Abstract
Procollagen N-proteinase (EC 3.4.24.14) cleaves the amino-propeptides in the processing of type I and type II procollagens to collagens. Deficiencies of the enzyme cause dermatosparaxis in cattle and sheep, and they cause type VIIC Ehlers–Danlos syndrome in humans, heritable disorders characterized by accumulation of pNcollagen and severe skin fragility. Amino acid sequences for the N-proteinase were used to obtain cDNAs from bovine skin. Three overlapping cDNAs had an ORF coding for a protein of 1205 residues. Mammalian cells stably transfected with a complete cDNA secreted an active recombinant enzyme that specifically cleaved type I procollagen. The protein contained zinc-binding sequences of the clan MB of metallopeptidases that includes procollagen C-proteinase/BMP-1. The protein also contained four repeats that are homologous to domains found in thrombospondins and in properdin and that can participate in complex intermolecular interactions such as activation of latent forms of transforming growth factor β or the binding to sulfatides. Therefore, the enzyme may play a role in development that is independent of its role in collagen biosynthesis. This hypothesis was supported by the observation that in some tissues the levels of mRNA for the enzyme are disproportionately high relative to the apparent rate of collagen biosynthesis.
Procollagen N-proteinases and procollagen C-proteinase are neutral metalloproteinases essential for the processing of the procollagen precursors of the three major fibrillar collagens, types I, II, and III (1–5). The processing of the procollagens by the N- and C-proteinases generates collagen monomers that spontaneously self-assemble into fibrils (6–8).
Recently, cDNAs for procollagen C-proteinase were cloned, and one cDNA was shown to code for a 70-kDa protein that was previously identified as BMP-1 (9, 10), one of a series of bone morphogenic proteins that induce new bone formation at ectopic sites (11). The structure of BMP-1/C-proteinase included a proenzyme N-terminal domain, a large metalloendoproteinase domain found in the superfamily of zinc metalloproteinases, an epidermal growth factor-like domain, and three regions of internal sequence homology referred to as CUB domains because they are found in the complement components C1r/C1s, a sea urchin protein called Uegf, and BMP-1 (12). The metalloproteinase domain was characteristic for the subfamily astacin in the family M12 that is part of the clan MB of metallopeptidases (13). The domain structure indicated that BMP-1/C-proteinase was analogous to proteins encoded by genes that were found to be essential for normal development in Drosophila, Xenopus, hydra, sea urchin, and fish (7, 14). A second cDNA for BMP-1/C-proteinase coded for an 80-kDa protein with the same first 712 amino acids as BMP-1 but with an additional C-terminal sequence of 120 amino acids rich in serine, threonine, and histidine. A third cDNA coded for a 100-kDa protein with the same N-terminal domain of 703 residues as BMP-1/C-proteinase but with a more extended C-terminal domain that increased the homology to the protein encoded by the Drosophila gene for tolloid (15). The results, therefore, suggested some still undefined role for BMP-1/C-proteinase in morphogenesis.
The type I procollagen N-proteinase (pNPI) has many similarities to the C-proteinase in that pNPI is a large neutral zinc metalloproteinase and is essential for the processing of type I and type II procollagens (1, 16). The polypeptide chain has a molecular mass of about 110 kDa by SDS/PAGE but it is frequently isolated in large complexes with type XIV collagen and other proteins. Deficiencies of pNPI activity in vivo cause dermatosparaxis in cows and sheep (17), and type VIIC Ehlers–Danlos in humans (18, 19). These inherited connective tissue disorders are characterized by a severe skin fragility because of the accumulation of pNcollagen and the formation of abnormal collagen fibers.
Recently, three of us (A.C., B.V.N. and C.M.L.) purified and defined a partial amino acid sequence of bovine pNPI (20). Here we report the structure of a complete cDNA and expression of the cDNA in a mammalian cell system. In addition, we have defined the tissue distribution of mRNAs for the enzyme.
MATERIALS AND METHODS
Construction and Analysis of the cDNA Library.
For cDNA synthesis (Great Lengths cDNA Synthesis kit; CLONTECH), specific oligonucleotide primers OP8 and OP11 (Fig. 1) were designed from partial amino acid sequences (17, 20). The cDNAs were ligated to an adapter, phosphorylated, size fractionated, and ligated into a λ vector (ExCell EcoRI/CIP; Pharmacia Biotech). The vector was packaged (Ready To Go λ Packaging Kit; Pharmacia Biotech) and used for lytic infection of NM522 Escherichia coli (Pharmacia Biotech). About 3.5 × 105 plaque forming units were screened with degenerated oligonucleotides OP1 and OP10 (Fig. 1) by Northern blot analysis. Three positive clones were used for in vivo phagemid release using E. coli according to manufacturer’s protocol (Pharmacia Biotech). Purified phagemids were sequenced with a DNA sequencing kit (Pharmacia Biotech) and vector-specific or pNPI sequence-specific primers.
