Abstract
The role of N-glycan trimming in glycoprotein fate and function is unclear. We have recently shown that hepatitis B virus (HBV) DNA is not efficiently secreted from cells in which α-glucosidase mediated N-glycan trimming is inhibited. Here it is shown that, in cells in glucosidase-inhibited cells, viral DNA, accompanied by envelope and core proteins, most likely accumulate within lysosomal compartments. Pulse–chase experiments show that although the viral glycoproteins (L, M, and S) are dysfunctional, in the sense that they do not mediate virion egress and are not efficiently secreted from the cell, they all still leave the endoplasmic reticulum (ER). Surprisingly, however, the glycoproteins retained within the cell were not rapidly degraded, appearing as aggregates, enriched for L and M, with intracellular half-lives exceeding 20 h. Moreover, by 24 h after synthesis, a substantial fraction of the detained glycoproteins appeared to return to the ER, although a considerable amount was also found in the lysosomes. To our knowledge, this is the first report that shows, as a consequence of inhibiting glycosylation processing, certain glycoproteins (i) become dysfunctional and aggregate, yet still depart from the ER, and (ii) have extended rather than shortened half-lives. Taken together, these data suggest that proper intracellular routing of HBV glycoproteins requires ER glucosidase function. It is hypothesized that failure to process N-glycan causes HBV glycoproteins to aggregate and that impaired protein–protein interactions and trafficking are the result of misfolding.
Enveloped viruses have been used to explore mammalian host secretory pathways, often yielding substantial insights (1–3). For successful morphogenesis, hepatitis B virus (HBV) envelope proteins must properly fold, and form elaborate intra- and interprotein crosslinks (4, 5). In addition, the envelope proteins must meet the other viral proteins in appropriate cell compartments. Secretion of virus requires that the envelope proteins become N-glycosylated and that they be trimmed (6, 7).
In mammalian cells, N-linked oligosaccharides are transferred to polypeptides as a 14 sugar structure of Glc3Man9GlcNAc2 (2). After transfer, more than 13 different enzymes mediate the temporally and spatially ordered removal (trimming) of terminal sugars and eventual reconstruction of the oligosaccharide in the endoplasmic reticulum (ER) and Golgi compartments (2). The reason that the cell first adds the oligosaccharide macromolecular structure, only to deconstruct and then rebuild the sugar subunits, is an enigma. Because the pathway is so highly ordered and so many different enzymes are invested, the trimming reaction is assumed to be important. Recently, for example, it has been suggested that for some, but not all, glycoproteins, proper folding and quality control is mediated by binding lectin-like host chaperons that recognize the monoglucose (trimmed) form of the oligosaccharide side chain (8, 9). Glucosidases I and II mediate removal of the terminal glucose residue from (glucose)3(mannose)9(N-acetylglucosamine)2 oligosaccharide, thus contributing the first steps in the trimming pathway (2). Cells in which glucosidase has been inhibited are unable to efficiently fold or secrete certain glycoproteins (10, 11). The effect upon influenza hemagglutinin and vesicular stomatitis virus G folding has been studied in particular detail (12). However, the long-term fate of those proteins and their functional properties were not evaluated in depth. Indeed, it is often assumed that defective proteins will be rapidly degraded (13, 14).
We have been using the assembly and secretion properties of HBV to study the role of glycan processing in glycoprotein fate and function. HBV is the human member of the hepadnaviridae, a family of small DNA containing liver tropic viruses that are the etiological agents of most liver cancer (reviewed in ref. 15). The three carboxy-coterminal envelope proteins, called L, M, and S, are collectively called “HBs (hepatitis B surface) proteins” (in the particulate form). They are all specified by the same gene, which is composed of a single continuous 400-codon ORF (5). HBV-infected liver cells secrete infectious, nucleocapsid-containing virions, as well as an excess of non infectious “subviral” particles that do not contain DNA. All particles are thought to bud from an ER or post-ER compartment (3, 16), but virions contain all three viral glycoproteins, while subviral particles such as spheres have less L (17). Being physically and functionally well characterized, they offered an ideal opportunity to study the functional consequences of preventing glycan trimming. We have previously shown that the glucosidase inhibitor, n-butyl-deoxynojirimycin (NB-DNJ), prevents HBV virion but not subviral particle secretion from stably transfected HepG2 (6, 7). Moreover, the NB-DNJ inhibition of virion secretion was due to the inhibition of the ER glucosidase (7). To determine which envelope protein functions require glycan processing and to understand the consequences to a glycoprotein of bearing unprocessed glycan, it was of interest to know the specific steps in the virus life cycle that were upset in cells in which ER glucosidase was inhibited.
MATERIALS AND METHODS
Cells, Media and Sources of Human HBV Subviral Particles.
2.15 cells, kindly provided by George Acs (Mt. Sinai Medical College, New York) were clonally isolated from HepG2 cells, following cotransfection of a plasmid containing a dimer of the HBV genome and a G418 resistance marker (18). HBV subviral particles were partially purified from human carrier serum by centrifugation through discontinuous sucrose gradients as described (19). Mouse mAbs specific for HBV surface antigens (HBs proteins) and core (hepatitis B core antigens; HBcAgs) antigens were kindly provided by Wolfram Gerlich (Giessen, Germany). The mAbs preS1 (L) specific (mAb MA/18); preS2 (M) specific (E), and the S domain (C20-02) have been used previously (17, 19, 20). The conformation-independent, HBsAg-specific mAb is called H166 (7). The HBcAg-specific mAb is called 42B/12. Rabbit antibody was made hyperimmune against either human albumin (Cappel) or α1-antitrypsin (Boehringer Mannheim). Mouse mAb specific for p58 was purchased from Sigma.
Cell Fractionation, Percoll and Sucrose Gradient Centrifugation.
