Abstract
RANK, the receptor activator of NF-κB, and its ligand RANKL (initially termed TRANCE, also termed ODF and OPGL), are a TNF superfamily receptor-ligand pair that govern the development and function of osteoclasts, lymphoid tissue, and mammary epithelium. While TNF family cytokines share a common structural scaffold, individual receptor-ligand pairs associate with high specificity. Given the low level of amino acid conservation among members of the TNF superfamily, the means by which these molecules achieve specificity cannot be completely understood without knowledge of their three-dimensional structures. To determine the elements of RANKL that mediate RANK activation, we have crystallized the ectodomain of murine RANKL and solved its structure to a resolution of 2.6 Å. RANKL self-associates as a homotrimer with four unique surface loops that distinguish it from other TNF family cytokines. Mutagenesis of selected residues in these loops significantly modulates RANK activation, as evidenced by in vitro osteoclastogenesis, thereby establishing their necessity in mediating the biological activities of RANKL. Such structural determinants of RANKL-RANK specificity may be of relevance in the pharmacologic design of compounds to ameliorate osteopenic disorders of bone.
Introduction
RANKL is a member of the TNF superfamily of cytokines. The protein was initially identified as a cytokine with an ability to stimulate T cell and dendritic cell function and was termed TRANCE–TNF-related activation-induced cytokine (1), or alternatively as RANKL (2). Cloned independently by two groups for its ability to stimulate osteoclast differentiation, activity, and survival, it was given the names ODF, osteoclast differentiation factor (3), and OPGL, osteoprotegerin ligand (4). TNF family members mediate a panoply of biological phenomena, modulating essential aspects of inflammation, organogenesis, host defense, autoimmunity, and apoptosis. In general, TNF family cytokines demonstrate pleiotropic capabilities in coordinating the development and function of many disparate tissues and cell lineages. RANKL, in particular, assumes prominent roles in bone, the immune system, and mammary epithelium.
RANKL is an essential cytokine in osteobiology. It mediates the differentiation of bone-resorbing osteoclasts from monocyte-macrophage precursors and modulates the survival and function of mature osteoclasts (5, 6). Mice deficient in RANKL, or its receptor RANK, exhibit a complete lack of osteoclasts, resulting in severe osteopetrosis and hypocalcemia. Clinically, RANKL has been implicated in the pathogenesis of postmenopausal osteoporosis. In this disorder, estrogen deficiency leads to enhanced expression of TNF and RANKL, as well as decreased production of osteoprotegerin (OPG, a soluble TNF receptor [TNFR] family decoy receptor for RANKL) (7, 8). Together, these alterations in cytokine expression induce bone loss by stimulating osteoclast formation. We have demonstrated recently that TNF synergizes with RANKL both in vitro and in vivo to drive cells of the monocyte-macrophage lineage to differentiate along the osteoclastogenic pathway (9). This synergism is achieved at the level of AP-1 and NF-κB activation, two downstream transcriptional pathways activated by both TNFR and RANK.
RANKL also plays an essential role in immunobiology. Expressed on T cells, dendritic cells, and their precursors, RANKL mediates the differentiation of T and B lymphocytes, as well as the survival of dendritic cells in the immune system. Hematopoietic CD4+ precursors require autocrine RANKL-RANK signaling for survival and differentiation while migrating from the marrow to primitive lymphoid tissue anlagen. Mice lacking RANKL or its receptor RANK exhibit defective peripheral and mesenteric lymph node organogenesis (10–12). Clinically, the expression of RANKL by CD4+ T cells mediates bone loss and joint destruction in animal models of arthritis and inflammatory osteolysis (13, 14).
In addition to its regulatory roles in bone and the immune system, RANKL-RANK signaling also governs the differentiation of mammary epithelium (15). Pregnant mice lacking RANKL fail to form lactating breast tissue, with a complete absence of competent lobulo-alveolar glands. Mammary epithelial precursors lacking RANK undergo accelerated apoptosis due to a defect in Akt activation. Furthermore, the osteoclast deficit in these animals obviates mobilization of calcium from bone to the milk. Collectively, these observations illustrate the prominent role of RANKL-RANK in coordinating the normal development and function of lymphoid and mammary tissue, as well as the homeostatic regulation of calcium metabolism and bone mass.
Structurally, TNF family cytokines are expressed as type II transmembrane proteins, each containing a membrane-anchoring domain, a connecting stalk, and a receptor-binding ectodomain. All TNF cytokines share a common structural core (reviewed in refs. 16, 17)—a scaffold of ten hydrogen-bonded strands, in two sheets, that assume a characteristic jellyroll β-sandwich fold. Individual TNF family cytokines differ primarily in the length and composition of the surface loops that connect these β-strands (18). All known TNF cytokine family members self-assemble into noncovalently associated trimers. The core β-strand topology underlies the intrinsic nature of these monomers to oligomerize around an axis of threefold rotational symmetry. While initially tethered to the membrane, biologically active trimers exist as both membrane-bound and soluble cleaved forms.
Although the receptors for RANKL, RANK and OPG, have been categorized as members of the TNFR superfamily according to sequence homology, no three-dimensional structures have been reported to date. Expressed as type I transmembrane proteins, all TNFRs are thought to employ a similar tertiary folding pattern to form elongated structures that project from the cell membrane. The extracellular ligand-binding domains are constructed primarily of three types of small, modular cysteine-rich repeats (19). While ligand binding is generally thought to occur via the second and third cysteine-rich domains (CRD), it has been suggested that the amino-terminal CRD may function as a pre-ligand assembly domain (PLAD). Ligation of TNF family receptors by their cognate ligands induces the cytoplasmic tails to form signaling complexes, with a characteristic threefold stoichiometry, allowing TNF receptor-associated factors (TRAFs) or Fas-associated with death domain (FADD) proteins to activate downstream effector pathways, including MAP (mitogen-activated protein) kinases and NF-κB (reviewed in refs. 16, 17).
