Abstract
The inhibitor of apoptosis protein survivin has been implicated in both cell cycle control and apoptosis resistance. To discriminate between these different roles, we used transgenic expression of survivin in the skin as a model for cell proliferation, differentiation, and apoptosis. Transgenic mice expressing survivin under the control of a keratin-14 promoter developed normally, without histologic abnormalities of the skin or hair, epidermal hyperplasia, or developmental abnormalities of basal or suprabasal epidermis. Keratinocyte proliferation assessed under basal conditions, or after ultraviolet-B (UVB) irradiation, or phorbol ester stimulation was unchanged in survivin transgenic mice. In contrast, survivin expression inhibited UVB-induced apoptosis in vitro and in vivo (i.e., sunburn cell formation), whereas it did not affect Fas-induced cell death. When crossed with p53 knockout mice, transgenic expression of survivin in a p53+/– background substituted for the loss of a second p53 allele and further inhibited UVB-induced apoptosis. These data provide the first in vivo evidence that survivin inhibits apoptosis and suggest that this pathway may oppose the elimination of cancerous cells by p53.
Introduction
The epidermis is a self-renewing stratified squamous epithelium that forms the outermost component of the skin (1). A balance of proliferation, differentiation, and cell death preserves epidermal homeostasis. In the basal layer, epidermal stem cells and transient amplifying cells give rise to keratinocytes that undergo multiple rounds of proliferation before terminal differentiation in the spinous and granular layers (2) and formation of the stratum corneum (3). Many features of the keratinization process are consistent with apoptosis, or programmed cell death, including DNA laddering (4), internucleosomal DNA fragmentation (5), and caspase activation (6, 7). Apoptosis also plays a critical role in maintaining normal hair growth, because follicular keratinocytes undergo apoptotic cell death in the catagen (involution) phase of the hair cycle (8).
Two apoptotic pathways of physiologic importance have been identified in skin. The first is triggered by ultraviolet-B (UVB) irradiation, which causes upregulation of the tumor suppressor p53 (9), thus potentially coupling to activation of caspase-9 (10). This process that eliminates keratinocytes harboring DNA damage, i.e., sunburn cells, absolutely requires p53 (11), and defects in this pathway have been associated with accelerated malignant transformation (12). Secondly, keratinocytes may undergo apoptosis after ligation of the cell surface death receptor Fas, which results in activation of caspase-8 (13), and dysregulation of this pathway has been implicated in skin diseases, including cutaneous graft versus host disease (14), toxic epidermal necrolysis (15), and eczematous dermatitis (16).
A potential modulator of epidermal functions has been identified recently as survivin (17), a structurally unique member of the inhibitors of apoptosis (IAP) gene family (18). In addition to cytoprotective bcl-2 proteins, certain IAP molecules have been shown to target a downstream step in apoptosis by associating with initiator and effector caspases and preventing their proteolytic processing and catalytic activity (18). Undetectable in normal skin (19), survivin was found abundantly expressed in fetal skin (20), transformed keratinocytes (19), and most cutaneous premalignant and malignant keratinocytic neoplasms (19). Moreover, interference with survivin expression/function using antisense or a dominant negative mutant caused spontaneous apoptosis in keratinocytes (19). Although these data, together with similar findings in other cell types (21), suggest a role of survivin in preserving cell viability, the precise role of survivin in the modulation of the caspase cascade has not been completely elucidated (22, 23). Adding to the complexity of the survivin pathway, recent findings obtained in gene deletion experiments demonstrated that, consistent with its cell cycle–dependent expression in mitosis (24), survivin played an essential role in cell division (25) and that survivin-like molecules in model organisms, i.e. Caenorhabditis elegans similarly participated in mitosis, but not in apoptosis control (26).
In this study, we took a transgenic approach using the skin as a molecular model to define differential role(s) of survivin in apoptosis versus cell division control. A similar transgenic strategy has been used in the past to investigate the contribution of bcl-2 family members (27, 28), and p53 regulators (29) in epidermal proliferation, differentiation, and apoptosis. We found that transgenic keratinocytes expressing survivin exhibit no changes in cell proliferation or differentiation, but are resistant to UVB-induced apoptosis, and that this pathway may contribute to malignant transformation by impairing p53-dependent apoptosis.
Methods
Keratin 14-survivin transgenic mice.
The survivin transgene was constructed using a pGEM2-based plasmid containing 2.1-kb upstream elements of the keratin-14 (K14) gene and 2.2 kb of the human growth hormone (HGH) gene and polyadenylation sequence (Figure 1a). This expression vector was kindly provided by William Philbrick (Yale University School of Medicine, New Haven, Connecticut, USA) with permission of Elaine Fuchs (University of Chicago, Chicago, Illinois, USA) and has been described previously (30, 31). A cDNA encoding the 140 amino acids and stop codon of mouse survivin (32) was cloned into the BamHI site between the K14 and HGH sequences (Figure 1a), and the internal and 3′ flanking EcoRI sites in the survivin gene were simultaneously disrupted by site-directed mutagenesis using the respective oligonucleotides 5′-CCGTCAGTGAgTTCTTGAAAC-3′ and 5′-CTTGATATCGAgTTCCTGCAGC-3′ and the GeneEditor system (Promega Corp., Madison, Wisconsin, USA) according to the manufacturer’s instructions. Loss of these EcoRI sites and the integrity of the survivin cDNA and flanking sequences were confirmed by restriction mapping and DNA sequencing. Following purification by ion-exchange chromatography (QIAGEN Inc., Valencia, California, USA), the K14/survivin/HGH fusion product was released with EcoRI, separated on a 0.9% agarose gel, and electroeluted into 12,000 molecular weight cutoff dialysis tubing. The fragment was further purified on an Elutip-d column (Schleicher & Schuell Inc., Keene, New Hampshire, USA), according to the manufacturer’s instructions, and dialyzed against injection buffer (10 mM Tris, 0.1 mM EDTA, pH 7.4) by floating the sample on 0.05-μm VMWP 01300 filters (Millipore Corp., Bedford, Massachusetts, USA). Finally, the fragment was diluted in injection buffer to 5 ng/μl, sterile-filtered (0.45 μm), and microinjected into B6C3F2 embryos that were implanted into B6C3F1 pseudopregnant females.
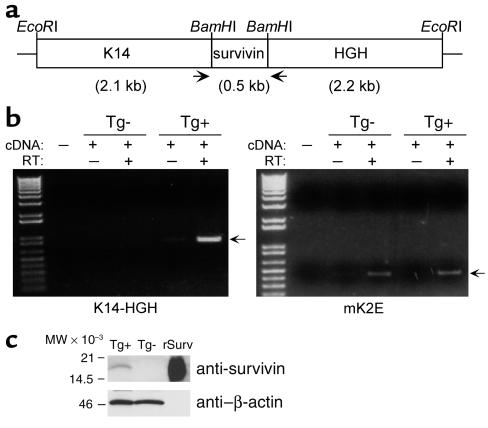
Figure 1.
