Abstract
Megalin is a large cell surface receptor that mediates the binding and internalization of a number of structurally and functionally distinct ligands from the lipoprotein and protease:protease inhibitor families. To begin to address how megalin is able to bind ligands with unique structurally properties, we have mapped a binding site for apolipoprotein E (apoE)-β very low density lipoprotein (βVLDL), lipoprotein lipase, aprotinin, lactoferrin, and the receptor-associated protein (RAP) within the primary sequence of the receptor. RAP is known to inhibit the binding of all ligands to megalin. We identified a ligand-binding site on megalin by raising mAb against purified megalin, selected for a mAb whose binding to megalin is inhibited by RAP, and mapped the epitope for this mAb. mAb AC10 inhibited the binding of apoE-βVLDL, lipoprotein lipase, aprotinin, and lactoferrin to megalin in a concentration-dependent manner. When cDNA fragments encoding the four cysteine-rich ligand-binding repeats in megalin were expressed in a baculovirus system and immunoblotted with AC10, it recognized only the second cluster of ligand-binding repeats. The location of the epitope recognized by mAb AC10 within this domain was pinpointed to amino acids 1111–1210. From these studies we conclude that the binding of apoE-βVLDL, lactoferrin, aprotinin, lipoprotein lipase, and RAP to megalin is either competitively or sterically inhibited by mAb AC10 suggesting that these ligands bind to the same or closely overlapping sites within the second cluster of ligand-binding repeats.
Megalin (gp330) is a cell surface receptor (1, 2) originally identified as a primary antigen in the renal autoimmune disorder known as Heymann nephritis (3–5). Immunocytochemical studies have shown that megalin is abundantly expressed in the kidney, specifically in the cells of the proximal tubule and glomerular epithelium (2, 4–6), but is also found to a lesser extent in a variety of epithelia including, epididymis, type II pneumocytes, labyrinth epithelium of the ear, ciliary body of the eye, lacrimal gland, thyroid, parathyroid, and the choroid plexus (7–9). Megalin is located in clathrin coated pits (5, 6) where it serves as an endocytic receptor for a number of structurally and functionally diverse proteins from the lipoprotein and protease:protease inhibitor families (1, 2). The list of ligands includes apolipoproteins B100 (10, 11), E (12), and J (13), lipoprotein lipase (9), lactoferrin (12), plasminogen (14), plasminogen activators and their inhibitor, type-I plasminogen activator inhibitor (12, 15–18), aprotinin (16), polybasic drugs such as polymyxin B and gentamycin (19), and several polypeptide hormones found in the glomerular filtrate (20). Recent studies suggest that megalin plays a direct role in a number of important receptor-mediated processes including the reabsorption of filtered proteins (15, 19, 20) and calcium (21) by cells of the proximal tubule, endocytosis of proteins by yolk sac epithelia (22), and clearance of proteases and protease inhibitor complexes from the alveolar space by type II pneumocytes (16, 23). Megalin may also function as a calcium sensor in the parathyroid (24).
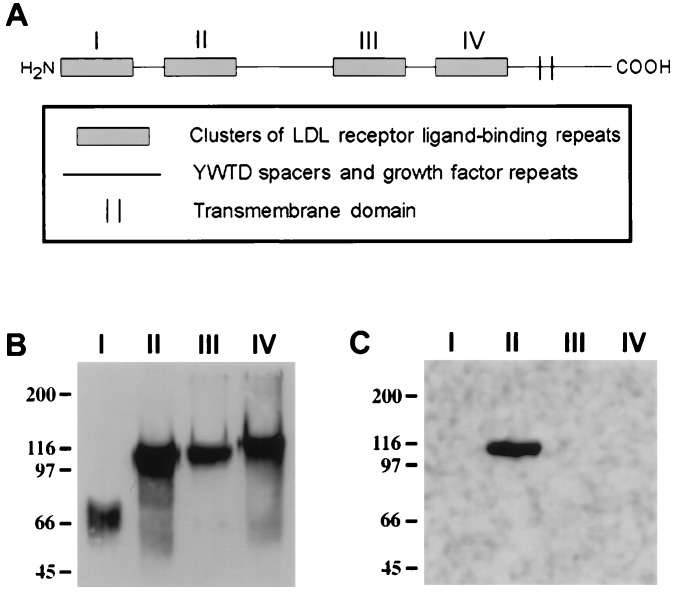
Sequence analysis of the cDNA for megalin (25) reveals that it is a type I transmembrane protein containing an extracellular region (4400 amino acids), a single transmembrane domain (22 amino acids), and a C-terminal cytoplasmic tail (213 amino acids). The extracellular domain of megalin contains the structural motifs characteristic of members of the low density lipoprotein (LDL) receptor family including four clusters of cysteine-rich LDL receptor ligand-binding repeats, growth factor repeats, an EGF repeat, and YWTD spacer regions.
The ability of megalin to bind such a diverse group of ligands raises several important questions concerning the mechanism of receptor–ligand interactions. Do the putative LDL receptor ligand-binding domains in megalin serve as sites for ligand binding? Do the ligands interact with distinct domains or bind to a common site on megalin? Do some or all of the ligands bind to more than one site on the receptor? To address these questions and to develop a better understanding of the molecular interactions between megalin and its ligands we generated a megalin-specific mAb that inhibited ligand binding and used it to map a common binding site on the receptor for several ligands.
MATERIALS AND METHODS
Reagents.