Cells (106), growing in 75 cm2 flasks, were harvested by scraping, sedimented by centrifugation at 1000 × g in a table-top Braun refrigerated microcentrifuge, and resuspended in 1.5 ml 5 mM Hepes (pH 7.0), 0.25 M sucrose, and 20 μM phenylmethylsulfonyl fluoride (SH buffer). Cells were disrupted with 8–10 strokes in a Dounce homegenizer, confirming the degree of cell disruption by microscopic examination. After separating the nuclei by centrifugation at 3000 × g, supernatants were applied to a step gradient of Percoll (Pharmacia), as in ref. 11, and resolved by ultracentrifugation for 1.33 h at 20,000 rpm at 10°C in an SW41 rotor (Beckman L8 ultracentrifuge). The gradient used in these studies was constructed by making successive layers of Percoll of 2 ml at 1.05 g/ml; 2 ml at 1.04 g/ml; 3 ml at 1.03 g/ml; and 2.5 ml at 1.02 g/ml. Samples reconstructed with chromogenic density markers (Pharmacia), which are visible in room light, were always included for comparison. Following centrifugation, 0.4 ml samples were collected by fractionation. The position of the density markers was noted, and density curves were generated. Each fraction was tested for the appropriate cellular or viral marker. Subcellular markers were as follows: p58; glucose 6 phosphate dehydrogenase (G6PD); hexosaminidase (Hex), and for binding of wheat germ agglutinin (WGA). Assay details are described elsewhere in the text. Selected fractions (200 μl) were disrupted by sonication and layered on to a gradient of sucrose and resolved by centrifugation at 36,000 rpm in an SW41 rotor for 16 h at 10°C, as described (19). Fractions were collected at tested for the presence of HBsAg polypeptides by the “antigen capture” method as indicated elsewhere.
Radiolabeling of Cells, Antigen Capture, WGA Assay, and Immunoprecipitation.
After growth in culture medium lacking or containing 1000 μg/ml (≈4 mM) NB-DNJ (provided by Monsanto/Searle), cells were pulsed with RPMI 1640 medium without methionine and containing 800 μCi/ml [35S]methionine (1000 Ci/mM; 1 Ci = 37 GBq; ICN) for 20 min. Medium containing label was removed and cells were either immediately harvested or culture was continued with normal growth medium for the indicated periods of time. Cells were harvested into SH buffer and disrupted by homogenization (as above). The concentration of NB-DNJ in the culture medium was maintained throughout labeling and chase. For the antigen capture and WGA assay, medium and cell solutions were made 0.1% Nonidet P-40 and incubated in wells of a 96 well tray (ELISA strip; Nunc) that had been precoated with specific antibody or WGA (Boehringer Mannheim) by incubation at 4°C overnight with an amount of antibody such that the immobilized antibody was experimentally determined to be in excess to antigen in the sample (this was 1 μg/ml for most of the HBV-specific mAbs and “blocked” with 2% BSA in PBS). After incubation with experimental sample for 2 h at 37°C, followed by five vigorous washes in PBS/Tween 20 (0.5%), wells were separated and the amount of radiolabel was determined by liquid scintillation.
Immunoprecipitation and SDS/PAGE.
For immunoprecipitation of fractions of the Percoll gradient, Percoll was sedimented from the fractions of the gradient by centrifugation. The Percoll-free supernatant was sonicated and incubated with the indicated mAb or rabbit antibody (≈1 μg/ml) for 2 h at 37°C. For clarified culture medium, antibodies were added directly to the sample. The 1 ml samples received 10 mg of protein A Sepharose (Pharmacia) that had been precoated with 2% BSA in PBS, and the reaction was incubated for an additional 1 h at 4°C. The immunoprecipitates were sedimented and washed as in ref. 19 and dissolved in SDS lysis buffer and resolved through 13% SDS/polyacrylamide gels as in ref. 20.
Enzyme Assay for G6PD and Hex.
Measurement of G6PD was based upon the assay described in ref. 21. Product was detected by spectroscopy at 660 nm. Hex activity was determined by methods described by Boehringer Mannheim. Product is detected by spectroscopy at 405 nm.
RESULTS
Inhibition of ER Glucosidase I and HBV DNA Particle Secretion in 2.15 Cells.
The fate and function of HBV DNA and envelope proteins was evaluated in 2.15 cells in the absence and presence of an inhibitor of ER glucosidase. 2.15 cells are derived from HepG2 cells, following the stable transfection of a plasmid bearing a dimer of the HBV genome and persistently secrete infectious HBV (18). NB-DNJ is an effective inhibitor of ER glucosidase (22). 2.15 cells treated with 1000 μg/ml (4.2 mM) NB-DNJ for 5 days are as viable as untreated cells, in spite of the inhibition of glucosidase (7). Consistent with the inhibition of glucosidase, NB-DNJ-treated 2.15 cells fail to secrete enveloped HBV and secrete antitrypsin at reduced rates. As in refs. 10 and 11, antitrypsin accumulating within these cells was shown to migrate in SDS/polyacrylamide gels with a mobility consistent with the retention of unprocessed glycan (data not shown). Thus, it is reasoned that ER glucosidase was inhibited in the 2.15 cells treated with NB-DNJ without any detectable toxicity.
Kinetics of HBV Glycoprotein Synthesis and Secretion.