Construction and characterization of K14-survivin transgenic mice. (a) The 0.5-kb mouse survivin cDNA, including the stop codon, was cloned into a BamHI site flanked by 2.1 kb of K14 promoter sequences and 2.2 kb of HGH and polyadenylation sequences, and the expression cassette was released from the vector by EcoRI digest. Arrows indicate the approximate location of PCR primers used for genotyping and RT-PCR. (b) RNA isolated from epidermal cells of K14-survivin transgenic (Tg+) and nontransgenic (Tg–) mice was incubated in the presence or absence of reverse transcriptase (RT) and amplified by PCR with primers complementary to K14 and HGH (left) or mouse keratin 2E (right). Molecular-weight (MW) markers are in the far left lanes, and arrows designate predicted products. (c) Lysates (50 μg) prepared from epidermal cells of K14-survivin (Tg+) and nontransgenic (Tg–) mice or aliquots (5 ng) of recombinant survivin (rSurv) were electrophoresed, transferred to nylon membranes, and blotted with Ab’s to survivin or β-actin, as indicated. Molecular-weight markers are shown on the left.
Of the five mice (out of 26 potential founders) that carried the transgene by tail DNA genotyping, one developed hydrocephalus at 3 weeks of age and was euthanized and two did not transmit the transgene to offspring in Mendelian fashion. One founder (no. 19) of the two remaining mice yielded offspring with detectable transgene expression, and these were propagated by repeated mating with C57BL/6NCR mice (National Cancer Institute, Bethesda, Maryland, USA). Litters from the second and third backcrosses were used for these studies.
Genotyping.
Potential founders and littermates were screened for the transgene by PCR. Genomic DNA was prepared by incubation of 2 mm of tail in 0.3 ml digestion buffer (50 mM NaCl, 50 mM Tris, pH 8, 100 mM EDTA, 1% SDS) containing 133 μg/ml proteinase K (Sigma Chemical Co., St. Louis, Missouri, USA) at 56°C for 4 hours, followed by addition of RNase A (100 μg/ml, Roche Molecular Biochemicals, Indianapolis, Indiana, USA) and further incubation at 37°C for 1 hour. Following repeated extraction with phenol/chloroform/isoamyl alcohol (25:24:1; Roche Molecular Biochemicals), genomic DNA was precipitated with isopropanol, washed in 70% ethanol, and resuspended in 10 mM Tris, 1 mM EDTA, pH 8. The transgene was amplified in 25 μl with 0.6 U Taq polymerase (Roche Molecular Biochemicals) from 0.1 μg genomic DNA using 200 μM 2′-dinucleoside 5′-triphosphates (Roche Molecular Biochemicals) and primers (10 pmol of each) corresponding to the flanking K14 (5′-TAGCCTGTGGGTGATGAAAG-3′) and HGH (5′-CTGAGATTGGCCAAATACTGG-3′) sequences. PCR reactions were carried out as follows: 2 minutes of denaturation at 94°C; 30 cycles of 94°C for 30 seconds, 56°C for 1 minute, and 72°C for 1 minute; and a 7-minute extension at 72°C. Five microliters was sufficient to visualize the 0.75-kb product on ethidium bromide–stained 1% agarose gels.
Preparation of keratinocytes.
Epidermal keratinocytes were isolated from 7- to 9-week-old mice as described elsewhere (33), with modifications. Hair was immediately removed from killed mice using Oster A5 clippers and a #40 blade (VWR Scientific, Bridgeport, New Jersey, USA), followed by a Braun 3612 shaver (Braun GmbH, Kronberg, Germany). Shaved skin was excised, residual subcutaneous tissues were scraped away, and skin was cut into 0.5-cm strips. After floating dermal side down in trypsin buffer (148 mM NaCl, 5.4 mM KCl, 5.5 mM glucose, pH 7.6) containing 2.3 mg/ml trypsin (T-8003; Sigma Chemical Co.) at 37°C for 1 hour, the epidermis was peeled off and keratinocytes were dissociated in trypsin buffer containing 12,000 U per milliliter DNase I (Calbiochem-Novabiochem Corp., La Jolla, California, USA) by vigorous agitation at 37°C for 7 minutes and then placed on ice. Cells were diluted 1:2 in ice-cold complete medium consisting of Dulbecco’s minimal Eagle’s medium (GIBCO-BRL; Life Technologies Inc., Grand Island, New York, USA) containing 10% FCS (Gemini Bio-Products, Woodland, California, USA), 2 mM L-glutamine (BioWhittaker Inc., Walkersville, Maryland, USA), 100 U/ml penicillin (BioWhittaker Inc.), 100 μg/ml streptomycin (BioWhittaker Inc.), and 1,200 U/ml DNase I and filtered through autoclaved Nitex nylon mesh (Safer America Inc., Depew, New York, USA). Cells were pelleted and suspended in complete medium, centrifuged over Histopaque-1083 (Sigma Chemical Co.) to remove dead cells, and then washed in PBS, pH 7.4. For Western blot analysis, keratinocytes were not subjected to gradient centrifugation.
RT-PCR and Western blot analysis.
Total RNA was purified from epidermal cells using TriReagent (Molecular Research Center Inc., Cincinnati, Ohio, USA). RT-PCR was performed with 0.5 μg RNA and the Superscript Preamplification System (GIBCO-BRL; Life Technologies Inc.) according to the manufacturer’s instructions. Oligo-dT was used to prime reverse transcription, and the K14 and HGH primers noted above were used for PCR. Primers 5′-TTGCCAGATCTCCTGCAGGTC-3′ and 5′-CCACCTCCAAATCGGCCTCCG-3′, corresponding to the mouse keratin-2E gene, were used as a positive control for nontransgenic animals. Reverse transcriptase was not added to some reactions as a negative control. In another series of experiments, epidermal cell lysates isolated from K14-survivin transgenic animals or control littermates were subjected to Western blot analysis using a rabbit polyclonal Ab raised against full-length recombinant human survivin (currently available from NOVUS Biologicals Inc., Littleton, Colorado, USA), which cross-reacts with mouse survivin, followed by chemiluminescence, as described (34).
Immunohistochemistry and immunofluorescence microscopy.