Rabbit apolipoprotein E (apoE) enriched β very low density lipoprotein (βVLDL) was a generous gift from Joseph Witztum (University of California, San Diego). Bovine lactoferrin, aprotinin, and lipoprotein lipase were purchased from Sigma. The anti-megalin polyclonal and monoclonal (mAb D155) antibodies were generated against megalin as described (5). Alkaline phosphatase-conjugated goat anti-mouse and horseradish peroxidase-conjugated goat anti-rabbit IgG were obtained from Bio-Rad.
Generation of mAb.
Rat microvilli were prepared as previously described (5), and proteins were extracted by incubating microvillar fractions for 1 h with 1% deoxycholate in 50 mM Tris·HCl, pH 8.0, followed by centrifugation (10,000 × g, 20 min, 4°C), the supernatant was adjusted to 5 mM CaCl2 and incubated with benzamidine-Sepharose (Sigma). The column was washed with 50 mM Tris (pH 8.0), 0.1% deoxycholate, and bound proteins were eluted with 50 mM Tris·HCl (pH 8.0), 0.1% deoxycholate, 10 mM EDTA, 8 M urea. Eluates were dialyzed exhaustively against phosphate buffered saline, and proteins were precipitated with acetone. The presence of megalin in the precipitate was confirmed by SDS/PAGE and immunoblot analysis with anti-megalin antibodies. Balb/C mice were immunized by intraperitoneal injections with 80 μg of the megalin-enriched preparation emulsified in complete Freund’s adjuvant (Sigma). Following two boosts, sera were obtained from tail veins and checked for immunoreactivity against megalin by immunoblotting. Hybridomas were generated by standard procedures (26). IgG was purified from ascites by protein A-affinity chromatography as described by the manufacturer (Bio-Rad).
Megalin Purification and Radioiodination.
Megalin was purified to homogeneity by mAb affinity chromatography as previously described (27), and radiolabeled using Na125I (Amersham) and the Iodo-Bead method according to the manufacturer’s instructions (Pierce). Specific activities were routinely 10,000–20,000 cpm/ng protein.
ELISA Procedure.
96-well microtiter plates (Corning) were coated with 1 μg mAb affinity-purified megalin per well for 16 h at 4°C. Wells were rinsed (3×) with phosphate buffered saline, and immobilized megalin was incubated with selected hybridoma supernatants in the presence or absence of 25 μg receptor-associated protein–glutathione S-transferase (RAP–GST) for 2 h at 23°C. After rinsing to remove unbound proteins, bound mAbs were detected with an alkaline phosphatase goat anti-mouse conjugate according to the manufacturer’s instructions (Bio-Rad).
Immunoblotting.
Proteins were added to Laemmli sample buffer containing 4% 2-mercaptoethanol, separated by SDS/PAGE, and electrotransferred to polyvinylidene difluoride (PVDF) membranes (Millipore) using a wet tank transfer method (Bio-Rad). Membranes were blocked with PBS, 0.1% Tween-20, 3% BSA for 1 h at 23°C, and incubated with primary antibodies for 2 h at 23°C. Membranes were then washed 3× (10 min), and bound primary antibodies were detected with either horseradish peroxidase- or alkaline phosphatase-conjugated secondary antibodies (40 min) followed by chemiluminescence (Pierce) or colorimetric (Bio-Rad) detection, respectively.
Solid-Phase Assay.
Ligands were diluted into buffer A (20 mM Hepes, pH 7.4/150 mM NaCl/2 mM CaCl2) and immobilized onto 96-well microtiter plates by incubation at 4°C for 16 h. Ligand coating concentrations were as follows: apoE-βVLDL, 100 μg/ml; lactoferrin, 50 μg/ml; aprotinin, lipoprotein lipase, and RAP–GST, 20 μg/ml. Nonspecific sites were blocked by adding buffer A containing 10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane-sulfonate, 3% BSA to wells for 1 h at 23°C. Radioiodinated megalin (5 × 106 cpm/ml) was preincubated with the indicated concentrations of mAb AC10 for 1 h at 23°C, then incubated with the immobilized ligands for an additional 3 h at 23°C. After rinsing, bound 125I-megalin was released with 2 N NaOH and quantitated by gamma counting.
Recombinant Protein Expression.
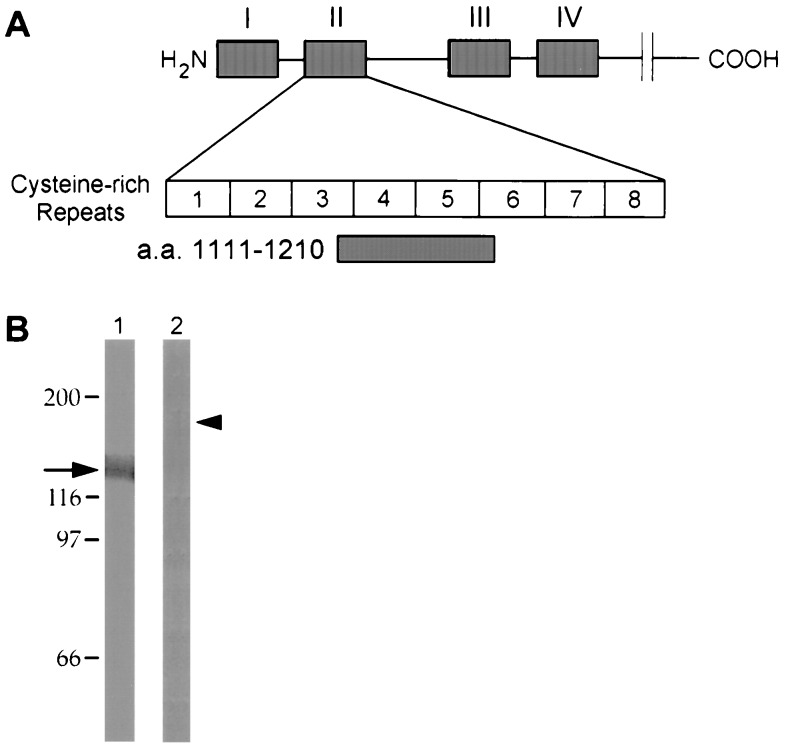
RAP–GST was isolated from a prokaryotic expression system by glutathione affinity chromatography (28). Recombinant baculoviruses expressing cDNA fragments encoding the first, second, third, or fourth clusters of ligand-binding repeats (I–IV) (Fig. 4A) were prepared and expressed in Sf9 insect cells as described (29). The expressed proteins were affinity-purified by Ni2+ chromatography (29). β-Galactosidase (β-gal) fusion proteins containing the complete RAP sequence (C14) or megalin amino acids 1111–1210 were generated as described (30).
