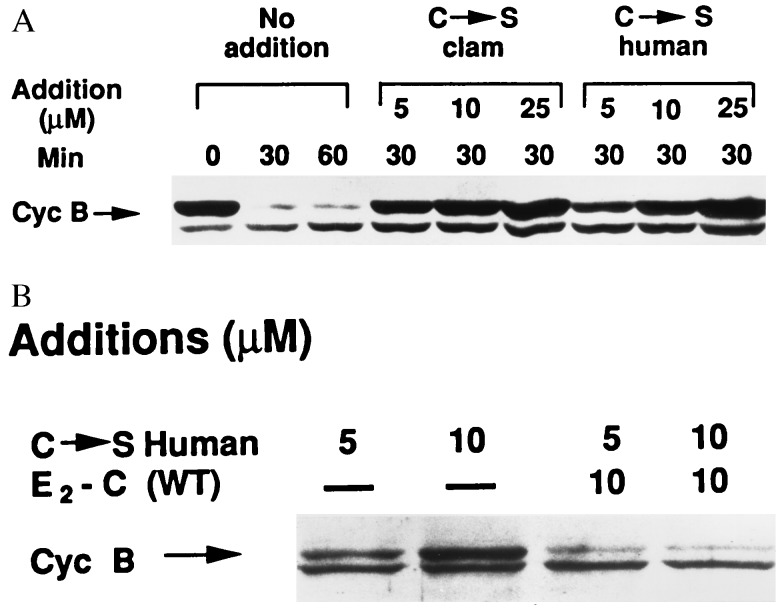
In all experiments described above, an N-terminal fragment of sea urchin cyclin B (24) was used as a convenient substrate, but this assay does not test the activity of UbcH10 toward natural, full-length cyclin B. In addition, the experiments were done using purified components in a reconstituted system, so it is possible that the effects of the C→S mutant could be influenced by additional factors present in unfractionated cytosol. Thus, we tested the effects of the C→S mutants on the degradation of endogenous cyclin B in crude extracts of clam oocytes, as monitored by immunoblotting. In the control incubation, degradation of endogenous cyclin B was essentially complete by 30 min; degradation was effectively blocked by increasing concentrations of either clam or human C→S derivatives (Fig. 4A). As with purified components, a large excess of the C→S mutant was required for complete inhibition of cyclin degradation (the concentration of endogenous E2-C in clam extracts is about 0.5 μM, data not shown). Similarly, inhibition of the degradation of endogenous cyclin B by the C→S mutant was overcome by the addition of excess wild-type human UbcH10 (Fig. 4B).
Figure 4.
The degradation of endogenous cyclin B in mitotic clam oocyte extracts is inhibited by C→S mutants of human UbcH10 and clam E2-C. Incubations contained in a volume of 12 μl: 10 μl extract of early M-phase clam oocytes (75 min postfertilization, see ref. 7), 2 mM MgCl2, 1 mM ATP, 20 mM creatine phosphate, 50 μg/ml creatine kinase, 0.5 mM DTT, 250 μM emetine, and recombinant E2-C derivatives as specified. After incubation at 16°C for the time periods indicated, samples of 2 μl were withdrawn into SDS electrophoresis sample buffer. The samples were separated on a 10% polyacrylamide gel, blotted on nitrocellulose, probed with an antiserum directed against clam cyclin B (25), and detected by ECL. (A) Effects of human UbcH10 and clam E2-C C→S mutants on the degradation of clam cyclin B. (B) Reversal of the effects of the C→S mutant by wild-type human UbcH10. The various recombinant Ubc derivatives were added at the concentrations indicated. The position of cyclin B is indicated. The identity of the crossreactive protein that migrates ahead of cyclin B is not known. WT, wild type.
UbcH10 C(114)S Is a Dominant-negative Inhibitor of Cell Cycle Progression in Vivo, Blocking Both Destruction of Mitotic Cyclins A and B and the Onset of Anaphase.