Freshly excised whole skin samples from K14-survivin transgenic mice or control normal littermates were embedded in OCT compound and frozen. Five-micrometer tissue sections were cut, fixed in ice-cold acetone for 10 minutes, air dried for 15 minutes, and rehydrated in Tris-buffered saline (TBS), pH 7.0, for 10 minutes. Slides were then placed over moistened paper towels in a covered tray. The sections were blocked with 0.5% normal goat serum (Vector Laboratories, Burlingame, California, USA) in TBS, pH 7.0, for 5 minutes. For immunocytochemistry, a polyclonal Ab raised against the survivin sequence A3-I19 and used in previous studies on mouse tissues (20) was applied at a concentration of 5 μg/ml in TBS, pH 7.0, containing 1% BSA and incubated overnight at 4°C. Binding of the primary Ab was visualized using a goat anti-rabbit polyclonal Ab (1:100) and Vectastain Elite ABC and 3-amino-9-ethyl carbazole (AEC) peroxidase substrate kits (Vector Laboratories) according to the manufacturer’s instructions. In control experiments, the survivin Ab was replaced with a rabbit polyclonal Ab to enhanced green flourescent protein (EGFP). At the end of the incubation, slides were washed in water, counterstained for 5 seconds with Harris hematoxylin-2 (Sigma Chemical Co.), and washed again in water, prior to mounting and coverslipping. In another series of experiments, paraffin sections were cut from shaved formalin-fixed skin of 8-week-old K14-survivin transgenic mice or control normal littermates and treated as described (34), except without an antigen-retrieval step. Skin sections were incubated with 2 μg/ml nonimmune rabbit IgG (Sigma Chemical Co.) or rabbit Ab’s to mouse keratin-1 or keratin-14 (MK1, MK14; Covance Research Products Inc., Richmond, California, USA) for 1 hour at 22°C. Staining was visualized using a Histomouse-SP kit (Zymed Laboratories Inc., South San Francisco, California, USA) with AEC as the chromophore. For subcellular localization of transgenic survivin, keratinocytes from K14-survivin transgenic mice and control littermates were isolated as described above and incubated overnight in complete medium to adhere to 13 mm no. 1 optical glass coverslips (ProSciTech, Thuringowa Central, Queensland, Australia). Cells were fixed at –20°C in methanol for 5 minutes followed by acetone for 5 seconds and then air dried. Analysis for survivin expression by fluorescence microscopy was performed using the polyclonal rabbit Ab to full-length survivin (NOVUS Biologicals Inc.). Binding of the primary Ab was visualized using Texas red–conjugated goat anti-rabbit IgG (Molecular Probes Inc., Eugene, Oregon, USA), and cell nuclei were detected by simultaneous labeling with Hoechst 33342. Coverslips were analyzed on a Zeiss Axiophot microscope (Carl Zeiss GmbH, Jena, Germany) equipped with ×63/NA planopochromatic lens, and image files were collected and assembled in Photoshop 5.0.
UVB-induced apoptosis in vivo.
Mice 7–9 weeks of age were shaved to expose two 1 × 1.5-cm patches of dorsal skin and 24 hours later irradiated unrestrained in open cages at a dose of approximately 600 J/m2 (at a rate of 2.5 J/m2 per second) using a bank of three sun lamps (FS20T12-UVB; National Biological Corp., Twinsburg, Ohio, USA). These bulbs emit wavelengths between 250 and 420 nm (72.6% UVB, 27.4% UVA, 0.01% UVC), with peak emission at 313 nm, according to the manufacturer. A filter (Kodacel TA422; Eastman Kodak Co., Rochester, New York) was placed over the cages to block residual UVC. Dosimetry was determined using a newly calibrated UVB-500C meter (National Biological Corp.). Animals were killed 24 hours after irradiation, and the exposed patches of skin were excised and fixed in 10% buffered formalin. Skin was then cut into 1- to 2-mm strips, which were stacked on top of each other for embedding in paraffin and routine staining with hematoxylin and eosin. This technique allowed for histologic analysis of 6–7 linear centimeter of skin on a single slide. Apoptotic “sunburn” cells were identified within the epidermis by the presence of eosinophilic cytoplasm and condensed, fragmented, or absent nuclei, as described (11). TUNEL staining was carried out on unstained sections using the ApopTag kit (Intergen Co., Purchase, New York, USA), as described previously (34).
Keratinocyte proliferation and hyperplasia in vivo.
Mice were irradiated with 600 J/m2 UVB, and 72 hours later were injected intraperitoneally with 5-bromo-2′-deoxyuridine (BrdU, 50 mg/kg; Sigma Chemical Co.) in PBS, pH 7.4. After 2 hours, mice were killed, and both irradiated and unirradiated (unshaven) skin was excised and processed as above. In separate experiments, mice were shaved and 0.2 ml acetone or acetone containing 6.8 nmol PMA (Sigma Chemical Co.) was applied every 3 days for a total of four treatments. Forty-eight hours after the last application, mice were killed and skin was excised and processed. Tissue sections were stained for BrdU using a kit (Zymed Laboratories Inc.), as described previously (35). In sections stained with hematoxylin and eosin, epidermal thickness was measured from the top of the granular layer to the basement membrane at up to 20 sites in multiple strips of skin.
UVB-induced apoptosis in vitro.
Freshly isolated keratinocytes were resuspended in PBS, pH 7.4, gently spread as a thin film (50–100 μl) onto the center of 35 × 10–mm polystyrene dishes (Falcon no. 353001), and irradiated with UVB. Complete medium was then carefully added (2 ml per dish) from the side so as not to disrupt the loosely adherent keratinocytes, and dishes were placed in a humidified incubator (37°C, 5% CO2). In some experiments, the pan-caspase inhibitor ZVAD-fmk (40 μM; Enzyme Systems Products Inc., Livermore, California, USA) was included in the medium. In other experiments, medium containing recombinant mouse IFN-γ (Calbiochem-Novabiochem Corp.) and hamster anti-mouse Fas Ab (clone Jo2, PharMingen, San Diego, California, USA) was added to unirradiated cells. After 20–24 hours, the adherent cells were washed, fixed, stained with 4,6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.), and assessed for apoptotic morphology by fluorescence microscopy, as described previously (34).
Keratin 14-survivin p53+/– mice.
K14-survivin transgenic mice were crossed with p53-deficient (p53–/–) mice on identical background strain (Taconic Farms, Germantown, New York, USA) to generate transgenic and nontransgenic p53+/– animals. Littermates were genotyped for the presence of the survivin transgene by PCR as discussed above. No gross or histologic differences were noted between transgenic and nontransgenic p53+/– animals observed from birth to 4 months.
Statistics.
Data derived from multiple animals or determinations were subjected to an unpaired t test using Welch’s correction (Prism; Graphpad Software for Science Inc., San Diego, California, USA). P values less than 0.05 were considered statistically significant.
Results
Expression and characterization of transgenic survivin in the skin.