Figure 4.
The AC10 epitope is contained within the second cluster of ligand-binding repeats in megalin. Megalin contains four clusters (I–IV) of cysteine-rich ligand-binding repeats in its extracellular domain which represent putative ligand-binding sites (A). Each of the four clusters was expressed in Sf9 cells using a polyhistidine tag in a baculovirus system and purified to homogeneity using Ni-affinity chromatography. The recombinant proteins were separated by SDS/7.5% polyacrylamide gel, transferred to PVDF, and immunoblotted with an anti-megalin polyclonal antibody (B) or mAb AC10 (C). Bound antibody was visualized by chemiluminescence. The polyclonal antibody to megalin recognized all four fragments (I–IV) which migrated with relative electrophoretic mobilities of 70, 110, 105, and 115 kDa, respectively. AC10 recognized only fragment II, the second cluster of ligand-binding repeats.
RESULTS
Identification of a mAb Whose Binding to Megalin Is Inhibited by RAP.
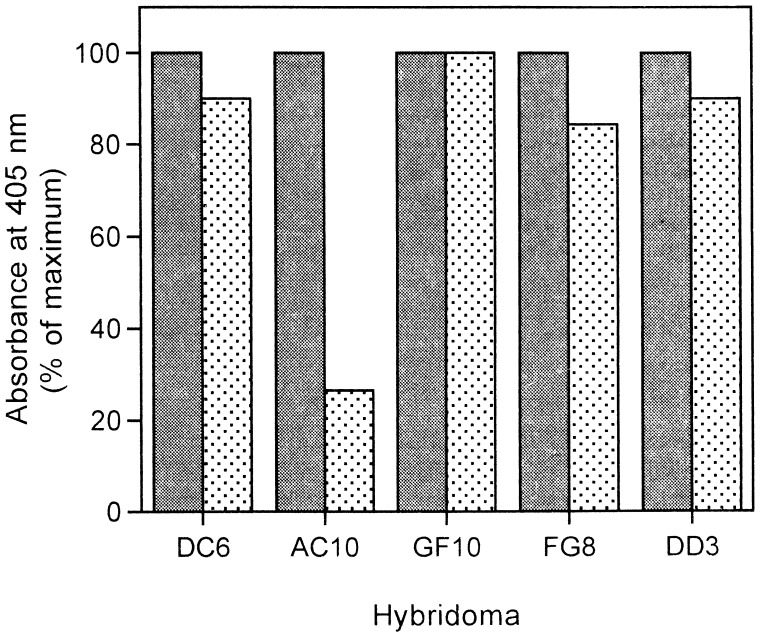
The binding of all known ligands to megalin is efficiently competed for by RAP (9, 15, 18, 31) suggesting that RAP and other ligands have the same or closely overlapping binding sites on megalin. To identify the binding site(s), we generated mAbs against purified megalin and selected for those whose binding to megalin is inhibited by RAP. Initially, we identified 170 hybridomas that produced antibodies specific for megalin based on immunoblotting. To identify those that compete for ligand binding, we performed an ELISA on immobilized megalin with hybridoma supernatants and quantitated the amount of mAb bound to the receptor in the presence or absence of purified RAP–GST (Fig. 1). Binding of most of the mAbs was unaffected by the presence of RAP–GST. However, the binding of one mAb, AC10, was inhibited ≈75% by RAP–GST, suggesting that this antibody specifically recognizes a RAP site on megalin.
Figure 1.
Identification of a mAb whose binding to megalin is inhibited by RAP. Purified megalin was immobilized onto microtiter plates and incubated with various hybridoma culture supernatants in the presence (stippled bars) or absence (solid bars) of RAP. Bound mAbs were quantitated by an alkaline phosphatase-conjugated secondary antibody followed by colorimetric detection. One mAb (GF10) bound efficiently to megalin, but its binding was unaffected by RAP. The binding of other mAbs (DC6, FG8, DD3) was only partially (10–15%) inhibited by RAP. However, the binding of one mAb (AC10) to megalin was reduced by ≈75% in the presence of RAP.
Megalin-Specific Ligands Compete for the Binding of AC10.