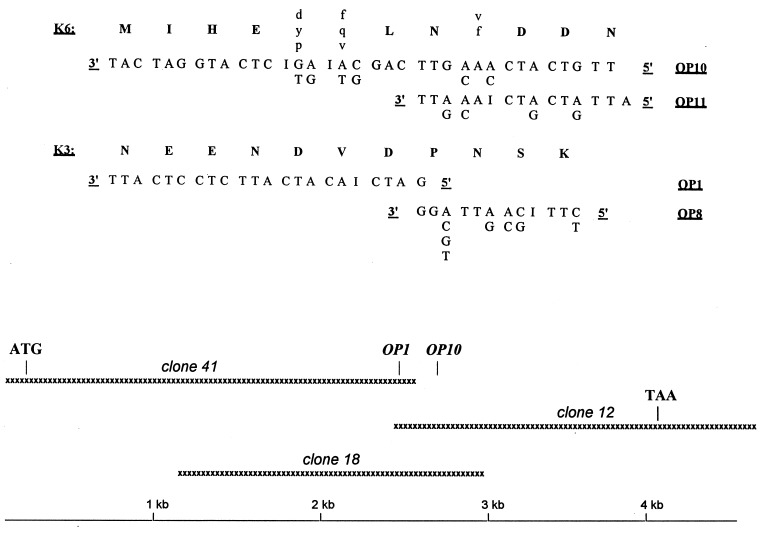
Figure 1.
(Upper) The oligonucleotide sequences deduced from two previously determined amino acid sequences (20). Unambiguously determined amino acids are in capital letters. (Lower) Three overlapping cDNAs provided a 4,580-nucleotide sequence containing a full-length ORF starting with an ATG codon (boldface type) in a perfectly conserved Kozak consensus sequence and ending with a TAA stop codon (boldface type) that is the beginning of an AATAAA polyadenylylation signal (italics).
Expression of the Recombinant Enzyme.
Three overlapping clones were assembled to generate a complete cDNA. The cDNA was then inserted into the NotI and XbaI sites of the pcDNA3 expression vector (Invitrogen) and used to prepare stably transfected clones of a human tumor cell line (HT-1080 or CCL-121; American Type Culture Collection) as described (6). Single G418-resistant clones were transferred to 24-well plates and grown to confluency. Each clone was transferred to 12-well plates in duplicate and cultured to confluency. Cells from one duplicate well were harvested and total RNA isolated (RNAeasy; Qiagen, Chatsworth, CA) for Northern blot assays with the full-length bovine cDNA coding sequence.
The duplicate wells from two clones expressing the mRNA were cultured for 24 h in serum-free high glucose DMEM. Proteins from the medium were precipitated overnight at 4°C with half of the sample volume of 25% polyethylene glycol (Mr = 8,000) in 0.1 M NaCl, 2 mM CaCl2, 0.015% Brij 30, and 0.01% NaN3 in 0.05 M Tris·HCl buffer (pH 7.4 at room temperature). The precipitate was pelleted by centrifugation and partially solubilized in 20 μl of a buffer containing 0.2 M NaCl, 7 mM CaCl2, 0.015% Brij 40, and 0.01% NaN3 in 0.025 M Tris·HCl buffer (pH 7.5 at room temperature). One microgram of 14C-labeled procollagen I purified from organ cultures of chicken tendons (15,000 cpm/μg) (16) was then added to assay for N-proteinase activity. After incubation of the samples at 35°C for 4 h, the reaction was terminated by addition of one-fourth volume of sample buffer containing 5% 2-mercaptoethanol, 11.5% SDS, 10% glycerol, and 0.5% bromphenol blue in 0.125 Tris·HCl (pH 6.8) and heated to 100°C for 5 min. The reaction products were separated by electrophoresis on a 3.5–12% gradient SDS/polyacrylamide gel (Mini Protean apparatus; Bio-Rad) and assayed with a PhosphorImager (Molecular Dynamics).
Expression of pNPI in Tissues.
For Northern blot analysis, 20 μg of poly(A)+ RNA or 40 μg of total RNA were separated by electrophoresis in 0.9% agarose/formaldehyde gel and transferred to a nylon membrane (Hybond N; Amersham). Filters containing total RNA were prehybridized at 42°C for 4 h in 5× SSPE (1× SSPE = 150 mM NaCl/1 mM EDTA/10 mM NaH2PO4, pH 7.4), 40% deionized formamide, 5× Denhardt’s solution, 0.1% SDS, 5% Dextran sulfate, and 100 μg/ml denatured salmon sperm DNA and then probed for 18 h in the same conditions with cDNA inserts labeled using [α32-P]dATP and a random priming labeling kit (Boehringer Mannheim). Blots were washed at 65°C in 0.3× SSC (1× SSC = 150 mM NaCl/15 mM Na citrate, pH 7.0), and 0.1% SDS before exposure to x-ray film. Filters containing poly(A)+ RNA were prehybridized for 2 h at 42°C in 2× SSC, 5× Denhardt’s solution, 5% Dextran sulfate, 0.5% SDS, and 200 μg/ml denatured salmon sperm DNA. They were then hybridized in the same conditions with 5 ng of a pNPI primer (OP1 or OP10, see Fig. 1) labeled with [α32-P]ATP and terminal transferase (Boehringer Mannheim). After 18 h incubation, the filters were washed at 57°C (OP1 probe) or 52°C (OP10 probe) in 2× SSC/0.1% SDS and exposed to x-ray film.