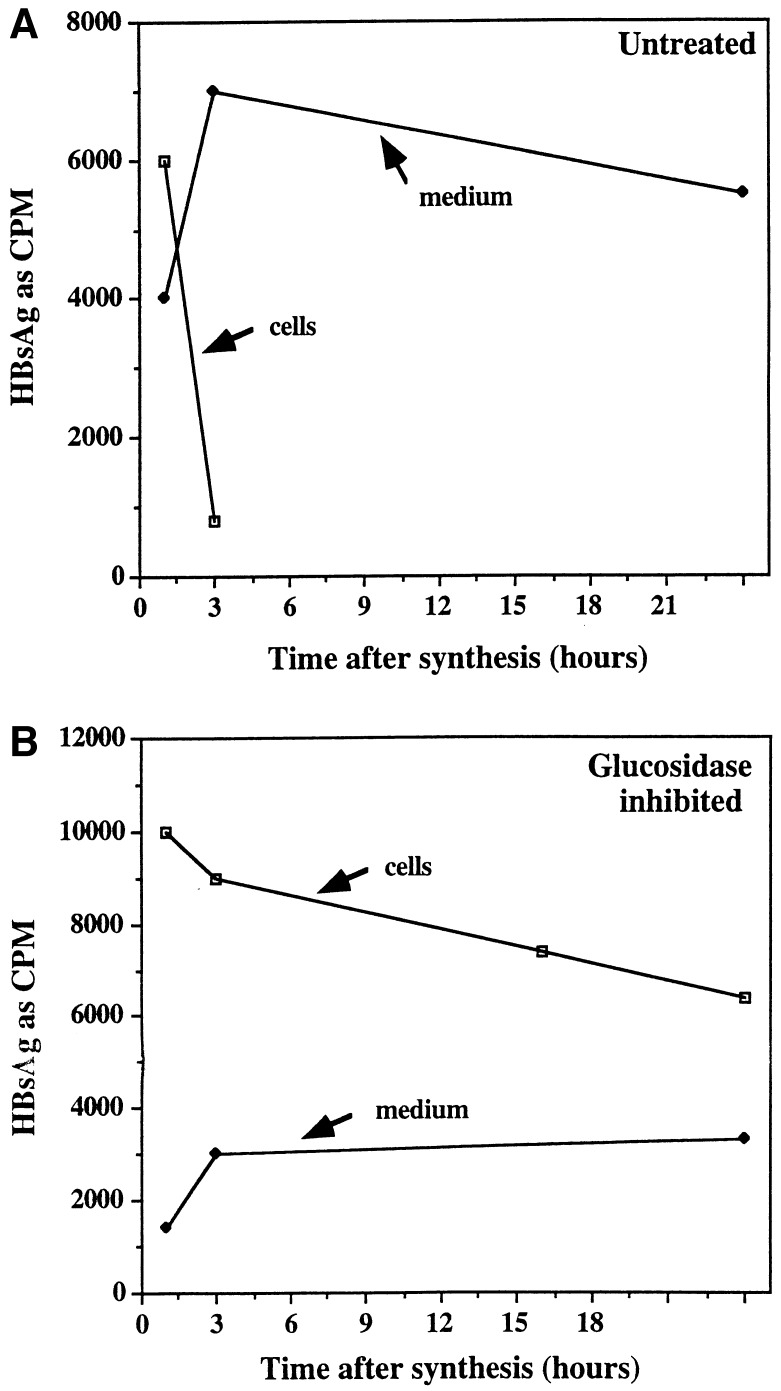
Since in glucosidase-inhibited cells secretion of HBV DNA-containing particles was inhibited and proper morphogenesis requires functional viral envelope proteins (23), it was of interest to know the kinetics of HBsAg secretion. The intracellular retention and half-lives of HBV glycoproteins in glucosidase-inhibited cells was determined by pulse–chase experiments. 2.15 cells, left untreated or preincubated with glucosidase inhibitor for 3 h, were incubated for 20 min with [35S]methionine, followed by replacement with medium without radiolabel. The amount of HBV glycoprotein in the cell and secreted into the culture medium was followed as a function of time following synthesis, using the solid-state mAb antigen capture detection system with an antibody (C20-02) that requires a common conformation on S, L, and M (see Materials and Methods). Fig. 1A shows that most detectable HBs proteins have left the intracellular compartment of untreated cells and are recovered in the culture medium, within 3 h after synthesis. Because, in other experiments, the amount of intracellular HBsAg in untreated 2.15 cells at 16 and 24 h after synthesis was approaching background levels, these points were not taken in the work shown. On the other hand, secretion of HBs proteins from inhibited cells is greatly impaired (Fig. 1B). Note that the term HBsAg is used to refer collectively to all HBV envelope proteins (L, M, and S). Moreover, and surprisingly, the intracellular HBsAg is still detectable using the conformation-dependent mAb (C20-02) at the longest time point shown, 24 h (Fig. 1B). This suggests that the retained HBs proteins are not rapidly degraded and have half-lives in excess of 24 h. Indeed, using mAbs that distinguish M from L and S, the secretion of M was much more severely impaired than that of S and L and the half-life of intracellular “M” was determined to be more than 24 h (unpublished data). This suggested that, with respect to secretion, material retained within the cell was either “discriminated” against or in compartments, making it unable to be secreted.
Figure 1.
Kinetics of HBsAg synthesis and secretion in glucosidase-inhibited cells. After 3 h in the absence or presence of glucosidase inhibitor (NB-DNJ), 2.15 cells were pulse labeled for 20 min with [35S]methionine and chased with medium lacking radioactivity. At the indicated times after labeling, the amount of HBsAg in cell lysates (cells) and culture medium (medium) was determined by the antigen capture assay described in Materials and Methods using mAb C20-02, which recognize L, M, and S. The amount of HBsAg bound to plates coated with antibody is presented as cpm.
Subcellular Fractionation of 2.15 Cells.