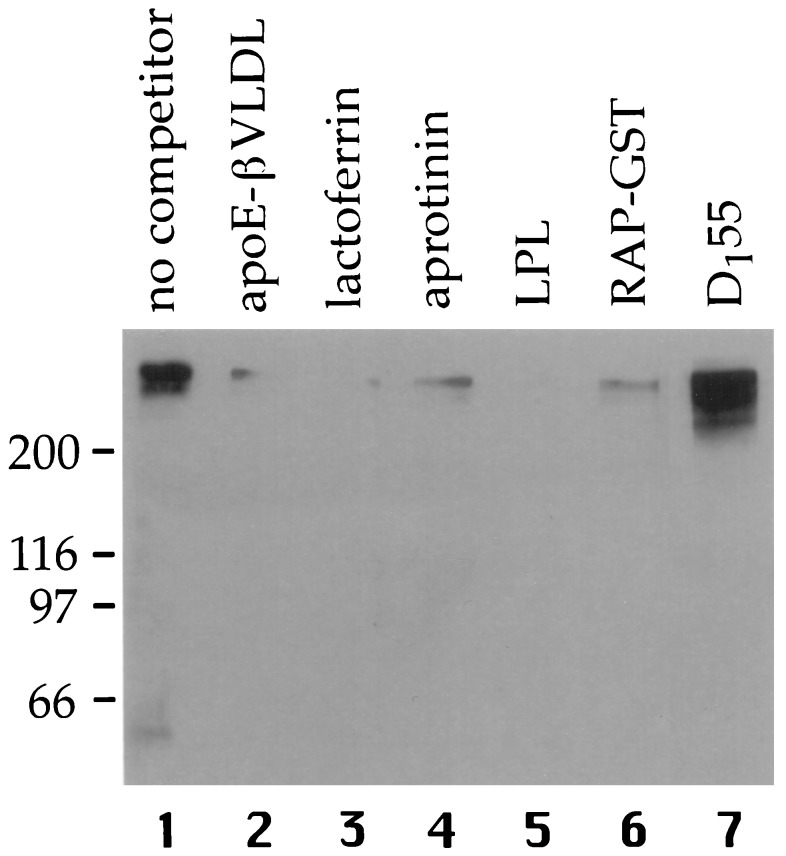
We next determined whether the binding of AC10 to megalin was inhibited by other megalin ligands. To this end, rat kidney microvillar proteins were separated by SDS/PAGE, transferred to PVDF membranes and incubated with AC10 IgG in the presence or absence of the indicated ligands (Fig. 2). In the absence of ligands, AC10 bound strongly to megalin (lane 1). However, apoE-βVLDL (lane 2), lactoferrin (lane 3), aprotinin (lane 4), lipoprotein lipase (lane 5), or RAP–GST (lane 6), efficiently competed (>90%) the binding of AC10 to megalin indicating that AC10 and all these ligands share similar or adjacent binding sites.
Figure 2.
Ligands inhibit binding of mAb AC10 to megalin. Microvillar proteins from rat kidney were separated by SDS/6% polyacrylamide gel, transferred to PVDF membranes, and immunoblotted with mAb AC10 IgG (10 μg/ml) in the presence of 100 μg/ml apoE-βVLDL (lane 2), lactoferrin (lane 3), aprotinin (lane 4), RAP–GST (lane 6), 20 μg/ml lipoprotein lipase (lane 5) or without competitor (lane 1). Bound AC10 was visualized by chemiluminescence using horseradish peroxidase-conjugated anti-mouse IgG. AC10 recognized megalin and no other microvillar proteins. The binding of AC10 to megalin was strongly inhibited by all of the ligands tested indicating that AC10 and these ligands bind to the same or adjacent sites on megalin. The electrophoretic position of megalin is indicated by the binding of anti-megalin mAb D155 (lane 7).
Inhibition of Ligand Binding by AC10 Is Concentration-Dependent.
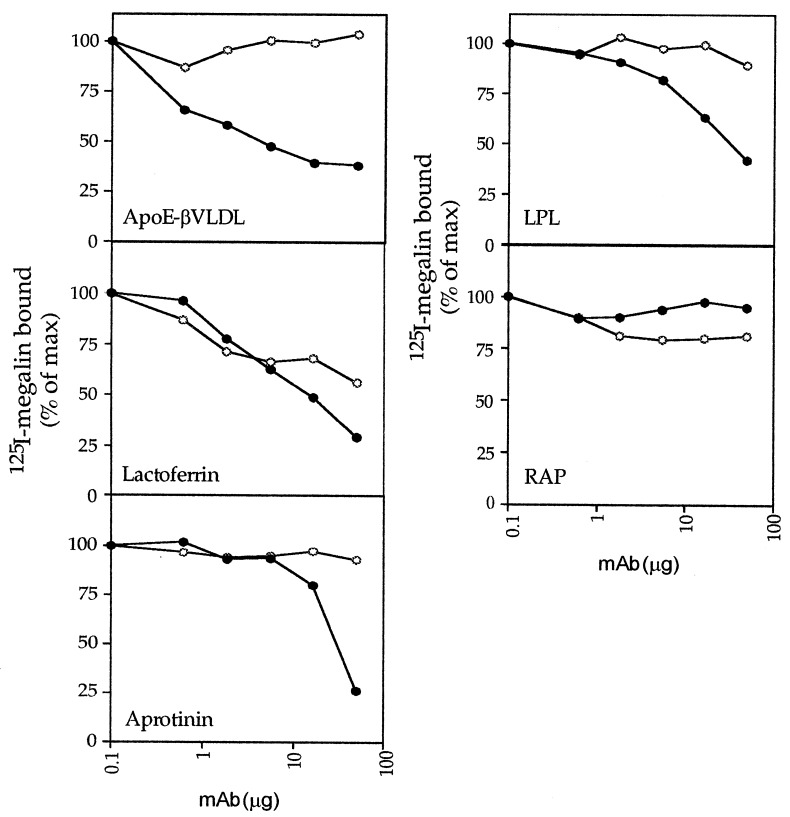
Megalin contains four putative ligand-binding domains (I–IV) (see Fig. 4A) each of which could contain one or several independent ligand-binding sites. To determine the number of binding sites for each ligand, purified ligands were immobilized onto 96-well plates and incubated with 125I-megalin in the presence or absence of varying concentrations of AC10. Bound 125I-megalin was quantitated, and the concentration of mAb IgG that caused 50% maximal inhibition (IC50) was determined (Fig. 3). AC10 inhibited 125I-megalin binding to all four ligands by 75–80% suggesting that each ligand has only one binding site on the receptor. If multiple binding sites were present, AC10 would only be able to partially (<50%) inhibit ligand binding. Alternatively, the partial inhibition of ligand binding by AC10 might also be due to the presence of a low-affinity binding site(s) located outside of the primary site recognized by AC10. The inhibition by AC10 of 125I-megalin binding to apoE-βVLDL, lactoferrin, aprotinin, and lipoprotein lipase was concentration-dependent with IC50 values of 40 nM, 1.1, 2.3, and 2.3 μM, respectively (Fig. 3). By contrast, AC10 did not inhibit binding of 125I-megalin to RAP in this assay indicating that megalin has at least one other high-affinity binding site for RAP that is spatially distinct from the epitope recognized by AC10.