To assay the enzyme activity in organs, tissues from a fetal calf at the third trimester stage were grounded in liquid nitrogen and suspended in the extraction buffer (1 M KCl/2 mM CaCl2/0.02% Brij 30/1.25 mM N-ethylmaleimide/0.25 mM phenylmethylsulfonyl fluoride in 50 mM Na cacodylate, pH 7.5). After shaking for 18 h at 4°C, the samples were centrifuged for 10 min at 15,000 g. The supernatants were collected and used to determine the enzymatic activity as described (20).
Reverse Transcription–PCR (RT-PCR) Assay.
The RT-PCR amplifications were carried out (GeneAmp Thermostable rTth Reverse Transcriptase RNA PCR kit; Perkin–Elmer) with three different pairs of [α32-P]-labeled primers (T4 polynucleotide kinase; Boehringer Mannheim). The primers amplified the region of the cDNA coding for part of the properdin repeat domain (A29, 5′-CACTCATAGCCCACAGAGTCGTCT-3′; A30, 5′-CCAACTCCAAGACCTTCATCGCCAT-3′), and the active Zn2+-binding site (A46, 5′-GCATGCAAGGCTATGCTCCTGTCA-3′; A47: 5′-GGCTGCAGCGGGACCAGTGGAA-3′). An external control RNA template (PAW 109; Perkin–Elmer) was added to the purified RNAs to monitor for amplification efficiency in each sample (A39, 5′-GTCTCTGAATCAGAAATCCTTCTATC-3′; A40: 5′-CATGTCAAATTTCACTGCTTCATCC-3′). The amount of total RNA was varied from 20 to 0.02 ng per tube to obtain comparable signals for all the tissues. The RT step was at 70°C for 15 min followed by a 2-min incubation at 95°C for denaturation of RNA–DNA heteroduplexes and then by a 30 cycles PCR amplification. The PCR conditions used were 94°C for 15 sec, 68°C for 20 sec, and 72°C for 10 sec for pNPI specific primers. Identical conditions were used for the external control template except the annealing temperature was 60°C. Samples were analyzed by electrophoresis through a 10% polyacrylamide gel and autoradiography. The films were assayed by laser densitometry (Ultroscan XL; LKB). Values for product bands were compared with standards and expressed in arbitrary units of absorbance per ng of total RNA.
RESULTS
Cloning and Sequencing.
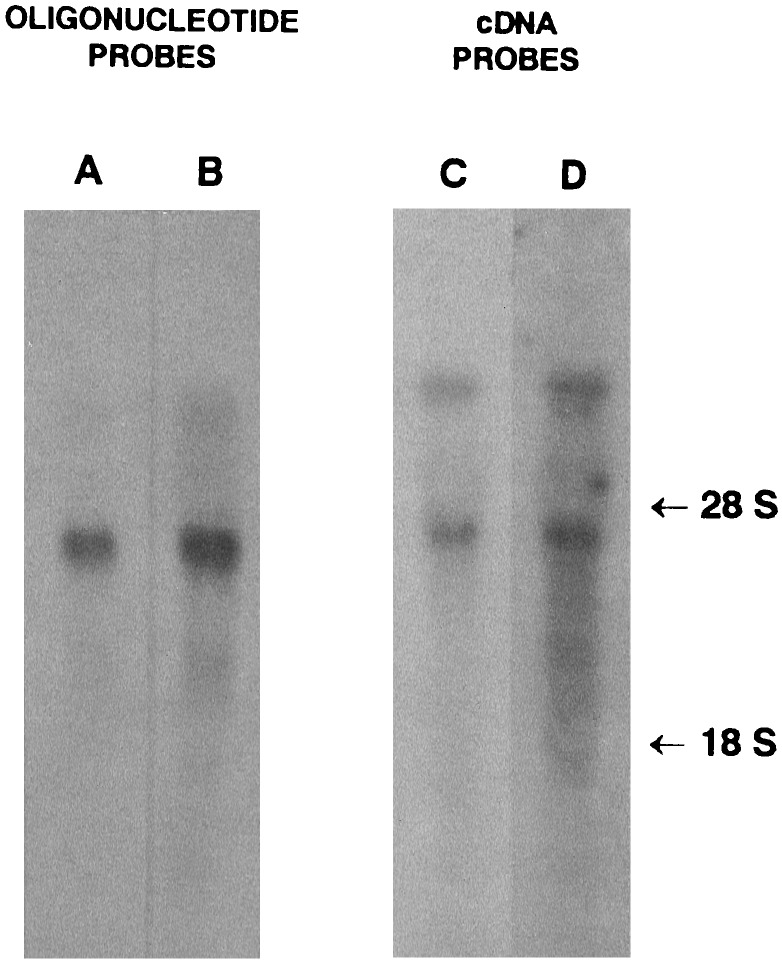
Degenerated oligonucleotide probes and primers (Fig. 1) were designed from two partial pNPI amino acid sequences (20) using some substitutions with a neutral base (inosine) or a guessmer (21, 22). For cDNA primers (OP8 and OP11), complete degeneracy was maintained at the 3′ end to allow a perfect annealing of a fraction of the primer pool to the pNPI mRNA (Fig. 1). The two independent oligonucleotide probes (OP1, OP10) hybridized mainly with mRNA of about 7, 4.8, and 3.7 kb in a Northern blot assay with bovine skin mRNA (Fig. 2, lanes A and B). Using the same conditions, we screened ≈3.5 × 105 plaque-forming units of a bovine skin library. Three positive clones were identified, one hybridizing with OP1 only and two with both probes. Purified inserts from two of these clones were labeled and hybridized to total RNA on Northern blot (Fig. 2, lanes C and D). Both revealed a pattern similar to that observed with the oligonucleotide probes. Nucleotide sequencing of the three overlapping clones (Fig. 1) revealed an ORF 3615 bp long beginning with an ATG codon in a perfectly conserved Kozak consensus sequence (23). The 3′ end contained a TAA stop codon that was part of an AATAAA polyadenylylation signal.