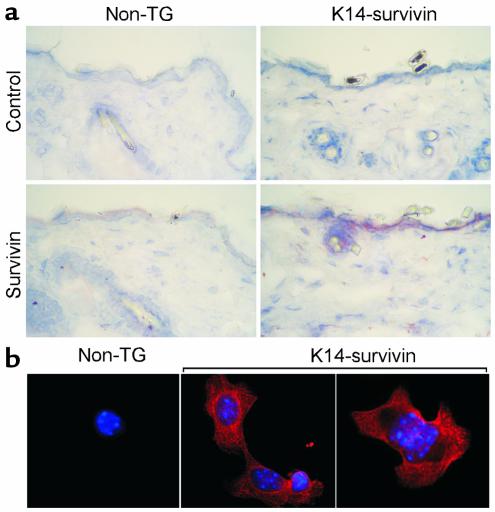
To investigate survivin function(s) in keratinocytes in vivo, transgenic mice were generated with wild-type mouse survivin under the control of a K14 promoter. This cassette (Figure 1a) has been used previously to direct expression of transgenes to the basal epidermis and outer root sheath of hair follicles (30, 31). Expression of the K14-survivin transgene by RT-PCR was demonstrated in keratinocytes derived from transgenic animals, but not from nontransgenic littermates (Figure 1b). Expression of the mouse keratin-2E gene served as a positive control and was detected in keratinocytes from both sets of animals (Figure 1b). In parallel experiments, a 16.5-kDa survivin band comigrating with recombinant survivin was demonstrated by Western blot analysis in epidermal lysates from K14-survivin transgenic mice, but not from nontransgenic littermates (Figure 1c). Consistent with these findings, selective expression of survivin in epidermal keratinocytes of K14-survivin transgenic mice was demonstrated by immunohistochemistry (Figure 2a). No survivin staining was seen in fibroblasts or other dermal cells or when a control Ab was used (Figure 2a). By contrast, epidermal keratinocytes of nontransgenic littermates did not demonstrate survivin staining (Figure 2a). Next, we investigated the subcellular distribution of transgenic survivin. Consistent with the data presented above, survivin expression was not detected in nontransgenic keratinocytes by immunofluorescence labeling (Figure 2b). By contrast, survivin expressed in keratinocytes of K14-survivin transgenic mice exhibited a cytoplasmic filamentous pattern, as seen by immunofluorescence labeling (Figure 2b).
Figure 2.
Immunohistochemical and subcellular localization of transgenic survivin expression. (a) Five-micrometer tissue sections were cut from fresh-frozen skin of K14-survivin transgenic mice (K14-survivin) or control nontransgenic (non-TG) littermates, fixed in ice-cold acetone, and analyzed by immunohistochemistry with Ab against survivin or a control Ab to EGFP. Binding of the primary Ab’s was visualized with a goat anti-rabbit polyclonal Ab using Vectastain Elite ABC and AEC peroxidase substrate kits. (b) Subcellular localization of transgenic survivin. Keratinocytes isolated from K14-survivin transgenic mice (K14-survivin) or nontransgenic littermates (non-TG) were adhered to glass coverslips, fixed in methanol-acetone, and incubated with an Ab to survivin, followed by Texas red–conjugated goat anti-rabbit Ab. Nuclei were stained with Hoechst 33342. Image merging analysis is shown.
Skin phenotype of K14-survivin transgenic mice.
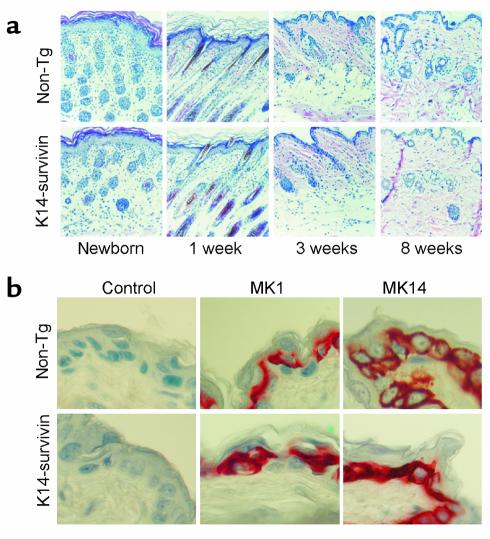
K14-survivin mice were clinically indistinguishable from nontransgenic littermates (not shown). Observation of transgenic animals from birth to 1 year of age revealed no abnormalities of skin or hair compared with nontransgenic littermates. Histologically, survivin expression did not perturb epidermal differentiation since epidermis from K14-survivin mice exhibited normal thickness, stratum corneum formation, timing of follicular development, and follicular morphology (Figure 3a). A potential effect of transgenic expression of survivin on normal skin differentiation was further investigated by immunohistochemical analysis of cytokeratins 14 and 1 as markers for basal and suprabasal epidermal cells, respectively. In these experiments, intense labeling with both cytokeratins was indistinguishably observed in K14-survivin and control nontransgenic littermates (Figure 3b), thus further demonstrating that transgenic survivin expression did not affect the normal skin differentiation program.
Figure 3.
Histologic analysis of epidermis in K14-survivin animals. (a) K14-survivin transgenic mice (K14-survivin) or nontransgenic littermates (non-TG) were genotyped at birth, sacrificed at the indicated time intervals, and dorsal skin was excised, processed, and stained with hematoxylin/eosin. (b) Skin sections isolated from K14-survivin or non-TG control littermates were analyzed for reactivity with Ab’s to mouse cytokeratin-1 (MK1) or -14 (MK14), or nonimmune rabbit IgG (control) by immunohistochemistry.
Effect of survivin expression on keratinocyte proliferation in vivo.
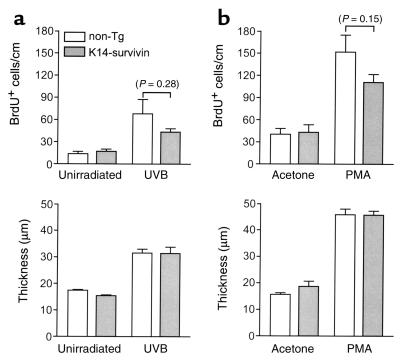
Given the proposed role of survivin as a critical regulator of cytokinesis (25) and the evolutionary conservation of this pathway (26), we initially examined the effect of transgenic expression of survivin on normal keratinocyte proliferation in vivo. Keratinocyte proliferation was induced by UVB irradiation or repeated application of PMA and assessed histologically by both BrdU staining and development of epidermal hyperplasia (thickness). As shown in Figure 4, survivin expression in skin did not influence basal proliferative rate or epidermal thickness. Moreover, neither UVB- nor PMA-induced proliferation or epidermal hyperplasia was significantly enhanced in K14-survivin mice compared with nontransgenic littermates (Figure 4).
Figure 4.