Figure 3.
Inhibition of the binding of ligands to megalin by AC10 is concentration-dependent. ApoE-βVLDL, lactoferrin, aprotinin, lipoprotein lipase (LPL), or RAP were immobilized onto 96-well plates and incubated with 125I-megalin (5 × 105 cpm/well) in the presence or absence of either mAb AC10 (•) or mAb D155 (○) at the indicated concentrations for 3 h at 23°C. Bound 125I-megalin was quantitated by gamma counting. mAb AC10 efficiently competed for 125I-megalin binding to all ligands except RAP with an IC50 of 40 nM for apoE-βVLDL, 1.1 μM for lactoferrin, 2.3 μM for aprotinin and lipoprotein lipase. Anti-megalin mAb D155 failed to inhibit 125I-megalin binding to apoE-βVLDL, aprotinin, lipoprotein lipase, or RAP, but it was able to compete for megalin binding to lactoferrin. The inhibition of megalin binding to lactoferrin by D155 may be due to either the antibody recognizing multiple homologous epitopes that are not shared by AC10 or by steric hindrance of the lactoferrin binding site of megalin.
AC10 Binds to an Epitope Located Within the Second Cluster of Ligand-Binding Repeats.
We next set out to identify the epitope in megalin that specifically binds AC10. We expressed sequences of megalin representing the four clusters of cysteine-rich ligand-binding repeats (I through IV) in a baculovirus system (Fig. 4A). We chose these regions because, in analogy to the LDL receptor, they represent putative ligand-binding domains. The purified expression products were immunoblotted with AC10 and a polyclonal anti-megalin antibody that recognizes multiple epitopes in megalin. All four megalin fragments were recognized by the polyclonal anti-megalin antibody (Fig. 4B). However, when the expression products were immunoblotted with AC10, only the second cluster of ligand-binding repeats was recognized (Fig. 4C). These results indicate that the binding site for AC10 is restricted to this domain. Since the antibody competes for ligand binding, ligand-binding sites must also be located within this domain.
The AC10-Specific Epitope Is Contained Within the Fourth and Fifth Ligand-Binding Repeats (Amino Acids 1111–1210) in Megalin.
We next attempted to further map the AC10 and ligand-binding site with the second cluster of ligand-binding repeats on megalin and determine if this epitope overlaps with that of the nephritogenic antibodies. We have previously mapped the binding site of nephritogenic antibodies to the fifth cysteine-rich repeat in this domain (29). For this we utilized a β-galactosidase fusion protein containing amino acids 1111–1210 from megalin that includes the fourth and fifth cysteine-rich repeats of the second putative LDL receptor ligand-binding domain (Fig. 5A). By immunoblotting, AC10 specifically recognized the β-gal fusion protein containing amino acids 1111–1210 and not a fusion protein containing an irrelevant polypeptide sequence (Fig. 5B). These data indicate that (i) the mAb epitope and binding site for apoE-βVLDL, lactoferrin, aprotinin, lipoprotein lipase, and RAP are located between amino acids 1111–1210, and (ii) the binding site for these ligands overlaps with the epitope to which nephritogenic antibodies bind.
Figure 5.
AC10 binds to amino acids 1111–1210 containing the fourth and fifth ligand-binding repeats in megalin. A β-gal fusion protein was generated that included amino acids 1111–1210 from megalin (A). This sequence represents the fourth and fifth cysteine-rich ligand-binding repeats of the putative second ligand-binding domain. (B) Megalin(1111–1210)-βgal (lane 1) and RAP-β-gal (lane 2) were separated by SDS/PAGE, transferred to PVDF membranes and immunoblotted with mAb AC10. Bound AC10 was determined with an alkaline phosphatase-conjugated anti-mouse IgG and colorimetric detection. mAb AC10 specifically recognized the megalin(1111–1210)-β-gal (arrow) indicating that its epitope is contained within the fourth and fifth cysteine-repeats of the second cluster of ligand-binding repeats. No interaction between mAb AC10 and RAP-β-gal (arrowhead) was detected indicating that AC10 is specific for megalin and does not bind to β-gal or an unrelated polypeptide sequence.
