Abstract
The spontaneous expression of heat shock genes during development is well documented in many animal species, but the mechanisms responsible for this developmental regulation are only poorly understood. In vertebrates, additional heat shock transcription factors, distinct from the heat shock factor 1 (HSF1) involved in the stress response, were suggested to be involved in this developmental control. In particular, the mouse HSF2 has been found to be active in testis and during preimplantation development. However, the role of HSF2 and its mechanism of activation have remained elusive due to the paucity of data on its expression during development. In this study, we have examined HSF2 expression during the postimplantation phase of mouse development. Our data show a developmental regulation of HSF2, which is expressed at least until 15.5 days of embryogenesis. It becomes restricted to the central nervous system during the second half of gestation. It is expressed in the ventricular layer of the neural tube which contains mitotically active cells but not in postmitotic neurons. Parallel results were obtained for mRNA, protein, and activity levels, demonstrating that the main level of control was transcriptional. The detailed analysis of the activity of a luciferase reporter gene under the control of the hsp70.1 promoter, as well as the description of the protein expression patterns of the major heat shock proteins in the central nervous system, show that HSF2 and heat shock protein expression domains do not coincide. This result suggests that HFS2 might be involved in other regulatory developmental pathways and paves the way to new functional approaches.
The spontaneous expression of heat shock proteins (HSPs) in the absence of stress can vary dramatically during development and in different in vitro differentiation systems. In Drosophila, this property results from the presence in the promoters of specific regulatory sequences, distinct from the sequences to which heat shock factor (HSF) binds (1).
The situation is far less clear in vertebrates for two reasons. First, they display additional forms of HSF. Two genes have been cloned in mouse and human, and three in chicken (refs. 2–5; and note added in proof), and it is likely that other members have not yet been identified. Second, the mechanisms leading to the differential expression of HSPs during development have been thoroughly analyzed in a limited number of cases. In early mouse embryogenesis, the transient expression of HSP70 and heat shock cognate (HSC) 70 during zygotic genome activation requires the heat shock element (HSE) sequences and the heat shock inducible factor (HSF1) (ref. 6; for an opposite point of view, see ref. 7). In the human K562 erythroleukemia cell line, hemin induces cell differentiation, transient expression of heat shock genes and activation of an additional HSF, HSF2, distinct from the main one involved in the control of the heat shock response (8, 9).
In addition to its expression during in vitro differentiation of the K562 cell line, HSF2 was previously shown to be active in a limited number of situations. No active HSF2 can be detected in adult mouse tissues (10), except testis (11). HSF2 might regulate the testis-specific hsp70 gene (hsp70.2). Mouse HSF2 is also found to be active for DNA binding in all embryonal carcinoma cell lines tested and in embryonal stem cells, under normal growth temperature (12, 13). We have recently shown that HSF2 is also specifically activated during preimplantation stages (12). A constitutive “HSF2-like” HSE-binding activity progressively develops around the 8-cell or morula stages (14) with a corresponding increase in HSF2 mRNA, as determined by reverse transcription–PCR (6).
The very unusual pattern of HSPs expression in early mouse embryo (15) led to the hypothesis that HSF2 might be the form of HSF involved in their developmentally controlled expression (16). Yet, the weakness of this hypothesis was the limited number of embryological data (2).
We address the developmental role of HSF2 (i) by following its expression and activity during mouse embryogenesis and (ii) by comparing these data with the expression of a reporter gene under the control of the hsp70.1 promoter and of other HSPs.
MATERIALS AND METHODS
Mouse Embryos.
For gel shift assays and Western blot analyses, embryos ranging from embryonic day (E) 8.5 to E15.5 were obtained from (BALB/c × SJL/J)F1 crosses. The day of copulary plug detection was considered as E0.5. Mothers were killed by cervical dislocation, and embryos were delivered by caesarian section, rapidly dissected in ice-cold phosphate buffer, quickly centrifuged, and immediately frozen in dry ice. The telencephalic vesicles, limbs or limb buds, and tail buds were dissected. For heat shock treatment, dissected structures were submitted to a 30-min heat shock at 43°C in PBS and then treated exactly as unshocked embryos.
For in situ hybridization (ISH) and immunocytochemical experiments, E9.5, E12.5, and E15.5 embryos were obtained from outbred OF1 mice (Iffa Credo) and fixed overnight by immersion in 4% paraformaldehyde in phosphate buffer (0.12 M, pH 7.2) at 4°C. The neural tube from E12.5 and E15.5 embryos was extracted. Embryos and neural tubes used for ISH were embedded in gelatin/albumin mix and vibratome-sectioned at 100–250 μm in sagittal and coronal orientations. Sections were gradually dehydrated in methanol/PBS/0.1% Tween 20 up to 100% methanol and stored at −20°C. For immunocytochemical experiments, embryos were embedded in 7.5% gelatin/15% saccharose in PBS, frozen at −56°C, and cryosectioned at 17 μm.