Effect of transgenic survivin expression on keratinocyte proliferation and skin hyperplasia in vivo. (a) Nontransgenic littermates (white bars) and K14-survivin mice (gray bars) were irradiated with 600 J/m2 UVB and 72 hours later injected with BrdU. After 2 hours, unshaven (unirradiated) and shaved (UVB-treated) dorsal skin was excised and processed. BrdU-labeled cells were detected by immunohistochemistry, and epidermal thickness was measured. Data are expressed as number of BrdU-positive cells per linear centimeter of interfollicular epidermis (top) and as epidermal thickness (bottom). Error bars indicate SEM for five mice per group. P value for nontransgenic and K14-survivin mice is noted above bars in parentheses. (b) Nontransgenic littermates (white bars) and K14-survivin mice (gray bars) were shaved and subjected to topical applications of acetone or acetone containing PMA every 3 days. Forty-eight hours after the fourth application, mice were injected with BrdU, and BrdU-labeled cells and epidermal thickness were assessed as above. Error bars indicate SEM for seven mice per group. P value for nontransgenic and K14-survivin mice is noted above error bars in parentheses.
Effect of survivin expression on keratinocyte apoptosis in vivo.
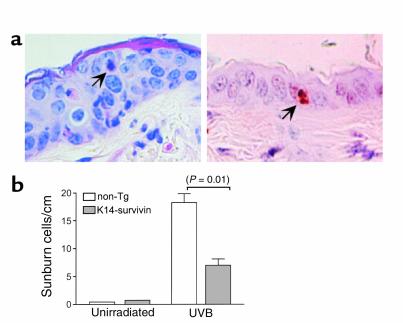
The antiapoptotic function of survivin in transgenic keratinocytes was next investigated on sunburn cell formation. Mice were shaved, exposed to a single dose (600 J/m2) of UVB, and skins were harvested 24 hours later for histologic examination. These experimental conditions have been optimized for sunburn cell detection in mouse skin (36). Sunburn cells were easily identified on hematoxylin/eosin–stained sections of UVB-irradiated skin and confirmed as apoptotic cells by internucleosomal DNA fragmentation by TUNEL staining (Figure 5a). Compared with nontransgenic littermates, sunburn cell formation in K14-survivin mice was reduced by 60% (P < 0.01) (Figure 5b). No reduction in sunburn cell formation was observed in transgenic animals derived from a nonexpressing founder line (not shown).
Figure 5.
Effect of transgenic survivin on keratinocyte apoptosis in vivo. (a) Sections of UVB-irradiated (600 J/m2) nontransgenic mouse skin were stained with hematoxylin/eosin (left) or for internucleosomal DNA fragmentation by TUNEL (right). Arrows indicate an apoptotic keratinocyte (sunburn cell). (b) Quantitation of UVB-induced apoptosis in vivo. K14-survivin transgenic mice (gray bars) or nontransgenic littermates (white bars) were irradiated with UVB, and 24 hours later exposed skin was excised and sunburn cells were identified and counted by light microscopy and hematoxylin/eosin staining. Data are expressed as number of sunburn cells per linear centimeter of skin analyzed. Error bars indicate SEM of 2 and nine mice per group of unirradiated and irradiated mice, respectively. P value for nontransgenic and K14-survivin mice is noted in parentheses.
Characterization of survivin-dependent cytoprotection in keratinocytes.
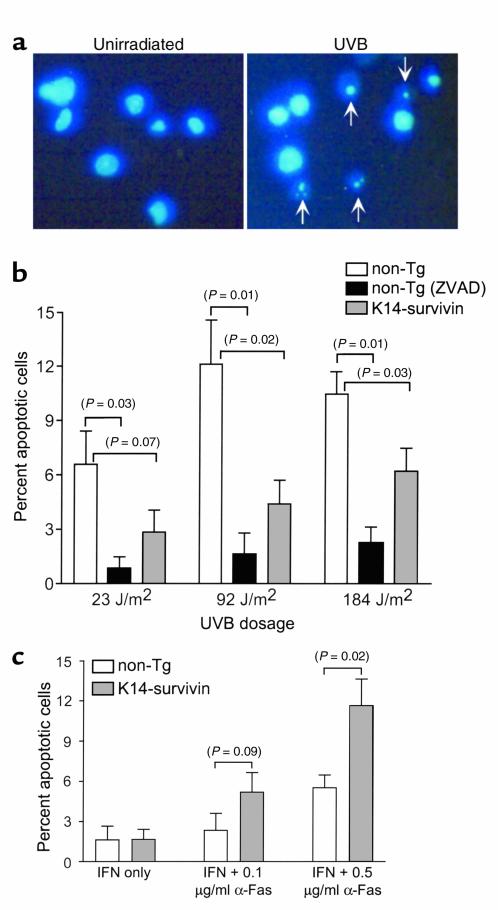
To further investigate the antiapoptotic pathway mediated by transgenic survivin in skin, we subjected freshly isolated keratinocytes to UVB irradiation followed by analysis of nuclear morphology of apoptosis using DAPI staining and fluorescence microscopy (34). As shown in Figure 6a, UVB irradiation resulted in chromatin condensation and nuclear fragmentation in isolated keratinocytes analyzed by DAPI staining 24 hours after UVB treatment (Figure 6a). Consistent with genuine caspase-dependent apoptosis, addition of the pan-caspase inhibitor Z-VAD-fmk entirely reversed UVB-induced keratinocyte apoptosis (Figure 6b). Under these experimental conditions, keratinocytes isolated from K14-survivin mice were significantly (P = 0.02) more resistant to UVB-induced apoptosis in vitro than those isolated from nontransgenic littermates (Figure 6b).
Figure 6.
Characterization of survivin-dependent cytoprotection in transgenic keratinocytes. (a) Isolated keratinocytes were untreated or irradiated with UVB (90 J/m2) in vitro, and 24 hours later fixed, stained with DAPI, and analyzed by fluorescence microscopy. Arrows indicate apoptotic keratinocytes with condensed and fragmented nuclei. (b) Keratinocytes isolated from nontransgenic littermates (white bars) or K14-survivin transgenic mice (gray bars) were exposed to the indicated UVB doses in vitro, then fixed and stained with DAPI. Keratinocytes from nontransgenic littermates were also separately incubated in the presence of the pan-caspase inhibitor, ZVAD-fmk (black bars). Data are expressed as percentage of apoptotic cells based on counting approximately 150 cells. (c) Effect of transgenic survivin on IFN-γ/anti-Fas–induced apoptosis. Keratinocytes isolated from nontransgenic littermates (white bars) or K14-survivin mice (gray bars) were cultured with 1,000 U/ml IFN-γ alone or in combination with 0.1 or 0.5 μg/ml anti-Fas. After 24 hours, cells were fixed, stained with DAPI, and analyzed for nuclear morphology of apoptosis. Data are expressed as percentage of apoptotic cells based on counting approximately 120–150 cells. For each condition, the background level of apoptosis (ranging from 5 to 10%) in unirradiated cells has been subtracted. Error bars indicate SEM of four independent experiments. P values are noted in parentheses.