The emerging complexity of the TNF superfamily and the essential role that RANKL plays in immunology and osteobiology raise functional questions that can be determined only by structural analysis. For instance, recognition between TNF family cytokines and their receptors is highly specific, with typical dissociation constants in the nanomolar to picomolar range. A number of crystal structures for TNF family cytokines, either alone or in complex with their receptors, have been reported: TNF-α (18, 20), TNF-β (21), TNF-β–TNFR complex (22), CD40L (23), ACRP30 (adipocyte complement–related protein) (24), TRAIL (25), and the TRAIL-DR5 complex (26–28). From these analyses, it is evident that the amino acid conservation among TNF cytokines is largely confined to internal hydrophobic residues involved in monomer folding or trimer assembly. In contrast, the unique regions form surface loops of varying composition and length that connect the core β-strands. Importantly, however, the overall lack of sequence conservation among both ligands and receptors has hindered meaningful structural prediction.
In order to identify the elements of RANKL that govern interaction with its receptor RANK, we have determined the crystal structure of murine RANKL to a resolution of 2.6 Å. We find that RANKL adopts the characteristic structural fold of a TNF family cytokine. Comparison of the RANKL structure with the available structures of other TNF family cytokines and receptors has identified elements of RANKL that it may use to interact with its receptors. Structure-based mutagenesis of these regions confirms their necessity in mediating the ability of RANKL to activate RANK.
Methods
Protein expression and purification.
The biologically active extracellular domain of RANKL was expressed in Escherichia coli as a glutathione S-transferase–fusion protein (GST-fusion protein) (9). Previous studies have confirmed that glycosylation of RANKL is not required for its biological activity (6). Briefly, a cDNA encoding the core ectodomain of murine RANKL residues 158–316 was cloned into pGEX-6P-1 (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) downstream of GST using the SalI and NotI restriction endonucleases. Following isopropylthio-β-D-galactoside–mediated (IPTG-mediated) induction of protein expression in protease-deficient BL21 (DE3) E. coli, cells were triturated in 150 mM NaCl, 20 mM Tris-HCl, pH 8.0, and 1 mM EDTA. Lysates were incubated with glutathione Sepharose (Amersham Pharmacia Biotech) for affinity purification of the fusion protein, followed by excessive washing with buffer comprising 150 mM NaCl and 20 mM Tris-HCl, pH 8.0. The GST-fusion protein was released from the affinity matrix by competitive elution with glutathione. The isolated protein was then subjected to anion exchange, followed by dialysis against 150 mM NaCl, 20 mM Tris-HCl, pH 7.2. Separation of RANKL from its GST fusion partner was accomplished by proteolytic cleavage with the type-14 human rhinovirus 3C protease (Amersham Pharmacia Biotech), performed for 4 hours at 4°C in 50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 10 mM EDTA, and 1 mM DTT (29). Uncleaved fusion protein, as well as the GST-tagged protease, was removed by passage over a glutathione affinity matrix. After cleaving off the GST epitope tag, 10 residues encoded by the vector (GPLGSPEFPR) remain fused to the N terminus of RANKL (residues 156–316, RGKPEAQPFA…QDID). The final product was further purified by size exclusion chromatography on Superdex 200 (Amersham Pharmacia Biotech), in which it migrated with a molecular weight of approximately 55 kDa, consistent with the size of a homotrimer. Protein identity was confirmed by mass spectrometry, with a mass of 19,034 Da conforming to predicted values plus or minus 1 Da. Purified RANKL was concentrated to 20 mg/ml in 20 mM NaCl, 20 mM Tris-HCl, pH 8.0.
Crystallization and data collection.
Crystals of RANKL were grown by vapor diffusion in hanging drops at 20°C. Diffraction-quality crystals were obtained with a precipitant comprising 80 mM HEPES, pH 7.5, 16 mM CaCl2, and 11–14% polyethylene glycol 4000. Two crystal forms predominated: primitive rhombohedral crystals (space group R3) growing to a typical size of 0.4 mm × 0.4 mm × 0.2 mm, with unit cell dimensions a = b = 150.6 Å, c = 139.5 Å, containing seven RANKL monomers in the asymmetric unit (ASU) and primitive orthorhombic crystals (space group P212121) growing to a typical size of 0.3 mm × 0.3 mm × 0.4 mm, with unit cell dimensions a = 65.3 Å, b = 82.0 Å, and c = 99.5 Å, containing three RANKL monomers in the ASU. Crystals were cryoprotected in 20% glycerol and flash cooled in liquid N2. Complete data to 3.0 Å resolution (fivefold redundant) was collected for the rhombohedral crystal and 2.6 Å (greater than elevenfold redundant) for the orthorhombic crystal by the oscillation method (1.5°), using a Rigaku rotating anode generator with CuKα radiation, equipped with Yale mirrors and an R-Axis IV detector. Data sets were processed with DENZO and SCALEPACK (30).
Structure determination and refinement.