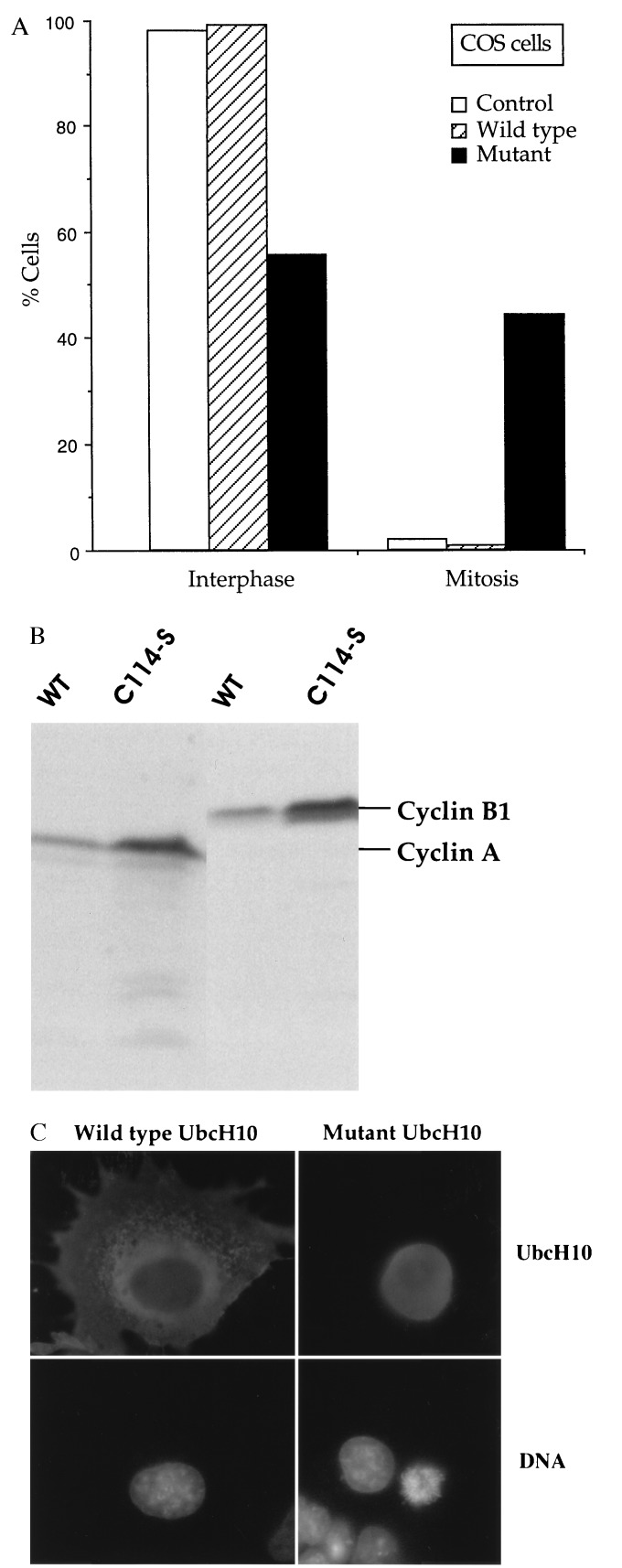
The ability of the C(114)S mutant to affect cell cycle progression in living cells was tested in two different systems: the somatic cell cycle of mammalian tissue culture cells and the rapid embryonic cell cycle of frog eggs. COS cells were transfected with AU1-tagged wild type or mutant UbcH10, and 48 h later the distribution of transfectants in interphase vs. mitosis was monitored by microscopy. Individual transfected cells, identified by staining with AU1 antibody, were scored as being in interphase (flattened cells, intact nucleus, decondensed chromatin) or mitosis (rounded cells, no obvious nuclear envelope, condensed chromosomes). About 1% of cells transfected with wild-type-UbcH10 were in mitosis, similar to 2% seen in mock transfected cultures (Fig. 5A). In striking contrast, nearly 50% of cells transfected with the C(114)S mutant had accumulated in mitosis (Fig. 5A), with most showing chromosomes in preanaphase arrays (Fig. 5C). Immunoblots showed that the C(114)S mutant increased the levels of both cyclin A and B, suggesting inhibition of their degradation (Fig. 5B). Because the mutant acts as a competitive inhibitor, requiring more than 10-fold excess to give 90% inhibition of cyclin ubiquitination in vitro, we cannot say whether cells remaining in interphase are ones that had proceeded through mitosis, because the amount of the C(114)S mutant was insufficient to give a complete mitotic arrest or because the mutant blocks a second cell cycle transition in interphase as well. Investigation of this question is underway.