We next investigated if transgenic expression of survivin in keratinocytes selectively influenced the intrinsic, caspase-9–dependent, or the extrinsic, caspase-8–dependent pathway of apoptosis. Fas is weakly expressed on cultured murine keratinocytes, but is induced by exposure to IFN-γ; the combined treatment of IFN-γ plus anti-Fas Ab has been shown to induce apoptosis in keratinocytes (37). For these experiments, keratinocytes were isolated from both transgenic and nontransgenic animals, treated with 1,000 U/ml IFN-γ and anti-Fas for 24 hours, and analyzed by DAPI nuclear staining. IFN-γ at this concentration alone did not induce apoptosis (Figure 6c), whereas culture periods greater than 24 hours were associated with high background levels of keratinocyte apoptosis (not shown). In contrast to the cytoprotective effect of survivin on sunburn cell formation, keratinocytes isolated from K14-survivin mice were highly susceptible to Fas-mediated apoptosis. In fact, IFN-γ and 0.5 μg/ml anti-Fas treatment resulted in twofold increased apoptosis (P = 0.02) in transgenic compared with nontransgenic keratinocytes (Figure 6c).
Survivin cooperates with loss of p53 in inhibiting apoptosis in vivo and in vitro.
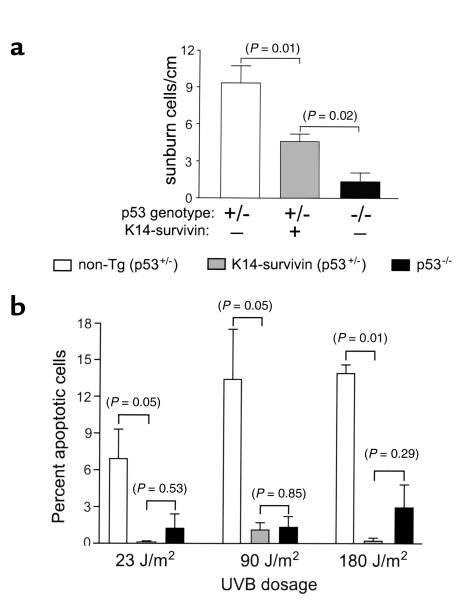
To investigate the impact of survivin cytoprotection on known molecular abnormalities of skin cancer, we crossed K14-survivin transgenic animals with p53-deficient (p53–/–) mice and generated K14-survivin transgenic and nontransgenic animals lacking a p53 allele (p53+/–). Groups of mice were irradiated with UVB, and 24 hours later skins were harvested for histologic examination. As shown in Figure 7a, loss of a p53 allele in nontransgenic animals was associated with a low level of sunburn cell formation in vivo, in agreement with previous observations (11). Loss of a p53 allele in K14-survivin transgenic animals was associated with a further substantial reduction (P < 0.01) in sunburn cell formation (Figure 7a). Although profoundly suppressed, the susceptibility of these animals to UVB-induced apoptosis remained higher (P = 0.02) than that of p53-deficient animals in which sunburn cell formation was completely abolished (Figure 7a).
Figure 7.
Survivin cooperates with loss of p53 in inhibiting UVB-induced apoptosis. (a) Nontransgenic (p53+/–; white bars), K14-survivin (p53+/–; gray bars), and p53 knockout (p53–/–; black bars) mice were irradiated with 600 J/m2 UVB, and sunburn cells were quantitated by hematoxylin/eosin staining. Data are expressed as number of sunburn cells per linear centimeter of skin analyzed. Error bars indicate SEM of 7, 11, and 3 mice per group of nontransgenic p53+/–, K14-survivin p53+/–, and p53–/– mice, respectively. (b) Keratinocytes isolated from nontransgenic p53+/– (white bars), K14-survivin p53+/– (gray bars), and p53–/– (black bars) mice were irradiated with UVB in vitro at the indicated doses. After 24 hours, cells were fixed, stained with DAPI, and analyzed for nuclear morphology of apoptosis. Error bars indicate SEM of three independent experiments. P values are noted in parentheses.
However, when keratinocytes were isolated from nontransgenic p53+/–, K14-survivin transgenic p53+/–, and p53–/– mice and assessed for susceptibility to UVB-induced apoptosis, the combination of survivin expression and loss of a p53 allele resulted in complete resistance to UVB-induced apoptosis, in vitro (Figure 7b). As shown in Figure 7b, the resistance to apoptosis of keratinocytes from K14-survivin p53+/– mice was not significantly (P = 0.29, 0.53, 0.85) different from that of p53–/– keratinocytes. Thus, survivin expression under these conditions can substitute for loss of a second p53 allele in inhibition of apoptosis.
Discussion
In this study, we have shown that transgenic expression of survivin in the skin did not affect epidermal cell proliferation or differentiation, whereas it effectively inhibited UVB-induced apoptosis. Second, survivin-dependent cytoprotection selectively targeted the “intrinsic” pathway of apoptosis and was ineffective against Fas-induced cell death. Finally, expression of survivin cooperated with loss of p53 in preventing keratinocyte apoptosis in response to UVB-induced DNA damage.
The impetus for this study was twofold. First, we wished to determine the potential impact of survivin in pathogenetic mechanisms of skin cancer in vivo. This stemmed from the differential expression of survivin in various cutaneous neoplasms but not in normal epidermis (19), which paralleled a more general paradigm of survivin overexpression in many human cancers but not in normal tissues (21). Although a previous report suggested that survivin was present in normal skin (38), data from our previous work (19) and sensitive techniques of RT-PCR and Western blot analysis presented here confirmed that survivin was expressed in transgenic keratinocytes but was undetectable in normal epidermis. Second, we wished to discriminate between potentially distinct roles of survivin in cell division versus apoptosis inhibition. Although a pathway of survivin-dependent cytoprotection has been identified in several studies (39), data obtained with knockout mice suggested that survivin played an essential role in mitosis (25), which, at least in C. elegans, could be separated from apoptosis inhibition (26).