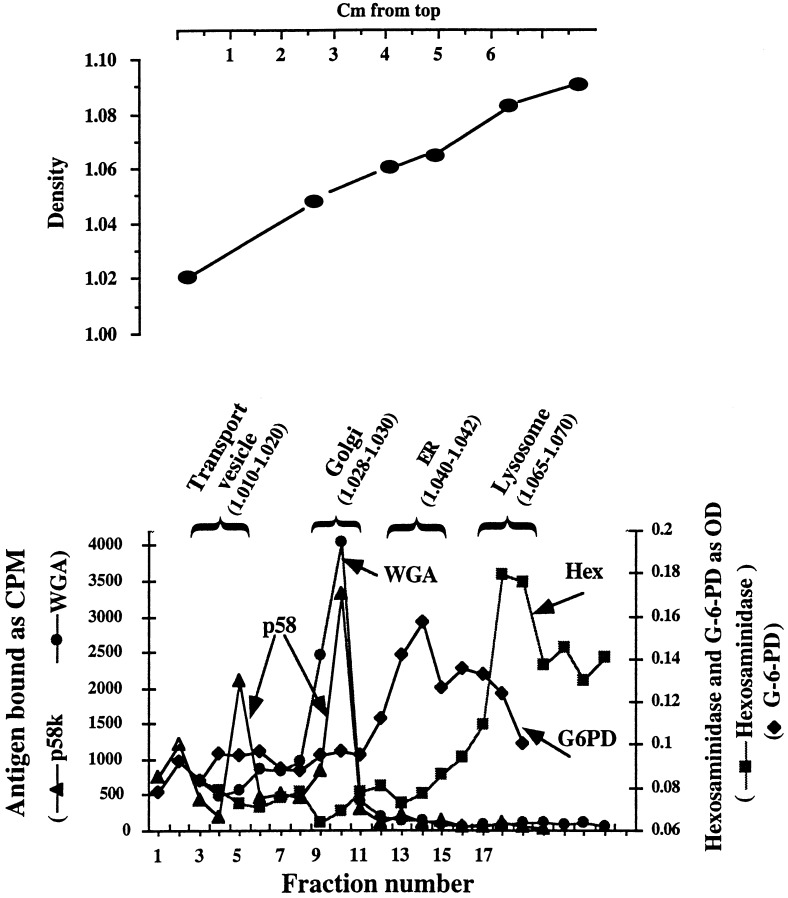
Because HBs proteins accumulate within glucosidase-inhibited cells, it was of interest to determine their intracellular distribution as a function of time following synthesis. Thus a method of subcellular fractionation was developed. After 3 h of either no treatment or glucosidase inhibition, 2.15 cell cytoplasms were resolved through Percoll gradients by centrifugation. As shown in Fig. 2, these gradients permit a reasonable separation of transport/intermediate compartment (p58), ER (G6PD), Golgi (binding of radioactive WGA) and lysosomal (Hex) compartments, as determined by the location of characteristic markers. Note that there are two peaks of p58 abundance at fractions 5 and 10. One of these peaks (fraction 10) is also the peak of WGA binding. Thus, on both the basis of density and the coincidence of WGA binding and p58, fraction 10 is assumed to contain the Golgi apparatus. The other peak of p58 (fraction 5) lacks WGA binding activity. Because p58 is associated with the intermediate compartment as well as the Golgi, and the intermediate compartment (and transport vesicles) migrates to lower densities in Percoll and related gradients (24, 25), fraction 5 is assumed to contain the peak of intermediate compartment/transport vesicle organelles. Also, distinct density markers cosedimented with the peaks of the ER, Golgi, and lysosomal fractions and were found to be useful indicators of the subcellular fractions (Fig. 2 Upper). It is emphasized that these subcellular fractionation experiments were repeated more than three times, yielding very reproducible results.
Figure 2.
Subcellular fractionation of 2.15 cells. Cytoplasmic lysates from 2.15 cells, maintained for 3 h in the absence and presence of glucosidase inhibitor, were prepared and resolved through Percoll gradients. Each fraction was tested by an antigen capture assay for the presence of p58, G6PD, Hex, and WGA binding activity. The densities of the peaks of each subcellular fraction is presented in parenthesis in g/ml. (Upper) Location within the Percoll gradient of density markers, which were visible under room light.
Intracellular Trafficking of HBs Proteins in Glucosidase-Inhibited Cells.
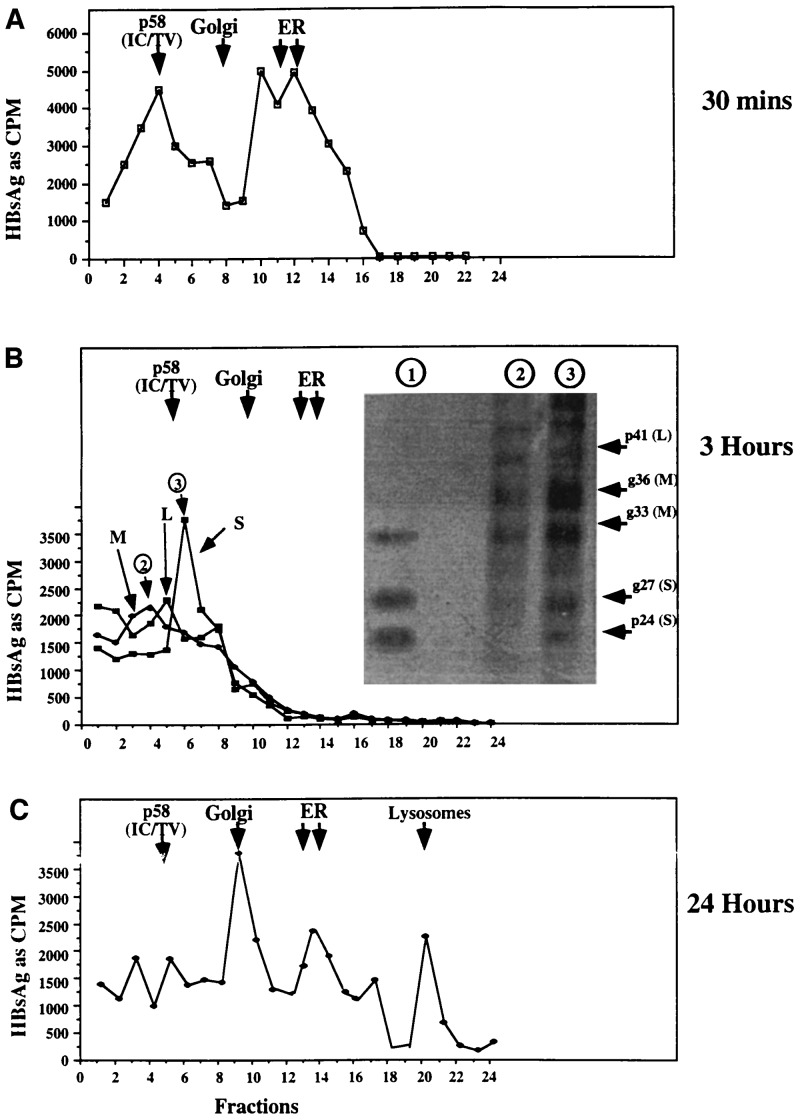
With a means of determing the presence of HBs proteins in different subcellular compartments, the fate of HBV glycoproteins in glucosidase-inhibited cells was studied as a function of time following protein synthesis. In untreated cells, HBsAg leaves the ER, travels to the Golgi, and is secreted within 3 h of synthesis (Fig. 1 and data not shown). The situation is quite different in glucosidase-inhibited cells. As shown in Fig. 3A, a substantial amount of HBsAg remains cosedimenting with the ER fractions, immediately following protein synthesis. Because the ER is the site of protein synthesis, this is not surprising. However, as shown in Fig. 3B, by 3 h, HBsAg has left the ER: analysis of each fraction with mAb that can distinguish L, M, and S, shows there is little detectable HBsAg