The RANKL structure was determined initially by molecular replacement using models constructed from TNF-β and TRAIL coordinates, Protein Data Bank (PDB) (www.rcsb.org/pdb/) codes 1TNR and 1D4V. These structures were combined into a chimeric search model by deletion of divergent regions and incorporation of RANKL sequence using the Insight II program module Homology (Molecular Simulations Inc., San Diego, California, USA). Rotation and translation function searches using AMORE (31) yielded seven RANKL monomers in the ASU of the rhombohedral crystal, arranged as two trimers with a seventh monomer situated on the crystallographic threefold axis of symmetry. Rigid body refinement of the chimeric model against the R3 data set gave an R factor of 46% for all data from 20- to 4-Å resolution. The initial RANKL model was built into sevenfold averaged electron density maps calculated at 3 Å resolution. Model building into composite 2Fo-Fc-omit maps were carried out using the program O (32). Iterative rounds of model building and refinement using both strict and restrained NCS operators were carried out using CNS (33). Phases for orthorhombic RANKL were obtained by molecular replacement with the rhombohedral coordinates. Translation functions were run in all possible primitive orthorhombic enantiomorphs, with the highest signal unambiguously in space group P212121 consistent with the systematic absences in the data. Rigid body refinement of the three monomers situated in the ASU yielded an R factor of 31% for data from 20- to 4-Å resolution. Model building was initiated on maps calculated using noncrystallographic symmetry (NCS) threefold averaging, and atomic refinement (33) was carried out using restrained NCS operators, excluding structurally diverse loop regions of each monomer. Structural coordinates have been deposited under accession code 1JTZ (www.rcsb.org).
Structural analysis.
Atomic accessible area was calculated with NACCESS v2.1.1 (from S.J. Hubbard and J.M. Thornton, University College, London, United Kingdom). Briefly, a 1.4-Å radius probe was rolled over the van der Waals’ surface of RANKL and other TNF family cytokine structures for calculation of solvent-accessible surface areas. HBPLUS v3.0 (34) was used to assess neighbor interactions and the geometry of hydrogen bonds.
Mutagenesis.
Site-directed mutagenesis of murine RANKL was performed by PCR. Mutagenized RANKL constructs were expressed in E. coli as described above, with confirmation of identity by nucleic acid sequencing and mass spectrometry. Homotrimerization, as indicated by migration on size-exclusion chromatography, was taken as evidence for proper protein folding and assembly. Lack of endotoxin contamination was confirmed by Limulus amebocyte lysate (LAL) assay (BioWhittaker, Walkersville, Maryland, USA).
Cell culture.
Pure populations of murine osteoclast precursors were isolated from bone marrow as we have described previously (9). Cells were cultured in α-MEM containing 10% FBS, with addition of 10 ng/ml M-CSF and various concentrations of RANKL to induce osteoclast differentiation. Cells were incubated at 37°C in a humidified atmosphere containing 6% CO2 and supplemented with fresh media and cytokines daily. Typical mature osteoclast formation was observed between culture days 5 and 9.
Cytochemistry.
Osteoclast formation was identified histochemically by the method of Burstone (35). Briefly, cellular acid phosphatase activity was employed to cleave the chromogenic substrate naphthol AS-BI phosphate in the presence of 0.05 M sodium tartrate. Quantitation of tartrate-resistant acid phosphatase (TRAP) activity was accomplished by addition of a colorimetric substrate, 5.5 mM p-nitrophenyl phosphate, in the presence of 10 mM sodium tartrate at pH 4.5. The reaction product was quantified by spectroscopic measurement of optical absorbance at 405 nm.
Results
Structure determination.
The biologically active ectodomain of murine RANKL was expressed in E. coli and found to crystallize in two forms: a rhombohedral crystal containing seven monomers in the ASU, arranged as two trimers with a seventh monomer situated on the crystallographic threefold axis of symmetry, and an orthorhombic crystal containing a single RANKL trimer in the ASU. The structure of RANKL was phased by molecular replacement and refined to a resolution of 2.6 Å using the orthorhombic data. With the exception of some mobile loop regions and residues 156–161 of the amino terminus, the entire RANKL ectodomain is well ordered in the final crystal structure (crystallographic R factor 23.5% and Rfree 28.6%, Table 1). The final model of the RANKL trimer has a total of four residues with Ramachandran angles that fall outside generously allowed regions, which constitute 1% of the non-glycine, non-proline residues of the trimer (Table 1). These four residues include Asn136 from all three monomers, which has strong electron density in all three monomers, and Ser206 in monomer Z, which is less well ordered than observed in the X and Y monomers. Overall, the stereochemical parameters for the RANKL trimer coordinates are better than average for a 2.6-Å protein crystal structure (PROCHECK G-factor of 0.2). Furthermore, the Ramachandran plot quality for RANKL is better than average for PDB structures of equal resolution.
Table 1.
Crystallographic data and refinement statistics for murine RANKL
The RANKL trimer.