DISCUSSION
In this study we have identified a binding site on megalin for apoE-βVLDL, lipoprotein lipase, aprotinin, lactoferrin, and RAP. To accomplish this we generated a mAb, AC10, that specifically inhibits ligand binding to megalin and used this mAb to map the binding site on the receptor (Fig. 6). We demonstrated that mAb AC10 inhibits the binding of apoE-βVLDL, lipoprotein lipase, aprotinin, and lactoferrin to megalin. This indicates that the ligands bind to the same or closely overlapping sites on megalin suggesting that AC10 is either a competitive inhibitor of ligand binding or blocks binding by steric hindrance. Our finding that AC10 is able to inhibit ≈75–80% of the binding of each of these ligands to megalin suggests that these ligands bind largely or exclusively to only one site on the receptor. By contrast, RAP appears to have more than one megalin binding site because AC10 only partially inhibits RAP binding to megalin.
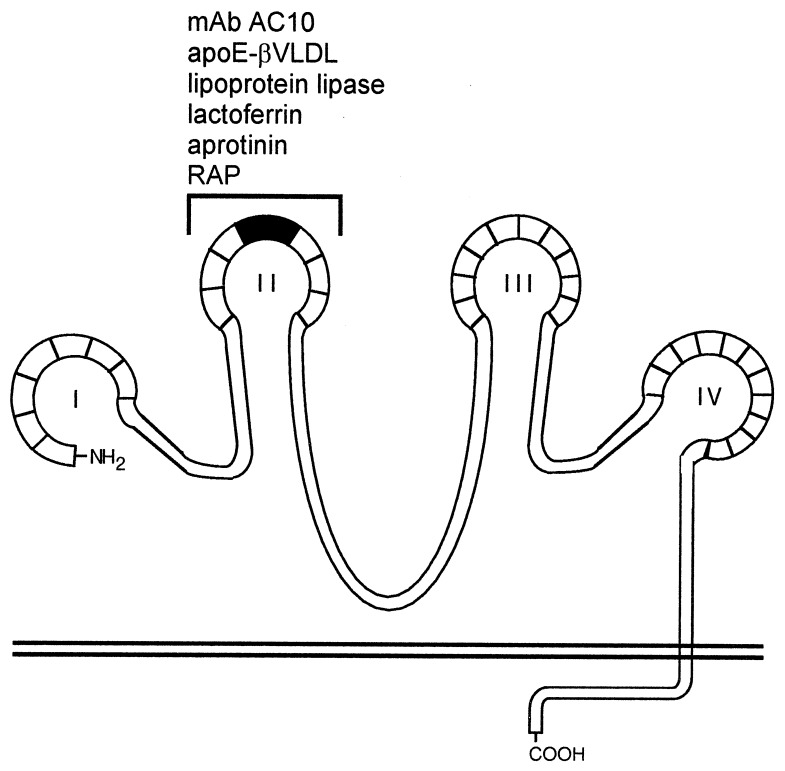
Figure 6.
Ligand binding map of megalin. Megalin contains four clusters (I–IV) of ligand-binding repeats that most likely attain a complex tertiary structure stabilized by numerous intramolecular disulfide bonds. Each of the four clusters are comprised of a specific number of homologous cysteine-rich repeats (boxed areas). In analogy to the LDL receptor, each cluster represents a putative ligand binding site containing both acidic and basic amino acid residues that may facilitate high-affinity, charge-dependent receptor–ligand interactions. The second cluster of ligand-binding repeats (bracket) contains a binding site (amino acids 1111–1210, shaded area) for apoE-βVLDL, lactoferrin, aprotinin, lipoprotein lipase, and RAP. This region also serves as a binding site for circulating autoimmune antibodies found in Heymann nephritis (29).
The fact that the ligand-binding site and the epitope recognized by mAb AC10 overlapped enabled us to identify the ligand-binding site by mapping the epitope on megalin for AC10. By immunoblotting of recombinant megalin fragments AC10 recognized only the second of the four clusters of cysteine-rich ligand-binding repeats. We further narrowed the location of the AC10 epitope to the fourth and fifth ligand-binding repeats (amino acids 1111–1210) in this fragment. Collectively, our results demonstrate that amino acids 1111–1210 represent a binding site on megalin for apoE-βVLDL, lipoprotein lipase, aprotinin, and lactoferrin, as well as RAP.
Structurally the second cluster of ligand-binding repeats in megalin closely resembles the ligand-binding domain of the LDL receptor. Within the second cluster there are eight cysteine-rich class A repeats each of which contains a highly conserved Ser-Asp-Glu sequence and a conserved spacing of six cysteine residues (32). In the LDL receptor, a disulfide bridge forms between cysteine(IV) and cysteine(VI) placing a cluster of negatively charged residues including the Ser-Asp-Glu sequence in a single loop (33) that is accessible for ligand binding. The Ser-Asp-Glu sequence has been shown to be important for high-affinity charge-dependent interactions between the LDL receptor and its ligands apoB100 and apoE (32, 34). This sequence is located between the fifth and sixth cysteines of each class A repeat in megalin (25) and, by analogy with the LDL receptor, may contribute to charge-dependent interactions between megalin and its ligands. Although it is not currently understood how megalin is capable of interacting with such a diverse group of ligands, the specificity of ligand binding may involve recognition of clusters of acidic residues on the receptor and basic residues on the ligand (19, 20, 32).
The second cluster of ligand-binding repeats is structurally and functionally an important region in megalin. This region not only contains a binding site for several ligands, but it also contains a binding site for circulating autoantibodies found in Heymann nephritis (29, 35), an experimental autoimmune disease in rats that closely resembles human membranous glomerulonephritis. Thus this site is exposed in vivo on the basal surface of the glomerular epithelium where it is competent to bind pathogenic antibodies as well as multiple ligands. In Heymann nephritis binding of circulating anti-megalin antibodies to the glomerular epithelium prevents the normal megalin-mediated clearance of lipoproteins that cross the glomerular basement membrane and results in their accumulation within immune deposits (unpublished data). Of the ligands used in this study, apoE-enriched βVLDL is of particular importance for the pathogenesis of Heymann nephritis because it was recently shown by Exner et al. (36) that apolipoproteins B and E accumulate within immune deposits in rats with Heymann nephritis and lead to extensive lipid peroxidation and damage to the glomerular basement membrane (37).