Figure 2.
Northern blot analysis of bovine skin RNA using pNPI specific probes. Twenty micrograms of poly(A)+ RNA (lanes A and B) or 40 μg of total RNA (lanes C and D) were separated by electrophoresis in agarose/formaldehyde gel, transferred onto a nylon membrane, and hybridized with pNPI-specific probes (lanes A and B, OP1 and OP10 oligonucleotide probes, respectively; lanes C and D, cDNA probes from clones 12 and 41). The membranes were exposed to x-ray film. Positions of 28S and 18S ribosomal RNA are indicated.
Expression of the Recombinant Enzyme.
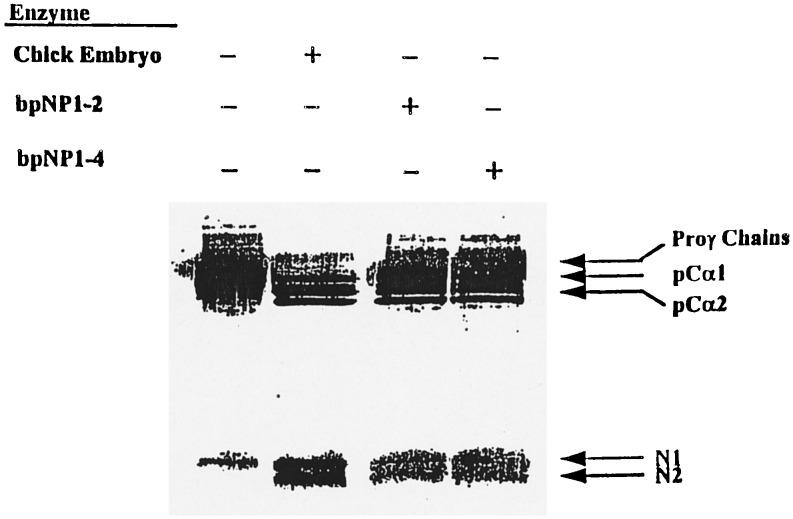
A full-length cDNA was used to prepare stably transfected HT-1080 cells (6), and clones were screened by Northern blot analysis. Media from two clones with high levels of the recombinant mRNA were concentrated and assayed for pNPI activity. Medium from both clones contained an enzymatic activity that specifically cleaved the proα1(I) and proα2(I) chains of type I procollagen to pCα1(I) and pCα2(I) chains (Fig. 3).
Figure 3.
Enzymatic activity of the recombinant bovine pNPI. Recombinant enzyme recovered from the conditioned medium of the two different HT-1080 clones expressing high level of pNPI mRNA were incubated with 14C-labeled procollagen substrate in the standard reaction buffer (16). After 4 h at 35°C, reaction products were separated by SDS/PAGE and visualized after autoradiography. bpNP1–2 and bpNP1–4, Enzyme from two different clones expressing a full-length cDNA for pNPI; Proγ chains, disulfide-linked proα chains of type I procollagen; pCα1 and pCα2, partially processed proα1(I) and proα2(I) chains of type I procollagen containing the C-propeptides but not the N-propeptides from proα1(I) and proα2(I) chains. N1 and N2, N-propeptides from proα1(I) and proα2(I) chains.
Protein Structure.
The ORF coded for a protein of 1205 amino acids (Fig. 4). The sequences included the three short amino acid sequences previously determined by microsequencing of the protein (20), an observation confirming that the cDNA coded for pNPI.