cosedimenting with the ER containing fractions. Note that the departure of HBs proteins from the ER by 3 h after synthesis was confirmed using a mAb that detects an epitope in a conformation-independent manner (data not shown); thus all detectable HBsAg leaves the ER by 3 hours! Moreover, it appears that the peaks of S, M, and L sediment separately (Fig. 3B). For example, although there is overlap, the peak of M is at least two fractions from the peaks of S. SDS/PAGE analysis of low density fractions (Fig. 3B Inset) showed an enrichment of M and possibly L polypeptide, relative to S; this is discussed further in the next section.
Figure 3.
Intracellular distribution of HBsAg L, M, or S in glucosidase-inhibited cells. Cytoplasmic lysates from 2.15 cells, maintained for 3 h in the absence or presence of glucosidase inhibitor, were labeled with [35S]methionine for 20 min, as in Fig. 1. At various times after labeling, immediately after the label (A; 30 min) or at 3 h (B) and 24 h (C), cytoplasmic lysates were prepared and resolved through Percoll gradients, as in Fig. 2. The positions of the peaks of the markers associated with the Golgi and ER (WGA and G6PD) are shown as arrows. Each fraction was probed for the presence of HBsAg by the antigen capture method using mAb. mAbs were used as follows: H166 (A); MA/18 (L), E (for M and L) and C20-02 for S (B). The amount of antigen bound to plates containing mAb was determined by cpm. (B Inset) Fractions 4 and 6 (indicated by circled 2 and 3) from the Percoll gradient shown in B were disrupted by sonication, and polypeptides were immunoprecipitated with an antibody that recognizes an epitope common to S, M, and L, and resolved by SDS/PAGE. An immunoprecipitate of HBV subviral particles prepared from the culture medium of control 2.15 cells 24 h after radiolabeling with [35S]methionine are also shown (lane 1). The autoradiographic image is presented. The relative positions of mono- and diglycosylated M (gp33 and gp36) as well as unglycosylated and monoglycosylated S (p24 and gp27) are also shown.
Analysis of the intracellular distribution of the HBsAg remaining within the cell 24 h after synthesis was also very informative (Fig. 3C). Note that, as shown, the detained HBs proteins are contained in fractions that cosediment with the ER, Golgi, and lysosomes! That is, although all HBsAg has left the ER by 3 h after synthesis (Fig. 3B), ≈29% of the intracellular HBsAg (Fig. 3C) has returned to these fractions at 24 h. In addition, ≈33%, 22%, and 16% of the surviving (detectable) intracellular HBsAg was recovered in the fractions cosedimenting with the Golgi, lysosomal, and intermediate compartment fractions, respectively (Fig. 4C). Although the size of the fraction volumes collected caused some markers to be contained mostly within a single fraction, these experiments were repeated at least three separate times, demonstrating the reproducibility of the results. Similar distributions were found at 16 and 18 h after synthesis (not shown).
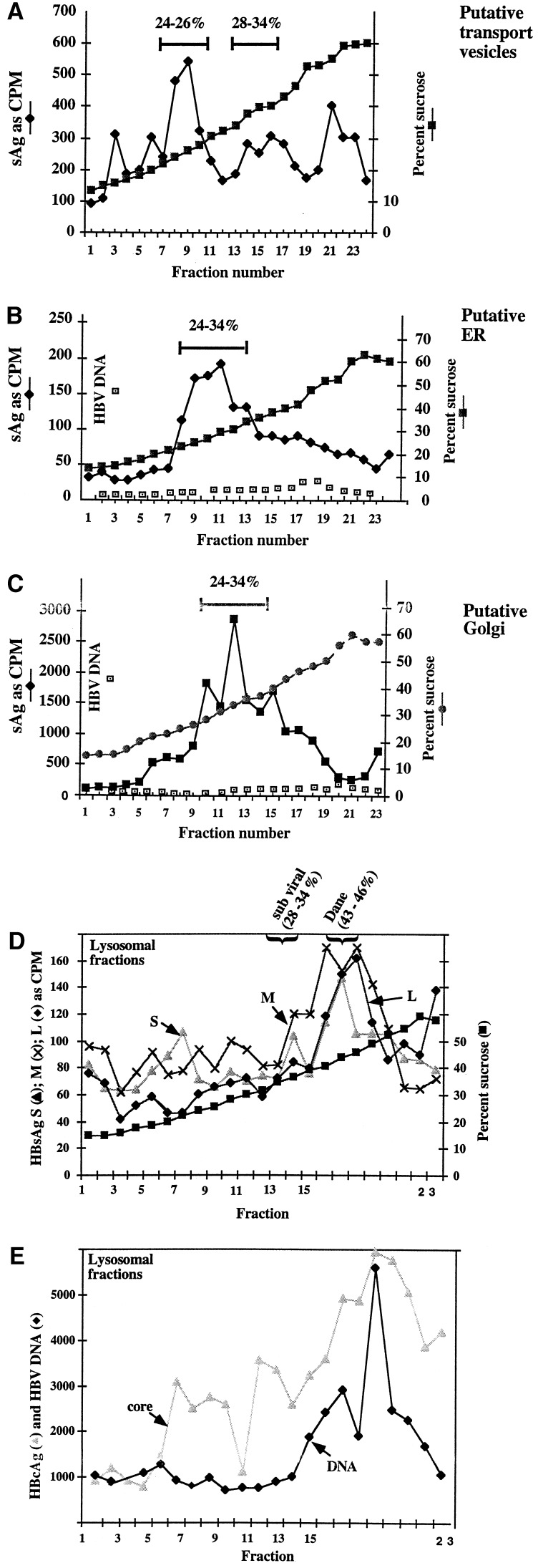
Figure 4.