The second cluster of ligand-binding repeats has also been shown to serve as a ligand-binding site on the LDL receptor-related protein (LRP)/α2-macroglobulin receptor using both combinatorial Fab cloning techniques (38) and ligand blotting to cyanogen bromide fragments from LRP (39). Lipoprotein lipase, urokinase-type plasminogen activator:plasminogen activator inhibitor type 1 complexes, pro-uPA, α2-macroglobulin, and RAP have been shown to bind to a 68-kDa fragment of the second cluster of ligand-binding repeats. Within the LDL receptor family LRP is structurally and functionally (10, 25, 32) the closest relative to megalin. The two molecules are similar in their overall structure and both possess four clusters of ligand-binding repeats, but they differ in the number of growth factor and epidermal growth factor repeats and in the sequence of their cytoplasmic tails that, except for the NPXY internalization sequences, show no homology. Within the second cluster of ligand-binding repeats megalin and LRP show 51% homology (25) and a high degree of conservation of the spacing of cysteine residues and the Ser-Asp-Glu motif, and bind some of the same ligands (i.e., lipoprotein lipase and RAP). However, other ligands bind only to LRP (i.e., α2-macroglobulin) (10) or to megalin (plasminogen) (14). In the future it will be of interest to identify which amino acid residues in the second cluster of repeats are necessary for general receptor–ligand interactions and which provide the basis for the differences in ligand specificities between LRP and megalin.
We have previously shown that there are two sites within the primary sequence of RAP capable of binding independently to megalin—one between amino acids 86–148 and the other between amino acids 178–248 (28). Currently it is not known if these sites on RAP bind in concert to adjacent regions in megalin or to two spatially distinct sites. The fact that the binding of mAb AC10 to megalin is competed by RAP, but the binding of RAP to megalin is only partially inhibited by AC10 suggests that RAP binds to more than one site on the receptor.
Acknowledgments
This work was supported by National Institutes of Health Grant DK17724 (to M.G.F.) and the SFB 05, Project 07 from the Fonds zur Forderung der Wissenschaftlichen Forschung (to D.K.). Hajime Yamazaki was supported by a fellowship from the Uehara Memorial Foundation (Toshimaku, Tokyo, Japan).
ABBREVIATIONS
- βVLDL
β very low density lipoprotein
- RAP
receptor-associated protein
- apoE
apolipoprotein E
- GST
glutathione S-transferase
- PVDF
polyvinylidene difluoride
- β-gal
β-galactosidase
- LDL
low density lipoprotein
- LRP
low density lipoprotein receptor-related protein
- uPA
urokinase-type plasminogen activator
References
- 1.Farquhar M G, Kerjaschki D, Lundstrom M, Orlando R A. Ann NY Acad Sci. 1994;737:96–113. doi: 10.1111/j.1749-6632.1994.tb44304.x. [DOI] [PubMed] [Google Scholar]
- 2.Farquhar M G, Saito A, Kerjaschki D, Orlando R A. J Am Soc Nephrol. 1995;6:35–47. doi: 10.1681/ASN.V6135. [DOI] [PubMed] [Google Scholar]
- 3.Kerjaschki D, Miettinen A, Farquhar M G. J Exp Med. 1987;166:109–128. doi: 10.1084/jem.166.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerjaschki D, Farquhar M G. Proc Natl Acad Sci. 1982;79:5557–5561. doi: 10.1073/pnas.79.18.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerjaschki D, Farquhar M G. J Exp Med. 1983;157:667–686. doi: 10.1084/jem.157.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerjaschki D, Noronha-Blob L, Sactor B, Farquhar M G. J Cell Biol. 1984;98:1505–1513. doi: 10.1083/jcb.98.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatelet F, Brianti E, Ronco P, Roland J, Verroust P. Am J Pathol. 1986;122:512–519. [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng G, Bachinsky D R, Stamenkovic I, Strickland D K, Brown D, Andres G, McCluskey R T. J Histochem Cytochem. 1994;42:531–542. doi: 10.1177/42.4.7510321. [DOI] [PubMed] [Google Scholar]
- 9.Kounnas M Z, Chappell D A, Strickland D K, Argraves W S. J Biol Chem. 1993;268:14176–14181. [PubMed] [Google Scholar]
- 10.Kounnas M Z, Steingrimur S, Loukinova E, Argraves K M, Strickland D K, Argraves W S. Ann NY Acad Sci. 1994;737:114–123. doi: 10.1111/j.1749-6632.1994.tb44305.x. [DOI] [PubMed] [Google Scholar]
- 11.Stefansson S, Chappell D A, Argraves K M, Strickland D K, Argraves W S. J Biol Chem. 1995;270:19417–19421. doi: 10.1074/jbc.270.33.19417. [DOI] [PubMed] [Google Scholar]