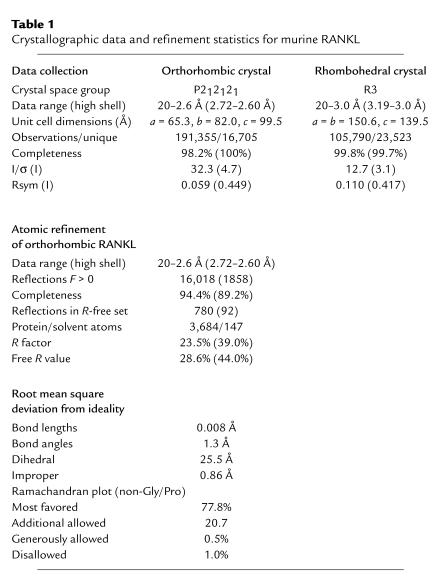
The ectodomain of RANKL adopts a fold that is characteristic of TNF family cytokines. Each RANKL monomer consists of a β-sandwich, composed of two, flat, antiparallel β-pleated sheets. The first sheet is formed by β-strands A′′, A, H, C, and F, while the second sheet is formed by β-strands B′, B, G, D, and E (Figure 1, a–d, Figure 2). The inner A′′AHCF β-sheet is involved in intersubunit association, whereas the B′BGDE β-sheet contributes largely to the outer surface. RANKL monomers self-associate along a threefold rotational axis of symmetry. The trimeric complex is best described as a truncated pyramid, with the membrane-proximal base wider than the distal apex. The RANKL trimer is 55 Å high, with approximate diameters of 55 Å at the base and 35 Å at the apex. The homotrimer is assembled such that one edge of the β-sandwich in each RANKL monomer, strands E and F, packs against the inner hydrophobic face of the AHCF β-sheet of the neighboring monomer (Figure 1, a and c). As such, the trimeric interface of the RANKL homotrimer consists of a platform of edge-to-face interactions. Viewed from the side, each monomer adopts a slight left-handed twist around the pseudo–three-fold rotational axis (Figure 1, a and b).
Figure 1.
Crystal structure of RANKL. (a) Ribbon diagram of the RANKL trimer, shown with the β-strands (green) and connecting loops (orange) of one RANKL monomer labeled according to standard TNF-β–sandwich nomenclature. The other two RANKL monomers are cyan and magenta, respectively. (b) In this view, oriented identically to a, the RANKL transmembrane stalk projects to the top of the image, while the membrane-distal region is toward the bottom. The homotrimer exhibits the shape of a truncated pyramid, being slightly wider at the membrane-proximal end. (c) Ribbon diagram of the RANKL trimer viewed down the axis of threefold symmetry, oriented with the membrane-distal face forward. The secondary structure of monomer X is labeled as in a. (d) The RANKL trimer, shown with the molecular surfaces of monomers X, Y, and Z colored in green, cyan, and magenta, respectively. The orientation of the molecule is identical to that in c. (e) Comparison of a single RANKL monomer with those of TNF and TRAIL. The β-strands of all three structures are colored green, while the connecting loops are colored orange for RANKL, gray for TNF (PDB code 1tnr), and blue for TRAIL (PDB code 1d4v). When the structures of these proteins are superimposed, it is apparent that the β-strands of RANKL superimpose almost identically with those of TNF and TRAIL. In contrast, the AA′′, CD, EF, and DE loops of RANKL exhibit unique topology when compared with those of other TNF family cytokines. (f) Electron density of the E-D-G β-strands. The structure is viewed from the solvent-accessible surface of the RANKL monomer, with an orientation similar to (e). Displayed in magenta is a 2.6-Å composite omit map (contoured at 1.2 ς) with the RANKL structure depicted in green (carbon), red (oxygen), blue (nitrogen), and yellow (sulfur).
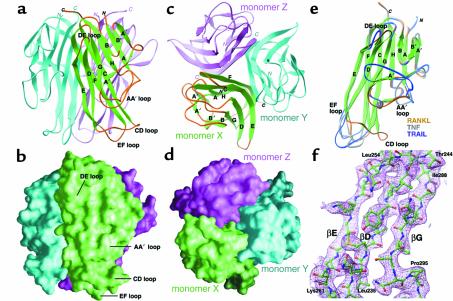
Figure 2.
Structure-based alignment of TNF family cytokines with RANKL. The sequence of the extracellular core domains of the TNF family cytokines TRAIL, CD40L, TNF-α, TNF-β, and ACRP30 are shown aligned to that of murine RANKL. Structural alignments to RANKL are based upon pairwise topological residue superposition of the crystal structure of RANKL with those of TRAIL (1d4v), CD40L (1aly), TNF-α (2tnf), TNF-β (1tnr), and ACRP30 (1c28). Residue numbers and secondary structure assignments for RANKL are depicted above the sequences. The ten β-strands that constitute the TNF family β-sandwich are drawn as green arrows, with standardized nomenclature. Solvent-accessible loops and coil regions connecting the β-strands are illustrated as brown lines. The AA′′ loop (orange line), connecting β-strands A and A′′, encompasses the A′ β-strand. Brown triangles above the residues indicate those involved in the trimeric interface of RANKL. The degree of homology at structurally equivalent positions among the family members (excluding ACRP30) is shown by colored circles below the sequences, 0–50% conservation (no circles), 50–90% (tan), >90% (brown). TRAIL residues boxed in green denote DR5 receptor contact sites on TRAIL. TNF-β residues boxed in magenta identify TNFR contact sites on TNF-β. By substituting RANKL in place of the ligands in the TRAIL:DR5 and TNF:TNFR co-crystal structures, we calculated potential contact sites for both of these receptors on RANKL. Boxed residues of RANKL indicate those calculated to physically contact DR5 (green), TNFR (magenta), or both (yellow) receptors during the docking analysis. Structural elements that are unique to RANKL are clustered in the solvent-accessible AA′′, CD, DE, and EF loops.