Physical analysis of HBs proteins derived from different subcellular compartments. Selected fractions from the Percoll gradient of glucosidase-inhibited cells, after labeling with [35S]methionine, shown in Fig. 3, were further resolved through sucrose gradients. (A) Sucrose gradient separation of fraction numbers 4–6 from the top (putative transport vesicles/intermediate compartments) of the Percoll gradient in Fig. 3B. The ER (from Fig. 3C, fraction numbers 14–16), Golgi (Fig. 3C, fraction numbers 8 and 9), and lysosomal (Fig. 3C, fraction numbers 19 and 20) containing regions from the Percoll gradient shown in Fig. 3C were also resolved through sucrose gradients and is shown in B, C, and D and E, respectively. Each fraction was assayed for the presence of HBsAg by the antigen capture method. The presence or absence of HBV DNA in each fraction was also determined in B, C, and E by a quantitiative PCR described in ref. 19. Values of intensity of the HBV-specific PCR product that were visualized following agarose gel electrophoresis, are in arbitrary units. In E, the presence of HBV core antigen was also determined using mAb 42B/12.
Evidence that HBs proteins were present within the ER (and other organelles) and were not merely cosedimenting with these compartments was obtained by two methods. For example, incubation of cells with DTT, at the time of labeling, caused a “build up” HBsAg within putative ER containing fractions (not shown). DTT is believed to prevent disulfide intramolecular bond formation, causing proteins to be retained within the ER (26). This is evidence that (i) the indicated fractions of the Percoll gradient do contain the ER and (ii) HBs proteins are present within the ER and are not merely cosedimenting with these organelles. Additional evidence that the HBs proteins are contained within the lumen of the organelles (and are not merely cosedimenting) comes from immuno-access analysis. That is, detection of HBsAg by the antigen capture assay required prior sonication and detergent (Nonidet P-40) treatment of each fraction. This suggests that the proteins are contained in vesicles. This also implies that, to a large measure, the integrity of the intracellular organelles (ER, Golgi, transport vesicles) has been maintained throughout the fractionation process and the HBs proteins are actually within these compartments, and not merely cosedimenting with them. Thus HBs proteins would be inaccessible to antibody during the antigen capture assay.
Size and Content of HBs Proteins Accumulating Within Glucosidase-Inhibited Cells.
The physical nature of the retained HBs proteins detained in glucosidase-inhibited cells was investigated by two methods. First, the ability of HBs proteins from the top of the Percoll gradient (putative transport vesicles/intermediate compartments) to sediment to characteristic positions within sucrose gradients, following appropriate ultracentrifugation, was examined (Fig. 4). Second, HBsAg polypeptides contained within HBsAg peak fractions from the Percoll gradient shown in Fig. 3, were immunoprecipitated and resolved through SDS/polyacrylamide gels (Fig. 3B Inset).
It was possible that the HBs proteins, detained in glucosidase-inhibited cells, were present as subviral particles. This possibility was tested by determining if HBs proteins isolated from Percoll gradients shown in Fig. 3 sedimented as large particles in sucrose gradients known to resolve subviral particles. Therefore, after sonication to disrupt organelle structure, fractions of the Percoll gradient prepared from glucosidase-inhibited cells 24 h after protein labeling (shown in Fig. 3) were resedimented through sucrose gradients. Fractions representing the Golgi, lysosomes, ER, and top of the Percoll gradient were used because HBsAg was shown to accumulate in these locations long after synthesis (see Fig. 3).
HBsAg in the transport vesicles and intermediate compartments.
Under the conditions used here, HBsAg derived from subviral particles and virions migrate to 32–42% sucrose following a rate zonal centrifugation (6). Fractions from the top of the Percoll gradient shown in Fig. 3B (presumably in or comigrating with transport vesicles), were recentrifuged through the sucrose gradient. Clearly, as shown in Fig. 4A, most of these HBs proteins (L, M, and S were not distinguished in this experiment) derived from the top of the Percoll gradient migrate well into the sucrose gradient, to between 24% and 26% sucrose. Lesser amounts migrate to positions expected of subviral particles (28–34% sucrose) and to uncharacterized dense regions (in excess of 50% sucrose). These data suggest that a substantial majority of the HBs proteins that are recovered from the top of the Percoll gradient in Fig. 3 actually exist as large particles that sediment well into sucrose after centrifugation. The large size of HBs proteins recovered from the top of the Percoll gradient 18 h after synthesis was confirmed by chromatography through Bio-Rad AG 1.5M. Briefly, HBs proteins from that time point eluted from the columns with a 2000-kDa marker, proving that the detained HBs proteins are very large (unpublished data).
HBsAg in the Golgi and ER.