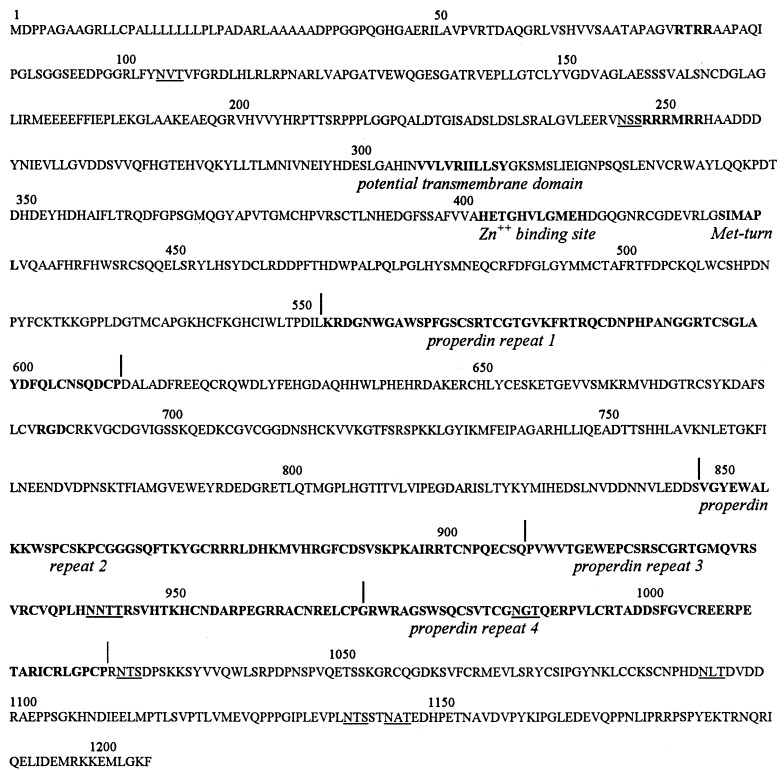
Figure 4.
Deduced amino acid sequence of pNPI. The 1205-amino acid protein contains a signal peptide region, composed mainly of leucine and alanine residues. Potential cleavage sites by mammalian subtilisins [RTRR (77–80)] and [RRRMRR (248,253)] and a RGD sequence (685–687) are indicated in boldface type. Potential glycosylation sites are underlined. The Zn2+-binding site, Met-turn, the potential transmembrane domain, and properdin repeats are indicated (below sequence).
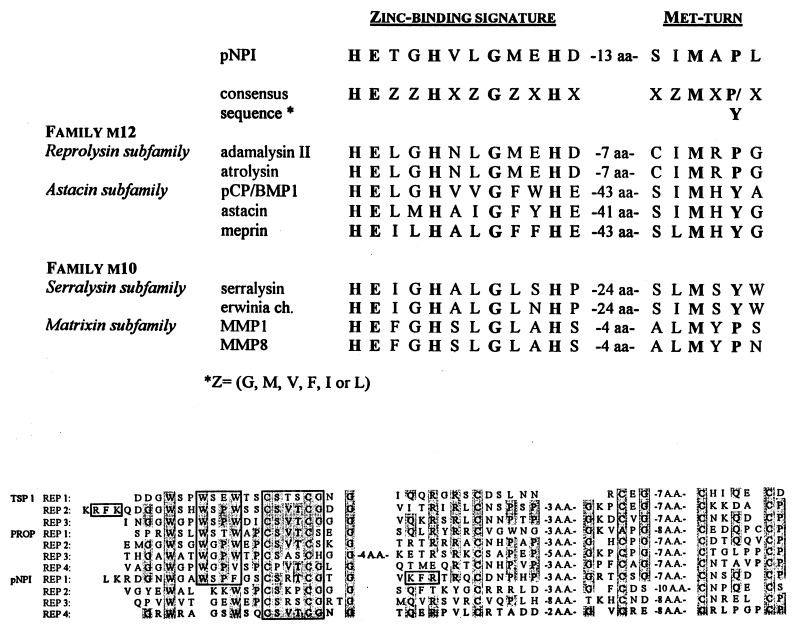
The structure of the protein (Fig. 4) included (i) a signal sequence of about 36 amino acids rich in Ala and Leu; (ii) one RGD sequence that is a potential integrin-binding site; (iii) 9 potential glycosylation sites (NXT/S), most at the C terminus of the protein; (iv) two sequences of K/R-X-K/R-R that are potential cleavage sites for mammalian subtilisins such as furins (24); and (v) a potential transmembrane domain. Further analysis of the sequence (Fig. 5) revealed the presence of a Zn2+-binding site followed by a Met-turn, features that are characteristic of the clan MB of metallopeptidases (13) also known as the metzincin superfamily (25). Sequence homology was highest with members of the reprolysin subfamily that forms with the astacin subfamily of the family M12 in the clan MB. Beyond the Met-turn were four properdin repeats (Fig. 4). The repeats showed a high homology with similar repeats in thrombospondin-1 and in properdin (Fig. 5). A short sequence in repeat 1 of pNPI (WSPFG) was similar to a well conserved sequence (W/Y-S-X-W/F-S/G) found in the cytokine receptor superfamily (26).
Figure 5.
Sequence comparisons. (Upper) The sequence of the Zn2+-binding domain of the pNPI is compared with sequences found in members of families M10 and M12 of the clan MB of metallopeptidases. Residues in boldface type indicate amino acids that are critical for Zn2+-binding and that represent the characteristic signature of the clan MB. (Lower) The four properdin repeats found in pNPI are compared with the three repeats present in human thrombospondin 1 (TSP1) and to four of the six repeats found in human properdin (PROP). The conserved residues are shaded. Subdomains of the second repeat in TSP1 that have previously been shown (27–29) to display biological activities and homologous sequences in other repeats are boxed.