Figure 5.
Effect of dominant-negative human UbcH10 on cell cycle progression in mammalian somatic tissue culture cells. (A) COS cells were mock-transfected (control) or transfected with AU1-tagged wild-type (WT) or dominant-negative (mutant) UbcH10, and after 48 h were fixed and stained with AU1 antibodies and Hoechst 33342.Two hundred cells each were scored for the percent cells in interphase vs. M phase. (B) Immunoblots of total protein extracts from cells transfected with either wild-type or dominant-negative AU1-tagged UbcH10, reacted with cyclin A or B antibodies. (C) Morphologies of cells transfected with either wild-type (Left) or dominant-negative UbcH10 (Right). Cells were stained with AU1 antibodies (Top) to identify transfected cells, and with Hoechst dye (Bottom) to monitor chromosomes.
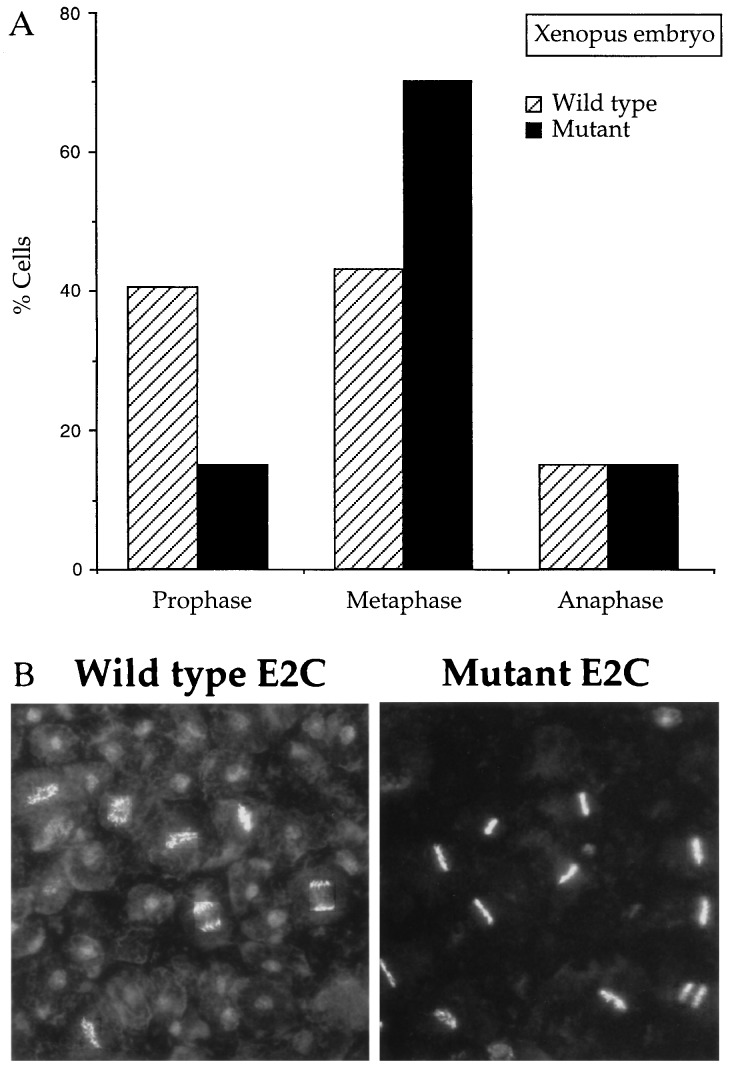
Injection of dominant-negative clam E2-C into one of the two cells of a dividing two-cell frog embryo slowed the rate of cell division (data not shown). The better chromosome morphology of these cells allowed us to examine the arrest point more carefully. Injected embryos were collected at mid-late blastula stages, fixed, stained with Hoechst 33342, and squashed to examine chromosome spreads. In embryos injected with wild-type E2-C, chromosomes in M phase showed the following distribution: 40% in premetaphase, 45% in metaphase, and 15% in anaphase. Embryos injected with the mutant E2-C showed a striking reduction in the percentage of premetaphase arrays coupled with a corresponding accumulation of metaphase figures (Fig. 6A). Typical fields of chromosomes are shown in Fig. 6B.