The first conclusion that can be drawn from our transgenic studies is that expression of survivin did not affect keratinocyte differentiation or normal epidermal maturation and did not increase basal or UVB/PMA-induced keratinocyte proliferation. These findings are at variance with the phenotype of skin hyperplasia observed after transgenic expression of other apoptosis inhibitors, including bcl-2 (28) or the p53 regulator MDM-2 (29), but are reminiscent of the role of bcl-XL (40) in controlling skin apoptosis without affecting cell proliferation. Thus, contrary to the speculations of knockout studies (25), our data demonstrate that overexpression of survivin in adult tissues, as seen typically in human tumors (21), does not confer, per se, a more pronounced proliferative phenotype. It is also clear that overexpression of survivin in cancer does not simply reflect a higher number of mitotic cells. As exemplified in our recent analysis of melanoma, survivin expression was detected in the vast majority of tumor cells, not only the mitotic fraction, and was indistinguishable in cases with low (1.8) or high (38.2) mitotic index (34). Accordingly, “cancer-specific” changes in survivin gene expression are beginning to be unraveled, and a recent study demonstrated that the human survivin promoter is methylated in noncancerous ovaries (90.6%), but unmethylated, thus transcriptionally active, in 79% of ovarian cancers expressing DNA demethylase (41).
Second, the data presented here identify for the first time a role of survivin in apoptosis inhibition in vivo. Although previous experiments from several groups implicated survivin in cytoprotection against various apoptotic insults (21, 39), the exact mechanism by which survivin couples to the caspase cascade has not been completely elucidated (22, 23) and may involve different structural requirements from those used by other cytoprotective IAP family members, i.e., XIAP (42). Here, survivin expression in the skin significantly inhibited UVB-induced keratinocyte apoptosis, and this pathway was further suppressed when survivin expression coincided with loss of a single p53 allele. p53-dependent apoptosis is the primary means of eliminating keratinocytes that have sustained UVB-induced DNA damage and represents an important defense mechanism against skin cancer (11). Accordingly, inactivating mutations of one p53 allele have been recognized as one of the earliest molecular abnormalities of UVB-induced skin carcinogenesis (43). Importantly, progression of skin cancer under these conditions is not associated with deletion of the second p53 allele (44), suggesting that the cooperation between survivin expression and loss of one p53 allele described here may further facilitate the development of skin cancer in susceptible patients. Although there are probably several mechanisms by which p53 activation couples to the apoptotic machinery, recent evidence suggests that the intrinsic initiator complex Apaf-1/caspase-9 may be an essential downstream component of this response (10). In this context, a physical complex between survivin and caspase-9 has been demonstrated in vivo, and targeting survivin with a phosphorylation-defective dominant negative mutant resulted in mislocalization of caspase-9 and caspase-9–dependent apoptosis (45). Although it remains to be elucidated how a survivin-caspase-9 complex preserves cytoprotection in vivo, the data presented here suggest that de novo expression of survivin may promote cell viability by interfering with initiation and/or amplification of the intrinsic caspase cascade. At least in keratinocytes, this pathway appears selective for caspase-9, given the inability of survivin to reduce Fas-induced, caspase-8–dependent apoptosis. Because oncogene-dependent apoptosis is mediated by p53 (10) and requires mitochondrial cytochrome c release and caspase-9 activation (46), it seems plausible that p53 and survivin may exert opposite effects on cytoprotection, consistent with the model proposed here of a functional synergism between survivin expression and loss of a p53 allele. The mechanism of increased sensitivity of transgenic keratinocytes to Fas-mediated killing is unclear, but may relate to an imbalance in apoptotic pathways resulting from forced expression of survivin (47). A similar differential sensitivity to apoptotic pathways was also noted in K14 transgenic mice expressing the p53 regulator, MDM2 (29).
In summary, these data suggest that p53 and survivin may represent opposing forces controlling the apoptotic cascade induced by UVB in keratinocytes and potentially culminating with caspase-9 activation. Overexpression of survivin, as commonly seen in skin cancer, would be expected to further dampen a protective activation of caspase-9, already partially compromised by the frequent loss of p53 during skin carcinogenesis (11). Whereas expression of survivin in adult tissues does not confer a proliferative advantage, the cooperation between survivin and loss of p53 described here may provide a general mechanism for aberrant inhibition of apoptosis that may favor the development and progression of neoplasia.
Acknowledgments
We thank Elaine Fuchs and William Philbrick for the K14 expression cassette, Jennifer McNiff for reviewing the skin histology, and Julie Lewis and Robert Tigelaar for assistance in isolating epidermal cells. We are also grateful to Lisa Chuba in Richard Flavell’s laboratory for performing the microinjections. This work was supported by NIH grants CA-78810 and CA-90917 to D.C. Altieri. D. Grossman was supported by NIH Dermatology Training Grant 5T32AR07016 and a Dermatologist Investigator Research Fellowship from the Dermatology Foundation. P.J. Kim was supported by a NIH Cancer Education Grant, Medical Student Research Training Fellowship, and by the Etta S. Chidsey Award in Cancer Research.
Footnotes
See the related Commentary beginning on page 965.
Douglas Grossman’s current address is: Huntsman Cancer Institute, Suite 5243, University of Utah, 2000 Circle of Hope, Salt Lake City, Utah, 84112, USA.
References
- 1.Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- 2.Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 3.Ishida-Yamamoto A, et al. Programmed cell death in normal epidermis and loricrin keratoderma. Multiple functions of profilaggrin in keratinization. J Investig Dermatol Symp Proc. 1999;4:145–149. doi: 10.1038/sj.jidsp.5640198. [DOI] [PubMed] [Google Scholar]
- 4.McCall CA, Cohen JJ. Programmed cell death in terminally differentiating keratinocytes: role of endogenous endonuclease. J Invest Dermatol. 1991;97:111–114. doi: 10.1111/1523-1747.ep12478519. [DOI] [PubMed] [Google Scholar]
- 5.Polakowska RR, Piacentini M, Bartlett R, Goldsmith LA, Haake AR. Apoptosis in human skin development: morphogenesis, periderm, and stem cells. Dev Dyn. 1994;199:176–188. doi: 10.1002/aja.1001990303. [DOI] [PubMed] [Google Scholar]
- 6.Weil M, Raff MC, Braga VM. Caspase activation in the terminal differentiation of human epidermal keratinocytes. Curr Biol. 1999;9:361–364. doi: 10.1016/s0960-9822(99)80162-6. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Ogo M, Hibino T. Partial purification and characterization of two distinct types of caspases from human epidermis. J Invest Dermatol. 1998;111:367–372. doi: 10.1046/j.1523-1747.1998.00295.x. [DOI] [PubMed] [Google Scholar]