Trimerization of RANKL monomers is associated with a decrease in solvent-accessible surface area of approximately 6,100 Å2 (2,042 Å2 for monomer X, 2,043 Å2 for Y, and 2,014 Å2 for Z), as compared with other TNF family cytokines (6,591 Å2 for TRAIL, 5,671 Å2 for CD40L, 5,601 Å2 for TNF-α, 5,827 Å2 for TNF-β, and 5,130 Å2 for ACRP30). The trimeric interface constitutes approximately 26% of the solvent-accessible surface of each RANKL monomer. There is currently no evidence that RANKL forms heterotrimers with other TNF family members. The selectivity of this interaction is most likely dictated by sequence-specific intersubunit hydrogen bonding. Each RANKL trimer contains 46 such buried polar interactions, approximately twice the number observed in a typical TNF family cytokine. Although hydrophobic interactions are not thought to engender specificity, they constitute a major driving force for self-association. Of the residues involved in RANKL trimerization, 40% are hydrophobic (Figure 2). The majority of the amino acid residues that participate in intersubunit interactions (32 out of 43 total) reside within the ten highly conserved β-strands, leaving the structurally divergent surface loops accessible for receptor-ligand interactions (Figure 2).
The solvent-accessible surface loops of RANKL are unique within the TNF family, displaying markedly divergent lengths and conformations: the AA′′ loop (residues 170–193, bridging β-strands A and A′′), the CD loop (residues 224–233), the DE loop (residues 245–251), and the EF loop (residues 261–269). This divergence is best appreciated by superimposing the structure of RANKL over those of TNF and TRAIL (Figure 1e). Representative electron density of the β-strand core of RANKL is presented in Figure 1f. The internal β-strands that constitute the TNF core fold exhibit nearly identical topology, while the loops that interconnect these strands fail to superimpose (Figure 1e).
Receptor-binding sites.
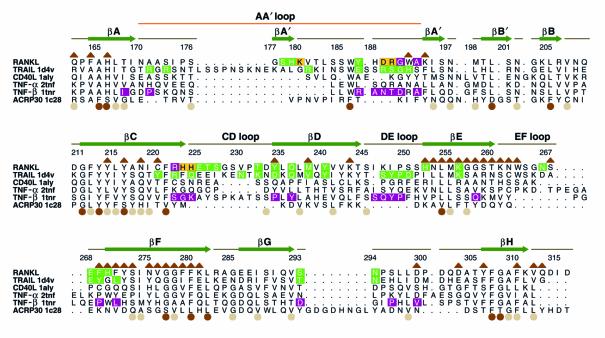
The two receptors for RANKL, namely RANK and OPG, belong to the TNFR family. Based on the two known crystal structures for TNF family receptor–ligand complexes, it is accepted that TNF family cytokines bind one elongated receptor molecule along each of the three clefts formed by neighboring monomers of the homotrimer. These intersubunit grooves can be appreciated looking downward along the threefold axis of rotational symmetry (Figure 1, c and d). From this perspective, it is apparent that the cleft is formed by two adjacent monomers of the RANKL homotrimer, thus ligand homotrimerization is requisite for receptor binding. In this manner, the RANKL trimer exhibits three spatially distinct, but equivalent, receptor-binding regions into which three receptor molecules triangulate without direct contact between receptor monomers themselves. Importantly, portions of the structurally unique AA′′, CD, DE, and EF loops of RANKL line the sides of the receptor-binding clefts (Figure 1c). Many naturally occurring as well as mutated TNF family cytokine variants exhibit altered affinities in receptor binding that result from mutations in these loops (Figure 2).
Not surprisingly, the TRAIL-DR5 (26–28) and TNF-β–TNFR (22) crystal structures reveal contact between the unique loops of the ligand that form receptor–binding grooves and the respective receptor molecules. These structures provide a framework for modeling the receptor-ligand interactions of other TNF family members. Although DR5 and TNFR share only 16 of 130 non-cysteine ectodomain residues, their ligand-binding domains occupy nearly equivalent positions in their respective complexes. Superposition of the Cα trace of the DR5 ligand-binding domain onto that of TNFR results in a root mean squared deviation of only 0.7 Å (26). Thus, superposition of RANKL in place of the ligands in these complexes should allow a reasonable prediction of interfacial residues on RANKL. Such analysis was used to identify residues of RANKL that are positioned within 4 Å of DR5 or TNFR (Figure 2 and Figure 3, a–c). Due to the remarkable structural congruence in the ligand-binding domains of these receptors, it is a reasonable inference that these residues may similarly serve as interfacial residues for RANK. When both the DR5 and TNFR contacts are mapped onto the surface of RANKL, it is apparent that prospective contact sites for both receptors (Figure 3d) encompass structurally unique elements of RANKL (in particular, residues from the AA′′, CD, and DE loops). Furthermore, when sequence conservation is also mapped onto the surface of RANKL, it can be appreciated that portions of the receptor contact patches employed by both receptors (yellow shading in Figure 3d) are not conserved across the family (white shading in Figure 3e). The lack of conservation at prospective contact points may underlie the highly specific recognition known to occur among TNF family cytokines and their receptors.
Figure 3.
Receptor contact regions of RANKL. (a) Schematic depiction of a RANKL trimer (green, cyan, and magenta monomers) docked with its receptor RANK (drawn here as four CRDs, yellow). (b) Surface representation of a RANKL trimer, oriented as in a, docked with a monomer of DR5, the TRAIL receptor (yellow Cα backbone worm). Areas on the surface of RANKL calculated to engage DR5 are shaded green, as is the sequence in Figure 2. (c) Surface representation of a RANKL trimer docked with a monomer of the TNFR (yellow Cα backbone worm). Areas on the surface of RANKL calculated to engage the TNFR are shaded magenta. (d) Comparison of DR5 (green shading) and TNFR (magenta shading) contact sites on the surface of RANKL. Overlapping areas that contact both receptors are shown in yellow. (e) Surface representation of a RANKL trimer with degrees of conservation mapped to the surface. The degree of conservation is directly proportional to intensity of colorization (white, no conservation; brown, conserved). Areas that contact both receptors show little to no sequence conservation across this family of cytokines (light tan to white shading). These unique regions provide specificity for receptor-ligand recognition, underlying the lack of cross-reactivity among receptors and ligands in this cytokine family. (f) Potential receptor contact areas of RANKL that were targeted for structure-based mutagenesis are shown in red.