Figure 6.
Hoechst-stained fields of cells from embryos injected with RNA encoding wild-type (striped bars) or dominant-negative mutant (black bars) clam E2-C. Fertilized Xenopus eggs were injected with 10 ng RNA encoding wild-type or dominant-negative clam E2-C and cultured to late blastula stage. Cells were fixed and stained with Hoechst 33342. (A) Two hundred cells in M phase were scored for percent in prophase, metaphase, and anaphase/telophase. (B) A field of Hoechst-stained chromosomes from embryos injected with wild-type (Left) and dominant-negative mutant (Right) RNA.
Previous work has established that cyclin destruction is required for cdc2 inactivation, which, in turn, leads to chromosome decondensation, spindle disassembly, and cytokinesis, but that anaphase onset can proceed independently of cyclin destruction. The results presented here establish conclusively that E2-C/UbcH10 is involved in cyclin destruction in vivo, both in somatic and embryonic cell cycles, and that it is required for a second, normally concurrent event that results in the onset of anaphase.
DISCUSSION
Three principal findings are reported here. First, we have cloned the human cyclin-selective ubiquitin conjugating enzyme UbcH10. Like its clam (E2-C) and frog (Ubc-x) counterparts, UbcH10 contains the distinctive, highly conserved N-terminal extension. Second, we have created mutant E2-C/UbcH10 proteins that function as competitive inhibitors of cyclin B ubiquitination and degradation in clam oocyte cell-free extracts and inhibit destruction of both cyclin A and B in vivo. Third, these mutant proteins act as dominant negative inhibitors of cell cycle progression in vivo, blocking or delaying anaphase onset in both the somatic cell cycles of mammalian tissue culture cells and the rapid, abbreviated embryonic cell cycles of fertilized frog eggs. Thus, dominant-negative E2-C/UbcH10 blocks both cdc2 inactivation and anaphase onset, two processes that are normally coordinated, but can be uncoupled experimentally (11–13) and in specialized cell cycles (reviewed in ref. 26).
E2-C (4–6) and the E3 (ubiquitin ligase) activity associated with the 20S cyclosome/APC particle (7, 8) were identified originally as components responsible for the highly selective and temporally regulated ubiquitination of mitotic cyclins. Cyclin destruction leads to the release of inactive, monomeric cdc2, and several, but not all, events of exit from mitosis. In both yeast and animal cells, expression of nondegradable mitotic cyclins prevents cdc2 inactivation, chromosome decondensation, spindle disassembly, and cytokinesis, but does not block anaphase onset (11–13, 24, 27, 28). Anaphase onset is thought to be regulated by proteolysis of unidentified target proteins that are needed to maintain sister chromatid cohesion (11). The fission yeast protein cut2 is one such candidate (16): cut2 is destroyed during anaphase and the expression of nondegradable cut2 blocks sister chromatid separation, but not cdc2 inactivation or cell division. Four results argue that degradation of cut2 in fission yeast and presumed functional homologs in other organisms is carried out by the cyclin destruction machinery and represents a second branch of the pathway regulated by this machinery. First, expression of a cyclin B N-terminal domain, which contains the sequences recognized by the cyclin destruction machinery, blocks cdc2 inactivation and delays anaphase onset (11). Second, this domain in cyclin can substitute for a related domain in cut2 (16). Third, cut2 degradation requires cut 9, the fission yeast homolog of cdc16, a component of the cyclosome/APC (8, 9, 16). Fourth, as shown here, both cyclin destruction and anaphase onset are blocked by expression of dominant-negative mutant E2-C/UbcH10, the other known component of the cyclin destruction machinery (4, 5). Taken together, these results argue that activation of the cyclosome/APC near the end of mitosis controls two separate and usually concurrent processes by regulated ubiquitination of two different types of target proteins, mitotic cyclins and proteins maintaining sister chromosome cohesion.