Tissue Distribution of Expression.
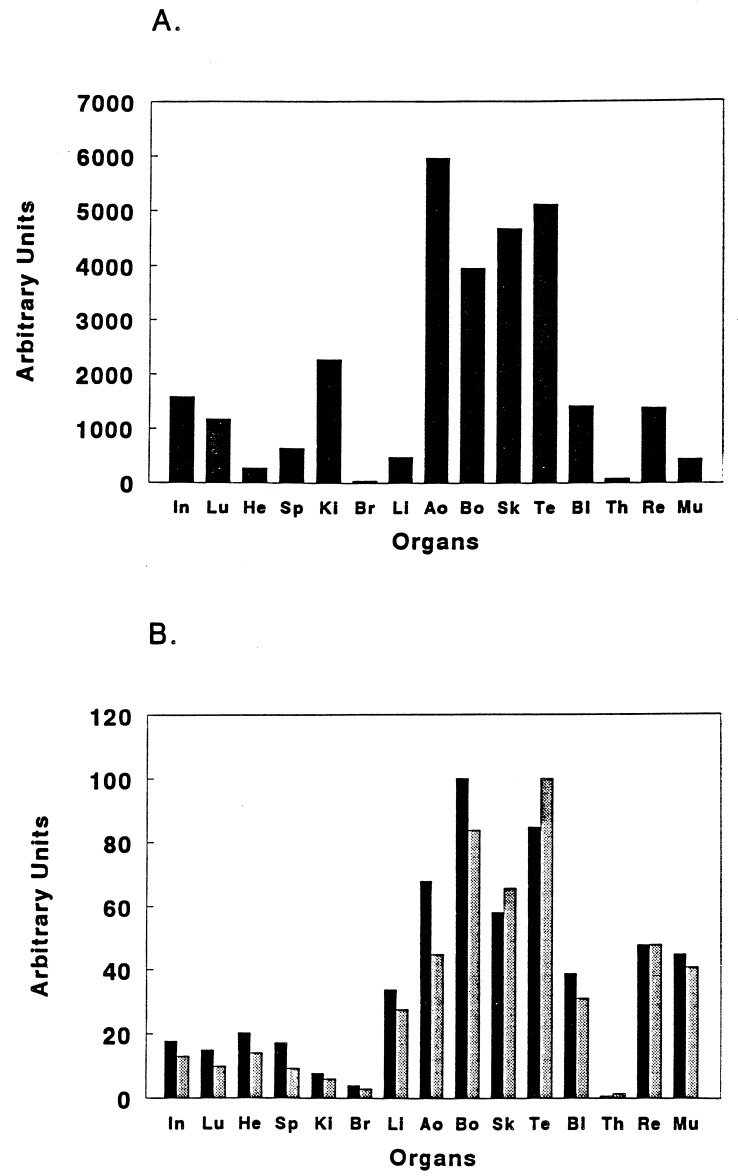
High levels of enzyme activity were detected in all type I collagen-rich tissues such as skin, bones, tendons, and aorta and trace amounts were observed in brain and thymus (Fig. 6A). Other organs displayed intermediate levels. Northern blot analysis of total RNA revealed that the mRNA for pNPI was barely detectable even in tissues such as skin, where it should be most abundant (Fig. 2). To increase the sensitivity of detection of mRNA for pNPI in tissues, a RT-PCR analysis was performed. Preliminary experiments were carried out to determine PCR conditions that allowed amplification to be directly proportional to the amount of specific RNA added (data not shown). Using these conditions, the signal from the external control template added to the RNA from different tissues varied less than by a factor of 2 (data not shown). The RT-PCR assays gave similar values with the two pairs of pNPI-specific primers. The tissue distribution of mRNA for the enzyme (Fig. 6B) paralleled the enzymatic activity measurements in most tissues in that strong signals were observed in skin, bone, tendon, and aorta, and very faint signals in brain and thymus. However, the mRNA level was disproportionately high in heart, liver, retina, and muscle. Also, the mRNA level was disproportionately low in kidney.
Figure 6.
Analysis of pNPI expression. (A) pNPI enzymatic activity was measured in extracts of various organs and expressed in arbitrary units. (B) pNPI mRNA steady-state level was measured by RT-PCR using two different pairs of [α32-P]-labeled primers, one allowing the amplification of part of the properdin repeat domain (A29-A30, ▪) and the other allowing the amplification of a region including the Zn2+-binding site (A46-A47, ░⃞). PCR-amplified products were separated on 10% polyacrylamide gels and assayed by autoradiography. Results obtained by quantification of the specific pNPI band by LASER scanning densitometry are expressed in arbitrary units. In, intestine; Lu, lung; He, heart; Sp, spleen; Ki, kidney; Br, brain; Li, liver; Ao, aorta; Bo, bone; Sk, skin; Te, tendon; Bl, bladder; Th, thymus; Re, retina; Mu, muscle.