The ER and Golgi containing fractions of the Percoll gradient shown in Fig. 3C contained HBs proteins which sedimented broadly, following recentrifugation through sucrose, from 24% to 34% sucrose, and may represent a combination of different sized HBsAg species, including intact and other forms (Fig. 4 B and C, respectively). This suggests that some intact subviral particles are still synthesized in glucosidase-inhibited cells. In control cells, HBV DNA was found in only the ER and Golgi fractions (data not shown). Because HBV is believed to bud from the ER or post-ER compartment and traffic to the Golgi, these are the expected intracellular locations of viral DNA. On the other hand, in glucosidase-inhibited cells, as shown in Fig. 4 B and C, the samples corresponding to the ER and Golgi contain very little, if any, detectable viral DNA.
HBsAg, core, and DNA in the lysosomes.
Although viral gene products migrated heterogenously throughout the sucrose gradient, Fig. 4 D and E show that the lysosomal fractions of the Percoll gradient contained HBV DNA, core, and surface antigen that sedimented to 42% sucrose. This is the density expected for intact HBV virions, which would be expected contain viral DNA, core, and HBsAg. It is unclear why particles sedimenting with the size of intact HBV are recovered from the lysosomes, but not the ER or Golgi. This may reflect the aberrant routing of HBV and its glycoproteins in these cells. SDS/PAGE analysis of HBsAg in the lysosomes revealed considerable degradation (not shown). Thus it appears that although (and remarkably) large particles of HBsAg and virion-like structures (which can sediment well into sucrose gradients) can be recovered in the lysosome, considerable degradation is occurring. It is suggested that this is the final point in the trafficking of HBV gene products in these cells.
HBs proteins detained within the transport vesicles/intermediate compartments of glucosidase-inhibited cells are enriched for M.
The HBsAg polypeptides migrating to fractions 4 and 6 of the Percoll gradient shown in Fig. 3B were immunoprecipitated with conformation-dependent antibody that recognizes all HBsAg (L, M, and S) polypeptides (mAb C20-02) and resolved by SDS/PAGE (Fig. 3B Inset). Most notable is the relative amount of M and S in the samples from glucosidase-inhibited cells. For example, as shown in lane 1 (Fig. 3B Inset), subviral particles are usually composed of a glycoprotein ratios of 80–90% S, 5–20% M, and 1% (if any) L (see ref. 6). The HBs proteins from 2.15 cells shown in Fig. 3B Inset, lane 1, contain ≈15% M, as determined by densitometry. On the other hand, the HBsAg from fractions 4 and 6 of the Percoll gradient shown in Fig. 3B contained much more M than S. The relative change in the amount of L in the detained particles was unclear from these results, but appears to be somewhat increased. Densitometry analysis indicated, for example, that the amount of M from fractions 4 and 6 was in excess of 70% of the resolved polypeptides. The aberrant ratio of L and M to S may account, in part, for the intracellular retention of these HBs proteins, although why such particles can depart from the ER is still unresolved. The reason for the enrichment of M is also not known.
DISCUSSION
These data show that, under normal circumstances, after synthesis, HBsAg leaves the ER, passes through the Golgi, and is secreted within 3 h (Fig. 1). In glucosidase-inhibited cells the situation is radically different, with a considerable fraction of the HBs proteins retained within the cell (Figs. 1 and 3). Moreover, the detained HBsAg is not rapidly degraded as has been observed for other glycoproteins detained within glucosidase-inhibited cells (13, 14). Indeed, HBs proteins detained within glucosidase-inhibited cells appear as aggregates greatly enriched for the M glycoprotein and have an intracellular half-life in excess of 24 h (Fig. 1). The detained HBsAg is “displaced” and recovered in the Golgi, as well as upper regions (presumably transport vesicles) of the Percoll gradient, and even returning to the ER long after initially leaving (Fig. 3).
In spite of the fact that HBV glycoproteins in glucosidase-inhibited cells do not function properly, being unable to be secreted or mediate virus egress, they have left the ER, thus escaping a step in the quality control pathway. Because they are not secreted, however, it is speculated that some form of post ER quality control mechanism must be at work. Since other work (A.M., X.L., T.M.B., B.S.B., and R.D., unpublished data) suggests that, in glucosidase-inhibited cells, S and L, but not M, can be properly processed in these cells, it is further hypothesized that an aberrant M can act in a dominant-negative manner, preventing the secretion of an entire particle that may contain properly folded proteins.
The “retrograde” transport of HBsAg from the Golgi back to the ER in glucosidase-inhibited cells is reminiscent of the recycling of misfolded mutant vesicular stomatitis virus (VSV) G protein in VSV productively infected cells, reported by Hammand and Helenius (27). Mutant viral protein in VSV-infected cells, in which host protein synthesis was inhibited, was shown to leave and then return to the ER (27). Thus, the work here extends previous findings to show that retrograde transport of HBsAg from the Golgi to the ER occurs in cells as a consequence of inhibition of glycosylation processing. Moreover, this occurred in cells that were otherwise viable. Also, there has been selectivity, since the S and L HBsAg are far less sensitive to glycosylation processing than is M. Finally, it is shown that the HBV glycoproteins are trafficking in the glucosidase-inhibited cells as large, presumably lipoprotein, particles rather than as single proteins. This may explain, to some extent, the sensitivity of HBs proteins, relative to host glycoproteins, to ER glucosidase function. The extreme sensitivity of HBV glycoproteins to glucosidase processing may also reflect an inability of viral glycoproteins to efficiently use the NB-DNJ-insensitive endomannosidase (28).
It is thus possible that, in the absence of functional glucosidase, a subpopulation of HBsAg bearing unprocessed glycan is not fully folded and is recycled to the ER. Proper folding and transport for some glycoproteins have been shown to involve cell chaperons that may require glycosylation processing of their substrates (29). If misfolding of HBsAg occurred in glucosidase-inhibited cells, it must be restricted to subdomains, since our detection scheme used conformation-specific mAbs. The possibility that the abberrant trafficking of HBs proteins is due to host proteins that have been rendered dysfunctional by glucosidase inhibition has not yet been excluded.