- 8.Weedon D, Strutton G. The recognition of early stages of catagen. Am J Dermatopathol. 1984;6:553–555. doi: 10.1097/00000372-198412000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Differential regulation of P53 and Bcl-2 expression by ultraviolet A and B. J Invest Dermatol. 1998;111:380–384. doi: 10.1046/j.1523-1747.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- 10.Soengas MS, et al. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science. 1999;284:156–159. doi: 10.1126/science.284.5411.156. [DOI] [PubMed] [Google Scholar]
- 11.Ziegler A, et al. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 12.Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
- 13.Wehrli P, Viard I, Bullani R, Tschopp J, French LE. Death receptors in cutaneous biology and disease. J Invest Dermatol. 2000;115:141–148. doi: 10.1046/j.1523-1747.2000.00037.x. [DOI] [PubMed] [Google Scholar]
- 14.Gilliam AC, Whitaker-Menezes D, Korngold R, Murphy GF. Apoptosis is the predominant form of epithelial target cell injury in acute experimental graft-versus-host disease. J Invest Dermatol. 1996;107:377–383. doi: 10.1111/1523-1747.ep12363361. [DOI] [PubMed] [Google Scholar]
- 15.Viard I, et al. Inhibition of toxic epidermal necrolysis by blockade of CD95 with human intravenous immunoglobulin. Science. 1998;282:490–493. doi: 10.1126/science.282.5388.490. [DOI] [PubMed] [Google Scholar]
- 16.Trautmann A, et al. T cell-mediated Fas-induced keratinocyte apoptosis plays a key pathogenetic role in eczematous dermatitis. J Clin Invest. 2000;106:25–35. doi: 10.1172/JCI9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 18.Deveraux QL, Reed JC. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 19.Grossman D, McNiff JM, Li F, Altieri DC. Expression of the apoptosis inhibitor, survivin, in nonmelanoma skin cancer and gene targeting in a keratinocyte cell line. Lab Invest. 1999;79:1121–1126. [PubMed] [Google Scholar]
- 20.Adida C, et al. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43–49. [PMC free article] [PubMed] [Google Scholar]
- 21.Altieri DC, Marchisio PC. Survivin apoptosis: an interloper between cell death and cell proliferation in cancer. Lab Invest. 1999;79:1327–1333. [PubMed] [Google Scholar]
- 22.Shin S, et al. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40:1117–1123. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 23.Verdecia MA, et al. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- 24.Li F, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 25.Uren AG, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- 26.Speliotes EK, Uren A, Vaux D, Horvitz HR. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- 27.Hockenbery DM, Zutter M, Hickey W, Nahm M, Korsmeyer SJ. BCL2 protein is topographically restricted in tissues characterized by apoptotic cell death. Proc Natl Acad Sci USA. 1991;88:6961–6965. doi: 10.1073/pnas.88.16.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Villanueva J, et al. Human keratin-1.bcl-2 transgenic mice aberrantly express keratin 6, exhibit reduced sensitivity to keratinocyte cell death induction, and are susceptible to skin tumor formation. Oncogene. 1998;16:853–863. doi: 10.1038/sj.onc.1201610. [DOI] [PubMed] [Google Scholar]
- 29.Ganguli G, Abecassis J, Wasylyk B. MDM2 induces hyperplasia and premalignant lesions when expressed in the basal layer of the epidermis. EMBO J. 2000;19:5135–5147. doi: 10.1093/emboj/19.19.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassar R, Fuchs E. Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 1991;5:714–727. doi: 10.1101/gad.5.5.714. [DOI] [PubMed] [Google Scholar]
- 31.Wysolmerski JJ, et al. Overexpression of parathyroid hormone-related protein in the skin of transgenic mice interferes with hair follicle development. Proc Natl Acad Sci USA. 1994;91:1133–1137. doi: 10.1073/pnas.91.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Altieri DC. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 1999;59:3143–3151. [PubMed] [Google Scholar]
- 33.Sullivan S, Bergstresser PR, Tigelaar RE, Streilein JW. FACS purification of bone marrow-derived epidermal populations in mice: langerhans cells and Thy-1+ dendritic cells. J Invest Dermatol. 1985;84:491–495. doi: 10.1111/1523-1747.ep12273454. [DOI] [PubMed] [Google Scholar]
- 34.Grossman D, McNiff JM, Li F, Altieri DC. Expression and targeting of the apoptosis inhibitor, survivin, in human melanoma. J Invest Dermatol. 1999;113:1076–1081. doi: 10.1046/j.1523-1747.1999.00776.x. [DOI] [PubMed] [Google Scholar]
- 35.Grossman D, Kim PJ, Schechner JS, Altieri DC. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc Natl Acad Sci USA. 2001;98:635–640. doi: 10.1073/pnas.230450097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto H, Mizuno K, Itoh T, Tanaka K, Horio T. Evaluation of apoptotic cells induced by ultraviolet light B radiation in epidermal sheets stained by the TUNEL technique. J Invest Dermatol. 1999;113:802–807. doi: 10.1046/j.1523-1747.1999.00757.x. [DOI] [PubMed] [Google Scholar]
- 37.Sayama K, Yonehara S, Watanabe Y, Miki Y. Expression of Fas antigen on keratinocytes in vivo and induction of apoptosis in cultured keratinocytes. J Invest Dermatol. 1994;103:330–334. doi: 10.1111/1523-1747.ep12394858. [DOI] [PubMed] [Google Scholar]
- 38.Chiodino C, et al. Communication: expression of the novel inhibitor of apoptosis survivin in normal and neoplastic skin. J Invest Dermatol. 1999;113:415–418. doi: 10.1046/j.1523-1747.1999.00711.x. [DOI] [PubMed] [Google Scholar]
- 39.Reed JC, Bischoff JR. BIRinging chromosomes through cell division: and survivin’ the experience. Cell. 2000;102:545–548. doi: 10.1016/s0092-8674(00)00076-3. [DOI] [PubMed] [Google Scholar]
- 40.Pena JC, Fuchs E, Thompson CB. Bcl-x expression influences keratinocyte cell survival but not terminal differentiation. Cell Growth Differ. 1997;8:619–629. [PubMed] [Google Scholar]
- 41.Hattori M, Sakamoto H, Satoh K, Yamamoto T. DNA demethylase is expressed in ovarian cancers and the expression correlates with demethylation of CpG sites in the promoter region of c- erbB-2 and survivin genes. Cancer Lett. 2001;169:155–164. doi: 10.1016/s0304-3835(01)00499-2. [DOI] [PubMed] [Google Scholar]
- 42.Sun C, et al. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature. 1999;401:818–822. doi: 10.1038/44617. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, et al. Role of PTCH and p53 genes in early-onset basal cell carcinoma. Am J Pathol. 2001;158:381–385. doi: 10.1016/S0002-9440(10)63980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang W, Ananthaswamy HN, Muller HK, Kripke ML. p53 protects against skin cancer induction by UV-B radiation. Oncogene. 1999;18:4247–4253. doi: 10.1038/sj.onc.1202789. [DOI] [PubMed] [Google Scholar]
- 45.O’Connor DS, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 47.Hill LL, et al. Fas ligand: a sensor for DNA damage critical in skin cancer etiology. Science. 1999;285:898–900. doi: 10.1126/science.285.5429.898. [DOI] [PubMed] [Google Scholar]