Structure-based mutagenesis.
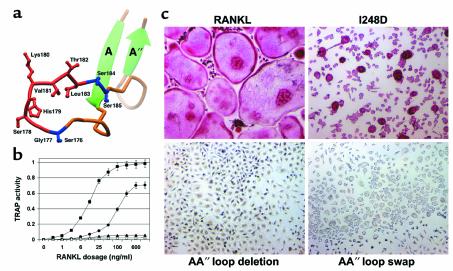
Comparison with known TNF-TNFR structures suggests that RANKL may bind its receptors through its unique solvent-accessible loops. We tested the biological relevance of these predictions by performing structure-based mutagenesis experiments. Three mutations were generated that impact the biological function of RANKL: a single amino acid substitution in the DE loop, replacing Ile248 with Asp (I248D); a deletion of the AA′′ loop, from Gly177 to Leu183 (AA′′ loop deletion); and a replacement of the AA′′ loop of RANKL (177–183) with that of TNF (AA′′ loop swap) (Figure 4a). RANKL, bearing either the AA′′ loop deletion or loop-swap mutations, fails to induce osteoclast precursors to differentiate in vitro, whereas the I248D substitution mutant demonstrates an eightfold decrease in potency relative to native RANKL (Figure 4b). Furthermore, the osteoclasts induced by I248D RANKL exhibit dysfunctional morphology, namely mononuclearity and small size relative to multinucleated, well-spread osteoclasts typically induced by native RANKL (Figure 4c). As such, the AA′′ loop deletion and AA′′ loop-swap mutants of RANKL fail to induce cellular differentiation by way of RANK. In contrast, construct I248D represents a novel form of RANKL that activates RANK, but induces comparable levels of cellular differentiation only when administered at eight times the concentration of native RANKL.
Figure 4.
Structure-based mutagenesis of RANKL. (a) Ribbon view of AA′′ loop illustrating the AA′′ loop deletion and AA′′ loop-swap mutants. For the AA′′ loop deletion, SGSHKVTLSS was mutated to SSS by removal of the segment depicted in red (GSHKVTL). For the AA′′ loop-swap mutant, SGSHKVTLSS was mutated to SLL by swapping the segments depicted in blue and red with the analogous segments from TNF. (b) Dose-response curves for native and mutant forms of RANKL (filled square, native RANKL; filled circle, I248D point substitution; filled triangle, AA′′ loop deletion; open triangle, AA′′ loop swap) are plotted for in vitro osteoclastogenesis. ED50 of native RANKL is ∼12 ng/ml, compared with ∼100 ng/ml for I248D RANKL. AA′′ loop deletion and AA′′ loop-swap mutants failed to induce detectable osteoclastogenesis at any dosage. (c) Osteoclastogenic cultures are depicted at identical magnification after 7 days of exposure to the various forms of RANKL. Cultures were stained for osteoclast-specific TRAP activity (red reaction product). Native RANKL induces the formation of large, multinucleated, TRAP-expressing osteoclasts. I248D RANKL generates TRAP-positive cells that lack the characteristic morphology of osteoclasts, with the majority of cells exhibiting mononuclearity. AA′′ loop deletion and loop-swap mutants fail to induce differentiation along the osteoclastogenic pathway.
Discussion
The potent biological effects of TNF and TNFR superfamily proteins are perturbed in many human disease states and thus serve as logical targets for the development of pharmaceuticals. Therapies modulating TNF family cytokines have been applied successfully in conditions such as rheumatoid arthritis, inflammatory bowel disease, and septicemia. Novel therapeutics targeting this family of cytokines are emerging for widespread diseases such as osteoporosis, atherosclerosis, cancer, autoimmune disorders, and graft rejection.
Osteoporosis, a disease endemic in Western society, reflects an imbalance in skeletal homeostasis that favors accelerated bone loss. Bone resorption is the unique function of the osteoclast, and the activity of this polykaryon is central to the pathogenesis of osteopenic disorders. In fact, postmenopausal osteoporosis occurs as a result of enhanced RANKL-stimulated osteoclastogenesis. Clinical progress achieved thus far in treating this disease reflects successes in modulating osteoclast number and activity.
Since its identification, RANKL has been characterized as a TNF family cytokine based solely on amino acid similarity. Unfortunately, low homology among TNF family members has hindered the generation of meaningful structural models. For instance, although CD40L is the most similar TNF family cytokine to RANKL, only 23% (37 of 159) of their ectodomain residues are identically conserved. A recently published review of the TNF and TNFR superfamilies (16) incorrectly aligned 30 residues of the D and E β-strands of RANKL, placing them out of register. Given the low level of amino acid conservation among members of the TNF superfamily, mechanisms by which these molecules achieve specificity cannot be understood without knowledge of their three-dimensional structures.