At first glance, this mechanism would appear to explain both the molecular and temporal coordination of cdc2 inactivation and anaphase onset, but this cannot be the whole story. In many systems, cyclin A disappears in advance of cyclin B (29–32) but, as shown here, introduction of dominant-negative E2-C/UbcH10 blocks destruction of both cyclin A and B. This apparent discrepancy could be resolved by the existence of further regulatory elements that control the exact timing of the ubiquitination or ubiquitin-dependent proteolysis of different target proteins.
Acknowledgments
We thank Ricardo Bastos and Changyu Wang for the HeLa cell cDNA library. We are especially grateful for Malcolm Whitman for assistance with the frog embryo injections. This work was supported by National Institutes of Health Grant HD-23696 (J.V.R.), the United States–Israel Binational Science Foundation and the Israel Science Foundation (A.H.), and a Wellcome International Prize Research Fellowship (REF 043779/JMW/MH) to F.M.T.
ABBREVIATIONS
- UBC
ubiquitin carrier protein
- APC
anaphase promoting complex
- C→S mutant
cysteine → serine mutant
Footnotes
References
- 1.Murray A. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 3.Deshaies R J. Curr Op Cell Biol. 1995;7:781–789. doi: 10.1016/0955-0674(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen L H, Luca F C, Ruderman J V, Eytan E. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- 5.Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman J V. Proc Natl Acad Sci USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, King R W, Peters J-M, Kirschner M W. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 7.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King R, Peters J, Tugendreich S M, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 9.Zachariae W, Nasmyth K. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahav-Baratz S, Sudakin V, Ruderman J V, Hershko A. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holloway S L, Glotzer M, King R W, Murray A W. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- 12.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rimmington G, Dalby B, Glover D M. J Cell Sci. 1994;107:2729–2738. doi: 10.1242/jcs.107.10.2729. [DOI] [PubMed] [Google Scholar]
- 14.Sigrist S, Jacobs H, Stratman R, Lehner C F. EMBO J. 1995;14:4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irniger S, Piatti S, Michaelis C, Nasmyth K. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 16.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Nature (London) 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 17.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sparkowski J, Anders J, Schlegel R. J Virol. 1994;69:6120–6123. doi: 10.1128/jvi.68.9.6120-6123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaBonne C, Burke B, Whitman M. Development. 1995;121:1472–1486. doi: 10.1242/dev.121.5.1475. [DOI] [PubMed] [Google Scholar]
- 20.Sung P, Prakash S, Prakash L. Proc Natl Acad Sci USA. 1990;87:2695–2699. doi: 10.1073/pnas.87.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan M L, Vierstra R D. J Biol Chem. 1991;266:23878–23885. [PubMed] [Google Scholar]
- 22.Sung P, Prakash S, Prakash L. J Mol Biol. 1991;221:745–749. doi: 10.1016/0022-2836(91)80169-u. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee A, Deshaies R J, Chau V. J Biol Chem. 1995;270:26209–26215. doi: 10.1074/jbc.270.44.26209. [DOI] [PubMed] [Google Scholar]
- 24.Glotzer M, Murray A W, Kirschner M W. Nature (London) 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 25.Westendorf J M, Swenson K I, Ruderman J V. J Cell Biol. 1989;108:1431–1444. doi: 10.1083/jcb.108.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray A, Hunt T. The Cell Cycle. New York: Freeman; 1993. [Google Scholar]
- 27.Murray A W, Solomon M J, Kirschner M W. Nature (London) 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 28.Luca F C, Shibuya E K, Dohrmann C D, Ruderman J V. EMBO J. 1991;10:4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans T, Rosenthal E T, Youngblom J, Distel D, Hunt T. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 30.Pines P, Hunter T. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pagano M, Pepperkok R, Verde F, Ansorje W, Draetta G. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girard F, Fernandez A, Lamb N. J Cell Sci. 1995;108:2599–2608. doi: 10.1242/jcs.108.7.2599. [DOI] [PubMed] [Google Scholar]