In untreated cells, HBV DNA is detected only in the ER and Golgi containing fractions of the Percoll gradient (not shown). In glucosidase-inhibited cells, HBsAg, core protein, and viral DNA were eventually recovered in lysosomal fractions. This suggests that particles are slowly but consistently being recognized as defective and are degraded. The long intracellular half-life of HBsAg in these cells is different from the short half-lives seen for other glycoproteins in cells in which glycan processing is inhibited (14). This too may be explained, however, by the physical nature of the detained HBV glycoproteins: they appeared to be recycling or trafficking throughout the cell as large aggregates, enriched for M and possibly L. In this system resolution and quantification of L was unsatisfactory, and thus unambiguous conclusions about it’s abundance in the detained particles cannot be made. Because an excess of L relative to S has been associated with retention of subviral particles (23), this in itself could account for inability of particle secretion. The question than becomes: why are M retained? Because the M within these aggregates had a slightly greater molecular weight, as detected by SDS/PAGE (unpublished data), than that of M bearing fully processed carbohydrate, we favor the hypothesis that M in these aggregates contained unprocessed glycan and this was responsible for retention.
This work thus dramatizes the consequences of inhibition of glucosidase to the function of a set of “reporter” glycoproteins. Understanding the reasons for secretion failure and mechanisms involved in the retention and return of these lipoprotein particles in glucosidase-inhibited cells will likely provide a new perspective upon the role of glycan trimming in glycoprotein behavior.
Acknowledgments
We thank Drs. Wolfram Gerlich (Giessen, Germany), William Mason (Fox Chase Cancer Center), Richard Peluso (Mt. Sinai Medical College), Fran Platt (Oxford University), and James Keen (Thomas Jefferson University) for reading this manuscript and their thoughtful comments. This work was supported with awards from the Hepatitis B Foundation, a gift from the Blanche and Irving Laurie Foundation, the North Atlantic Treaty Organization, U.S. Public Health Service Grant CA 06927 (National Institutes of Health), and an appropriation from the Commonwealth of Pennsylvania and Monsanto/Searle, Inc.
ABBREVIATIONS
- HBV
hepatitis B virus
- ER
endoplasmic reticulum
- NB-DNJ
n-butyl-deoxynojirimycin
- HBs protein
hepatitis B surface protein
- HBsAg
hepatitis B surface antigen
- HBcAg
hepatitis B core antigen
- G6PD
glucose 6 phosphate dehydrogenase
- Hex
hexosaminidase
- WGA
wheat germ agglutinin
References
- 1.Dwek R A. Science. 1995;269:1234–1235. doi: 10.1126/science.7652569. [DOI] [PubMed] [Google Scholar]
- 2.Elbein A D. Semin Cell Biol. 1991;2:309–317. [PubMed] [Google Scholar]
- 3.Huovila A P, Eder A, Fuller S. J Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganem D. Curr Top Microbiol Immunol. 1991;168:61–84. doi: 10.1007/978-3-642-76015-0_4. [DOI] [PubMed] [Google Scholar]
- 5.Gerlich W, Bruss V. In: Hepatitis B Vaccines in Clinical Practice. Ellis R, editor. New York: Dekker; 1992. pp. 41–82. [Google Scholar]
- 6.Block T, Platt F, Lu L, Gerlich W, Foster G, Blumberg B, Dwek R. Proc Natl Acad Sci USA. 1994;91:2235–2239. doi: 10.1073/pnas.91.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X, Mehta A, Butters T, Dwek R, Block T. Virology. 1995;213:660–665. doi: 10.1006/viro.1995.0038. [DOI] [PubMed] [Google Scholar]
- 8.Ou W J, Cameron P H, Thomas T Y, Bergeron J J M. Nature (London) 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 9.Hammond C, Braakman I, Helenius A. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodish H, Kong A. J Cell Biol. 1984;98:1720–1729. doi: 10.1083/jcb.98.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parent B, Yeo T-K, Olden K. Mol Cell Biochem. 1986;72:21–33. doi: 10.1007/BF00230633. [DOI] [PubMed] [Google Scholar]
- 12.Hammond C, Helenius A. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- 13.Kearse K P, Willimans D B, Singer A. EMBO J. 1994;13:3678–3686. doi: 10.1002/j.1460-2075.1994.tb06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore S, Spiro R. J Biol Chem. 1993;268:3809–3812. [PubMed] [Google Scholar]
- 15.Beasley R P. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 16.Patzer E J, Nakamura G R, Simonsen G C, Levinson A L, Brands R. J Virol. 1986;58:884–892. doi: 10.1128/jvi.58.3.884-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heerman H-H, Goldman I, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sells M A, Chen M L, Acs G. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X, Block T, Gerlich W. J Virol. 1996;70:2277–2285. doi: 10.1128/jvi.70.4.2277-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meisel H, Sominskaya I, Lu X, Spiller G H, Pushko P, Borisova G, Pumpens P, Grens E, Kruger D H, Gerlich W H. Intervirology. 1995;37:330–339. doi: 10.1159/000150397. [DOI] [PubMed] [Google Scholar]
- 21.Aronson N J, Touster O. Methods Enzymol. 1974;31:90–102. doi: 10.1016/0076-6879(74)31009-9. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson G B, Butters T D, Dwek R A, Platt F. J Biol Chem. 1993;268:570–576. [PubMed] [Google Scholar]
- 23.Bruss V, Ganem D. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plutner H, Davidson H W, Saraste J, Balch W E. J Cell Biol. 1992;119:1097–1116. doi: 10.1083/jcb.119.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweizer A, Fransen J A M, Matter K, Kreis T E, Ginsel L, Hauri H-P. J Cell Biol. 1990;53:185–196. [PubMed] [Google Scholar]
- 26.Braakman I, Helenius J, Helenius A. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammand C, Helenius A. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore S E, Spiro R G. J Biol Chem. 1990;265:1304–13112. [PubMed] [Google Scholar]
- 29.Helenius A. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]