DISCUSSION
The identification and cloning of the cDNA coding for the pNPI was performed by screening a bovine skin cDNA library with degenerated probes designed from previously reported partial amino acid sequences (20). Expression of the cDNA as active enzyme in stably transfected mammalian cells established that the cDNA encoded pNPI.
The 1205 residue protein encoded by the full-length cDNA is larger than the polypeptide chain of 107 kDa previously identified as active pNPI (20). The size discrepancy is even greater if the multiple potential sites in the protein are glycosylated. Therefore, the enzyme is probably similar to other metalloproteinases that are first synthesized as proenzymes (13). The presence of two sequences of RTRR and RRRMRR in the N-terminal domain suggests that the processing could occur through a furin-like enzyme (24) similar to the processing observed with other metalloproteinases such as meprin and stromelysin 3 zymogen (30, 31).
The structure of the zinc-binding domain of pNPI places it in the family M12 of the clan MB of metalloproteinases as defined by Rawlings and Barrett (13). BMP-1/C-proteinase is in the same family. However, pNPI is probably not in the same astacin subfamily as BMP-1/C-proteinase, since its zinc-binding sequence is different. Also, it contains a potential integrin-binding sequence of RGD that is found in the disintegrins of the reprolysin subfamily. However, the pattern of Cys distribution around the RGD sequence is different from that found in disintegrins and pNPI has little homology outside the catalytic domain with other members of the reprolysin subfamily. Therefore, pNPI may be a member of a separate and still undefined subfamily of the family M12 (13).
The large C-terminal domain of pNPI shows no homology with BMP-1/C-proteinase or related proteins such as tolloid. Instead, four repeats in the C-terminal domain are homologous in terms of size, N-terminal sequence, and pattern of cysteine residues to three domains found in thrombospondins-1 and -2 (32), to six domains in properdin (33), and to domains found in a diverse group of proteins such as some components of the complement (34) or several proteins from unicellular organisms (35). The only common property among all these proteins is their involvement in complex intermolecular interactions (36, 37). The properdin repeats have been extensively characterized for thrombospondin 1 (27–29). The second repeat of thrombospondin-1 was shown to (i) bind heparin at three sequences (KRFK, WSPW, CSVTCG), (ii) activate latent forms of transforming growth factor β via a nonproteolytic pathway by a cooperation of two sequences (K/R-F-K/R and WSPW), (iii) bind sulfatide, (iv) promote cell adhesion, and (v) inhibit angiogenesis through the CSVTCG sequence. The sequences of CSVTCG, KFR, and WSPF in pNPI may well have similar biological functions. Therefore, in addition to its pNPI activity, the enzyme may be involved in other physiological events during development and angiogenesis. This hypothesis is supported by recent data indicating that other members of the family M12 of the clan MB of metallopeptidases are essential for development and neurogenesis (38–40). The hypothesis is also supported by the observation here that the level of mRNA for pNPI is disproportionately high in heart, liver, retina, and muscle.
Acknowledgments
The skillful assistance of Mrs. Y. Scheen-Goebels is greatly acknowledged. A.C. is a Chargé de Recherche supported by the Belgian Fonds National de la Recherche Scientifique. This work was supported in part by Belgian Fonds National de la Recherche Scientifique Grant 3.4529.95 and by the Fonds de Recherche de la Faculté de Médecine de l’Université de Liége.
ABBREVIATIONS
- pNPI
procollagen N-proteinase that cleaves type I and type II procollagen
- RT-PCR
reverse transcription–PCR
Footnotes
References
- 1.Lapière C M, Lenaers A, Kohn L. Proc Natl Acad Sci USA. 1971;68:3054–3058. doi: 10.1073/pnas.68.12.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nusgens B, Goebels Y, Shinkai H, Lapière C M. Biochem J. 1980;191:699–706. doi: 10.1042/bj1910699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halila R, Apostolou S, Winqvist R, Callen D, Prockop D J, Peltonen L. Am J Hum Genet. 1992;51:128. [Google Scholar]
- 4.Hojima Y, van der Rest M, Prockop D J. J Biol Chem. 1985;260:15996–16003. [PubMed] [Google Scholar]
- 5.Kadler K E, Watson R B. Methods Enzymol. 1995;248:771–781. doi: 10.1016/0076-6879(95)48052-8. [DOI] [PubMed] [Google Scholar]