We have determined the three-dimensional structure of RANKL to a resolution of 2.6 Å. The x-ray crystal structure presented herein clearly places RANKL in the TNF family of cytokines. Like all TNF proteins examined to date, RANKL self-assembles into stable, noncovalently associated trimers. To facilitate discussion, we have indicated the residues involved in this trimerization in Figure 2. Surprisingly, 70% of the trimer contacts are nonconserved at the amino acid level. The majority of the unique trimerization residues of RANKL are polar in nature. RANKL, in comparison with other TNF family cytokines, employs an unusually large number of buried polar interactions between neighboring monomers at the trimer interface. These polar interactions are sequence specific and thus contribute to the specificity of self-association. Important polar interactions between neighboring monomers include Lys194-Asp301, Lys243-Asp299, and Lys256-Asp303. Another notable feature of the RANKL trimer interface is the presence of a conserved ring of hydrophobic side chains lining the core at the base of the molecule, a feature commonly observed in TNF family members that likely constitutes a major driving force for self-assembly. In RANKL, a number of conserved residues contribute to this tiling: Tyr214, Tyr216, Phe280, and Phe310. Nonconserved residues involved in RANKL trimerization that are likely to structurally preclude heteroassociation with other TNF family cytokines include Trp192 and Lys194 in the AA′′ loop; Lys261, Asn262, and Trp263 in the EF loop; and three residues of the F β-strand, Phe269, His270, and Phe271.
RANKL amino acid diversity is most extreme in the surface-exposed loops and flanking strands in which little sequence and structural homology to the family can be discerned. As can be appreciated in Figure 1e, the AA′′ loop of RANKL diverges from those of TRAIL and TNF. While the longer AA′′ loop of TRAIL traverses horizontally across the face of the monomer and the shorter AA′′ loop of TNF follows a tight hairpin turn to immediately join the A′′ β-strand, the AA′′ loop of RANKL projects toward the apex of the molecule. The AA′′ loop, together with lateral displacement of the CD loop, confers a unique surface to the upper third of the RANKL molecule. A more subtle outward shift of the DE loop shapes the receptor-binding groove at the base of the RANKL molecule. This observation suggests that sequence diversity is localized at the receptor-binding interfaces of the ligand and affirms the emergent paradigm that complementary diversity of interfacial residues in receptor-ligand contact regions serves as the major determinant of biological specificity among TNF family members.
The signaling receptor for the RANKL cytokine, RANK, is a classical TNFR family member expressed on a wide variety of different cell lineages. RANKL is also specifically engaged by OPG, a soluble decoy receptor that sequesters both membrane-associated and soluble versions of RANKL, preventing the activation of RANK. Although OPG appears to be a member of the TNFR family based upon the presence of conserved cysteine residues, it is unclear whether it engages RANKL by the same mechanism as RANK. This uncertainty arises due to the low degree of sequence homology between these two receptors, as well as reports that OPG exists as a disulfide-linked homodimer (36). An early study suggested that another TNF family cytokine, TRAIL, may also interact with OPG (37), although the physiological relevance of this observation is still unclear. RANKL binds to OPG with an affinity of 6.7 nM as measured by surface plasmon resonance (38). In contrast, the affinity of TRAIL for OPG is reported to be only KD = 400 nM (39), even though TRAIL binds to its membrane receptor DR5 with a KD = 2 nM under similar conditions. The significantly higher affinity of RANKL for OPG makes it unlikely that TRAIL/OPG interactions impact RANKL activity. Elucidation of the OPG-RANKL and RANK-RANKL co-complex crystal structures and their comparisons with other available structural information will provide insight into determinants of receptor-ligand specificity exhibited by these proteins.
The structures of the two known TNF family receptor-ligand complexes, TNF-β–TNFR and TRAIL-DR5, provide a biological context for modeling the interaction of RANKL and RANK. Docking analyses with these structures have identified prospective receptor contact residues: Lys180, Asp189, and Arg190 in the AA′′ loop; His223 and His224 in the CD loop; and Lys 247, Ile248, and Pro249 in the DE loop. We have confirmed the biological relevance of these predictions by performing structure-based mutagenesis experiments. A single amino acid substitution in the DE loop significantly lowers the potency of RANKL and substantially suppresses its ability to induce the differentiation of morphologically sound osteoclasts. It is apparent that both deletion of the AA′′ loop and its substitution with the analogous loop of TNF leave the core topology of RANKL intact, since this protein readily and spontaneously self-assembles as homotrimers. The inability of these RANKL mutants to demonstrate biological activity establishes the necessity of the AA′′ loop for RANK activation.
The crystal structure of RANKL identifies specific elements of this cytokine that are essential for the engagement and activation of its signaling receptor. Analysis of the structure of RANKL reveals structural determinants of specificity that inform current understanding of receptor-ligand interactions across the TNF family. The structure-based comparative and functional analysis presented herein provides a framework for the rational pharmacologic modulation of RANKL and its receptors for therapeutic endeavors.
Acknowledgments
We thank Michael Miley, Tom Brett, and other members of the Fremont Laboratory for assistance in data analysis and preparation of figures. Kyunghee Choi, Paul Allen, and Emil Unanue are gratefully acknowledged for advice and support. This study was supported by the Burroughs-Wellcome Fund (D.H. Fremont), the Kilo Diabetes and Vascular Research Foundation (D.H. Fremont), an Alpha Omega Alpha Medical Honor Society Research Fellowship (J. Lam), the Washington University Medical Center Alumni Association Distinguished Alumni Scholarship Fund (J. Lam), and the National Institutes of Health: grants CA-86803-01 and GM-62414 (D.H. Fremont); AR-07033 and GM-07200-25 (J. Lam); AR-32788, AR-46523, and DE-05413 (S.L. Teitelbaum); and AR-46852 (F.P. Ross). Structural coordinates of RANKL have been deposited under accession code 1JTZ (www.rcsb.org).