- 12.Willnow T E, Goldstein J L, Orth K, Brown M S, Herz J. J Biol Chem. 1992;267:26172–26180. [PubMed] [Google Scholar]
- 13.Kounnas M Z, Loukinova E B, Stefansson S, Harmony J A, Brewer B H, Strickland D K, Argraves W S. J Biol Chem. 1995;270:13070–13075. doi: 10.1074/jbc.270.22.13070. [DOI] [PubMed] [Google Scholar]
- 14.Kanalas J J, Makker S P. J Biol Chem. 1991;266:10825–10829. [PubMed] [Google Scholar]
- 15.Moestrup S K, Christensen E I, Nielsen S, Jorgensen K E, Bjorn S E, Roigaard H, Gliemann J. Ann NY Acad Sci. 1994;737:124–137. doi: 10.1111/j.1749-6632.1994.tb44306.x. [DOI] [PubMed] [Google Scholar]
- 16.Stefansson S, Kounnas M Z, Henkin J, Mallampalli R K, Chappell D A, Strickland D K, Argraves W S. J Cell Sci. 1995;108:2361–2368. doi: 10.1242/jcs.108.6.2361. [DOI] [PubMed] [Google Scholar]
- 17.Andreasen P A, Sottrup-Jensen L, Kjoller L, Nykjaer A, Moestrup S K, Petersen C M, Gliemann J. FEBS Lett. 1994;338:239–245. doi: 10.1016/0014-5793(94)80276-9. [DOI] [PubMed] [Google Scholar]
- 18.Moestrup S K, Nielsen S, Andreasen P, Jorgensen K E, Nykjaer A, Roigaard H, Gliemann J, Christensen E I. J Biol Chem. 1993;268:16564–16570. [PubMed] [Google Scholar]
- 19.Moestrup S K, Cui S, Vorum H, Bregengard C, Bjorn S E, Norris K, Gliemann J, Christensen E I. J Clin Invest. 1995;96:1404–1413. doi: 10.1172/JCI118176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlando R A, Rader K, Authier F, Bergeron J J, Farquhar M G. J Am Soc Nephrol. 1995;6:378. doi: 10.1681/ASN.V9101759. (abstr.). [DOI] [PubMed] [Google Scholar]
- 21.Christensen E I, Gliemann J, Moestrup S K. J Histochem Cytochem. 1992;40:1481–1490. doi: 10.1177/40.10.1382088. [DOI] [PubMed] [Google Scholar]
- 22.Leung C C, Cheewatrakoolpong B, Yan C L. Anat Rec. 1989;223:363–367. doi: 10.1002/ar.1092230403. [DOI] [PubMed] [Google Scholar]
- 23.Poller W, Willnow T E, Hilpert J, Herz J. J Biol Chem. 1995;270:2841–2845. doi: 10.1074/jbc.270.6.2841. [DOI] [PubMed] [Google Scholar]
- 24.Lundgren S, Hjalm G, Hellman P, Ek B, Juhlin C, Rastad J, Klareskog L, Akerstrom G, Rask L. Exp Cell Res. 1994;212:344–350. doi: 10.1006/excr.1994.1153. [DOI] [PubMed] [Google Scholar]
- 25.Saito A, Pietromonaco S, Loo A K, Farquhar M G. Proc Natl Acad Sci USA. 1994;91:9725–9729. doi: 10.1073/pnas.91.21.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goding J W. Monoclonal Antibodies: Principles and Practice. San Diego: Academic; 1986. [Google Scholar]
- 27.Orlando R A, Kerjaschki D, Kurihara H, Biemesderfer D, Farquhar M G. Proc Natl Acad Sci USA. 1992;89:6698–66702. doi: 10.1073/pnas.89.15.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlando R A, Farquhar M G. Proc Natl Acad Sci USA. 1994;91:3161–3165. doi: 10.1073/pnas.91.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito A, Yamazaki H, Rader K, Nakatani A, Ullrich R, Kerjaschki D, Orlando R A, Farquhar M G. Proc Natl Acad Sci. 1996;93:8601–8605. doi: 10.1073/pnas.93.16.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietromonaco S, Kerjaschki D, Binder S, Ullrich R, Farquhar M G. Proc Natl Acad Sci USA. 1990;87:1811–1815. doi: 10.1073/pnas.87.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herz J, Goldstein J L, Strickland D K, Ho Y K, Brown M S. J Biol Chem. 1991;266:13364–13369. [Google Scholar]
- 32.Krieger M, Herz J. Annu Rev Biochem. 1994;63:601–637. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- 33.Bieri S, Djordjevic J T, Daly N L, Smith R, Kroon P A. Biochemistry. 1995;34:13059–13065. doi: 10.1021/bi00040a017. [DOI] [PubMed] [Google Scholar]
- 34.Mahley R W. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 35.Raychowdhury R, Zheng G, Brown D, McCluskey R T. Am J Pathol. 1996;148:1613–1623. [PMC free article] [PubMed] [Google Scholar]
- 36.Exner M, Susani M, Witztum J L, Hovorka A, Curtiss L K, Spitzauer S, Kerjaschki D. Am J Pathol. 1996;149:1313–1320. [PMC free article] [PubMed] [Google Scholar]
- 37.Neale T J, Ojha P P, Exner M, Poczewski H, Ruger B, Witztum J L, Davis P, Kerjaschki D. J Clin Invest. 1994;94:1577–1584. doi: 10.1172/JCI117499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horn I R, Moestrup S K, van den Berg B M, Pannekoek H, Nielsen M S, van Zonneveld A J. J Biol Chem. 1995;270:11770–117705. doi: 10.1074/jbc.270.20.11770. [DOI] [PubMed] [Google Scholar]
- 39.Moestrup S K, Holtet T L, Etzerodt M, Thogersen H C, Nykjaer A, Andreasen P A, Rasmussen H H, Sottrup-Jensen L, Gliemann J. J Biol Chem. 1993;268:13691–13696. [PubMed] [Google Scholar]