- 6.Fertala A, Sieron A L, Hojima Y, Ganguly A, Prockop D J. J Biol Chem. 1994;269:11584–11589. [PubMed] [Google Scholar]
- 7.Prockop D J, Hulmes D J S. In: Extracellular Matrix Assembly and Structure. Yurchenco P D, Birk D E, Mechan R P, editors. New York: Academic; 1994. pp. 47–90. [Google Scholar]
- 8.Prockop D J, Kivirikko K I. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 9.Kessler E, Takahara K, Biniaminov L, Brusel M, Greenspan D S. Science. 1996;271:360–362. doi: 10.1126/science.271.5247.360. [DOI] [PubMed] [Google Scholar]
- 10.Li S W, Sieron A L, Fertala A, Hojima Y, Arnold W V, Prockop D J. Proc Natl Acad Sci USA. 1996;93:5127–5130. doi: 10.1073/pnas.93.10.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wozney J M, Rosen V, Celeste A J, Mitsock L M, Whitters M J, Kriz R W, Hewick R M, Wang E A. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 12.Bond J S, Beynon R J. Protein Sci. 1995;4:1247–1261. doi: 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawlings N D, Barrett A J. Methods Enzymol. 1995;248:183–241. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 14.Fukagawa M, Suzuki N, Hogan B L M, Jones C M. Dev Biol. 1994;163:175–183. doi: 10.1006/dbio.1994.1133. [DOI] [PubMed] [Google Scholar]
- 15.Takahara K, Lyons G E, Greenspan D S. J Biol Chem. 1994;269:32572–32578. [PubMed] [Google Scholar]
- 16.Hojima Y, Mörgelin M M, Engel J, Boutillon M-M, van der Rest M, McKenzie J A, Chen G-C, Rafi N, Romanic A M, Prockop D J. J Biol Chem. 1994;269:11389–11390. [PubMed] [Google Scholar]
- 17.Hanset R, Ansay M. Ann Med Vet. 1967;7:451–470. [Google Scholar]
- 18.Smith L T, Wertelecki W, Milstone L M, Petty E M, Seashore M R, Braverman I M, Jenkins T G, Byers P H. Am J Hum Genet. 1992;51:235–244. [PMC free article] [PubMed] [Google Scholar]
- 19.Nusgens B V, Verellen-Dumoulin G, Hermans-Le T, De Paepe A, Nuytinck L, Piérard G E, Lapière C M. Nat Genet. 1992;1:214–217. doi: 10.1038/ng0692-214. [DOI] [PubMed] [Google Scholar]
- 20.Colige A, Beschin A, Samyn B, Goebels Y, Van Beeumen J, Nusgens B V, Lapière Ch M. J Biol Chem. 1995;270:16724–16730. doi: 10.1074/jbc.270.28.16724. [DOI] [PubMed] [Google Scholar]
- 21.Lathe R. J Mol Biol. 1985;183:1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 11.9–11.10. [Google Scholar]
- 23.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 24.Barr P J. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- 25.Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Rüth F-X, Mc Kay D B, Bode W. Protein Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazan J F. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo N-H, Krutzsch H C, Nègre E, Vogel T, Blake D A, Roberts D D. Proc Natl Acad Sci USA. 1992;89:3040–3044. doi: 10.1073/pnas.89.7.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo N-H, Krutzsch H C, Nègre E, Zabrenetzy V S, Roberts D D. J Biol Chem. 1992;267:19349–19355. [PubMed] [Google Scholar]
- 29.Schultz-Cherry S, Chen H, Mosher D F, Misenheimer T M, Krutzsch H C, Roberts D D, Murphy-Ullrich J E. J Biol Chem. 1995;270:7304–7310. doi: 10.1074/jbc.270.13.7304. [DOI] [PubMed] [Google Scholar]
- 30.Milhiet P E, Chevallier S, Corbeil D, Seidah N G, Crine P, Boileau G. Biochem J. 1995;309:683–688. doi: 10.1042/bj3090683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pei D, Weiss S J. Nature (London) 1995;375:244–247. doi: 10.1038/375244a0. [DOI] [PubMed] [Google Scholar]
- 32.Laherty C D, O’Rourke K, Wolf F W, Katz R, Seldin M F, Dixit V M. J Biol Chem. 1992;267:3274–3281. [PubMed] [Google Scholar]
- 33.Nolan K F, Schwaeble W, Kaluz S, Dierich M P, Reid K B M. Eur J Immunol. 1991;21:771–776. doi: 10.1002/eji.1830210333. [DOI] [PubMed] [Google Scholar]
- 34.Haefliger J-A, Tschopp J, Vial N, Jenne D E. J Biol Chem. 1989;264:18041–18051. [PubMed] [Google Scholar]
- 35.Kobayashi S, Eden-McCutchan F, Framson P, Bornstein P. Biochemistry. 1986;25:8418–8425. doi: 10.1021/bi00374a014. [DOI] [PubMed] [Google Scholar]
- 36.Tuszynski G P, Rothman V L, Deutch A H, Hamilton B K, Eyal J. J Cell Biol. 1992;116:209–217. doi: 10.1083/jcb.116.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins J M G, Wiedemann H, Timpl R, Reid K BM. J Immunol. 1995;155:5777–5785. [PubMed] [Google Scholar]
- 38.Childs S R, O’Connor M. Dev Biol. 1994;162:209–220. doi: 10.1006/dbio.1994.1079. [DOI] [PubMed] [Google Scholar]
- 39.Rooke J, Pan D, Xu T, Rubin G M. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 40.Finelli A L, Xie T, Bossie C A, Blackman R K, Padgett R W. Genetics. 1995;141:271–281. doi: 10.1093/genetics/141.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]