Footnotes
Jonathan Lam and Christopher A. Nelson contributed equally to this work.
References
- 1.Wong BR, et al. TRANCE is a novel ligand of the tumor necrosis factor receptor family that activates c-Jun N-terminal kinase in T cells. J Biol Chem. 1997;272:25190–25194. doi: 10.1074/jbc.272.40.25190. [DOI] [PubMed] [Google Scholar]
- 2.Anderson DM, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature. 1997;390:175–179. doi: 10.1038/36593. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda H, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacey DL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 5.Li J, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc Natl Acad Sci USA. 2000;97:1566–1571. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 7.Cenci S, et al. Estrogen deficiency induces bone loss by enhancing T cell production of TNFα. J Clin Invest. 2000;106:1229–1237. doi: 10.1172/JCI11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofbauer LC, et al. Estrogen stimulates gene expression and protein production of osteoprotegerin in human osteoblastic cells. Endocrinology. 1999;140:4367–4370. doi: 10.1210/endo.140.9.7131. [DOI] [PubMed] [Google Scholar]
- 9.Lam J, et al. TNF-α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest. 2000;106:1481–1488. doi: 10.1172/JCI11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong YY, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 11.Dougall WC, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D, et al. Regulation of peripheral lymph node genesis by tumor necrosis factor member TRANCE. J Exp Med. 2000;192:1467–1478. doi: 10.1084/jem.192.10.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong YY, et al. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature. 1999;402:304–309. doi: 10.1038/46303. [DOI] [PubMed] [Google Scholar]
- 14.Teng Y, et al. Functional human T-cell immunity and osteoprotegerin ligand control alveolar bone destruction in periodontal infection. J Clin Invest. 2000;106:R59–R67. doi: 10.1172/jci10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fata JE, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell. 2000;103:41–50. doi: 10.1016/s0092-8674(00)00103-3. [DOI] [PubMed] [Google Scholar]
- 16.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 17.Jones EY. The tumour necrosis factor receptor family: life or death choices. Curr Opin Struct Biol. 2000;10:644–648. doi: 10.1016/s0959-440x(00)00149-4. [DOI] [PubMed] [Google Scholar]
- 18.Jones EY, Stuart DI, Walker NP. Structure of tumour necrosis factor. Nature. 1989;338:225–228. doi: 10.1038/338225a0. [DOI] [PubMed] [Google Scholar]
- 19.Naismith JH, Sprang SR. Modularity in the TNF-receptor family. Trends Biochem Sci. 1998;23:74–79. doi: 10.1016/s0968-0004(97)01164-x. [DOI] [PubMed] [Google Scholar]
- 20.Lewit-Bentley A, et al. Structure of tumour necrosis factor by X-ray solution scattering and preliminary studies by single crystal x-ray diffraction. J Mol Biol. 1988;199:389–392. doi: 10.1016/0022-2836(88)90323-3. [DOI] [PubMed] [Google Scholar]
- 21.Eck MJ, Ultsch M, Rinderknecht E, de Vos AM, Sprang SR. The structure of human lymphotoxin (tumor necrosis factor-beta) at 1.9 Å resolution. J Biol Chem. 1992;267:2119–2122. [PubMed] [Google Scholar]
- 22.Banner DW, et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 23.Karpusas M, et al. 2Å crystal structure of an extracellular fragment of human CD40 ligand. Structure. 1995;3:1031–1039. doi: 10.1016/s0969-2126(01)00239-8. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro L, Scherer PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr Biol. 1998;8:335–338. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- 25.Cha SS, et al. 2.8 Å resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity. Immunity. 1999;11:253–261. doi: 10.1016/s1074-7613(00)80100-4. [DOI] [PubMed] [Google Scholar]
- 26.Hymowitz SG, et al. Triggering cell death: the crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol Cell. 1999;4:563–571. doi: 10.1016/s1097-2765(00)80207-5. [DOI] [PubMed] [Google Scholar]
- 27.Mongkolsapaya J, et al. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol. 1999;6:1048–1053. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
- 28.Cha SS, et al. Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity. J Biol Chem. 2000;275:31171–31177. doi: 10.1074/jbc.M004414200. [DOI] [PubMed] [Google Scholar]
- 29.Cordingley MG, Callahan PL, Sardana VV, Garsky VM, Colonno RJ. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J Biol Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Navaza J, Saludjian P. Amore: an automated molecular replacement program package. Methods Enzymol. 1997;276:581–591. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 32.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 35.Burstone MS. Histochemical comparison of naphthol-phosphates for the demonstration of phosphatases. J Natl Cancer Inst. 1958;20:601–606. [PubMed] [Google Scholar]
- 36.Yamaguchi K, et al. Characterization of structural domains of human osteoclastogenesis inhibitory factor. J Biol Chem. 1998;273:5117–5123. doi: 10.1074/jbc.273.9.5117. [DOI] [PubMed] [Google Scholar]
- 37.Emery JG, et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–14367. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 38.Willard D, et al. Expression, purification, and characterization of the human receptor activator of NF-kB ligand (RANKL) extracellular domain. Protein Expr Purif. 2000;20:48–57. doi: 10.1006/prep.2000.1278. [DOI] [PubMed] [Google Scholar]
- 39.Truneh A, et al. Temperature-sensitive differential affinity of TRAIL for its receptors. DR5 is the highest affinity receptor. J Biol Chem. 2000;275:23319–23325. doi: 10.1074/jbc.M910438199. [DOI] [PubMed] [Google Scholar]