Whole-Cell Extraction from Embryos and F9 Embryonal Carcinoma Cells.
F9 embryonal carcinoma cells were grown, heat-shocked, and extracted as described (17).
Embryos were submitted to 2–4 rapid freeze-thaw cycles in 5 volumes of extraction buffer as F9 cells (17). The following numbers of embryos were used for extraction: 10 E8.5 embryos (about 10 somites), 6 embryos and yolk sacs at E9.5 (14 somites), 11 E9.75 embryos (24 somites), 8 embryos and 5 yolk sacs at E10.5, 2 embryos and 4 yolk sacs at E11.5, and 2 embryos at older stages.
Gel Mobility Shift Assay.
EMSA was performed as described (17) using two different double-stranded oligonucleotides, 32P-end-labeled by T4 polynucleotide kinase and [γ-32P]ATP. One, HSE2, contains several consensus sequences that constitute a strong binding site for the HSF family (17). The other binds proteins from the SP1 family (18). Typically, 27 μg of embryo extracts were used. The specificity of the complexes was checked by competition with an excess of unlabeled double-stranded probe (data not shown). The radioactivity present in the specific shifted complex was quantified by PhosphorImager (Molecular Dynamics) using imagequant version 3.3 software following overnight exposure with a PhosphorScreen (Molecular Dynamics). Polyclonal antibodies were used to detect the presence of HSF1 or HSF2 in the shifted complexes. These antibodies were prepared and used as described (19) with modifications as in Mezger et al. (12).
Western Blot Analysis.
Whole-cell embryo or F9 cell extracts (40 μg) were loaded on a 10% SDS/polyacrylamide gel and transferred to a nitrocellulose filter. The efficiency of protein transfer was checked in parallel by staining of the gel. Western blot analysis was performed with the ECL peroxidase detection system (Amersham). HSF2 polyclonal antibodies were used at 1:2500 dilution and anti-rabbit secondary antibody (Promega) at 1:500 dilution, as described. For HSC/HSP70 or HSP90 detection (data not shown), mouse mAbs (SPA820 and SPA830) were purchased from and used as described by StressGen Biotechnologies (Victoria, Canada).
ISH Analysis.
We subcloned a weakly conserved fragment of mHSF2 cDNA. A 418-bp SacI–EcoRI fragment (corresponding to the last 158 nt of the ORF plus the 260 nt of the 3′ untranslated region) was isolated from the C9-pGEM1 clone and subcloned into pBluescript SK(+). The HSF2 subclone was linearized with SacI and transcribed using T3 RNA polymerase, or with EcoRI and transcribed using T7 RNA polymerase, to generate the antisense or sense probes, respectively.
Digoxygenin-mediated ISH was performed as described by Wilkinson (20).
Immunocytochemistry.
Anti-HSP antibodies were used at the following dilutions: 1:100 for HSP25 rabbit polyclonal antibody (SPA801; StressGen Biotechnologies), 1:40 for HSP90α or β rabbit polyclonal antibodies (PA3–013 and PA3–012; Affinity BioReagents, Neshanic Station, NJ), 1:100 for HSC70 rat mAb (SPA815; StressGen Biotechnologies), and 1:200 for HSP70 mouse mAb (anti-HSP72; Amersham). Serial cryosections were incubated for 1 h with one of the five antibodies. Sections were incubated for 30 min with peroxidase-conjugated secondary antibodies: sheep anti-rat IgG (Boehringer Mannheim) at 1:200, goat anti-rabbit IgG (Promega) at 1:100, and goat anti-mouse IgG (Sigma) at 1:100. Peroxidase activity was revealed using 0.03% diaminobenzidine tetrahydrochloride (Sigma)/0.005% H2O2 in 0.1 M Tris·HCl (pH 7.6).
Transgenic Mouse Line and Luciferase Assays.
The hsp70.1 Luc transgene is a 6.6-kbp linear DNA construct carrying the coding region of firefly luciferase driven by 0.8 kbp from the promoter of the hsp70.1 gene (21). We used transgenic line 1 (described in ref. 22). Rapidly microdissected tissues were extracted and luciferase activity was measured as described (21). Protein concentrations in the extracts were determined using the Bradford method. Each value represents the mean of three experiments performed with different embryos.
RESULTS
A Constitutive HSE-Binding Activity Is Present and Regionalized in Postimplantation Embryos.
Gel shift analysis using a consensus HSE oligonucleotide was performed on dissected structures of embryos from different stages of organogenesis (ranging from E8.5 to E15.5).
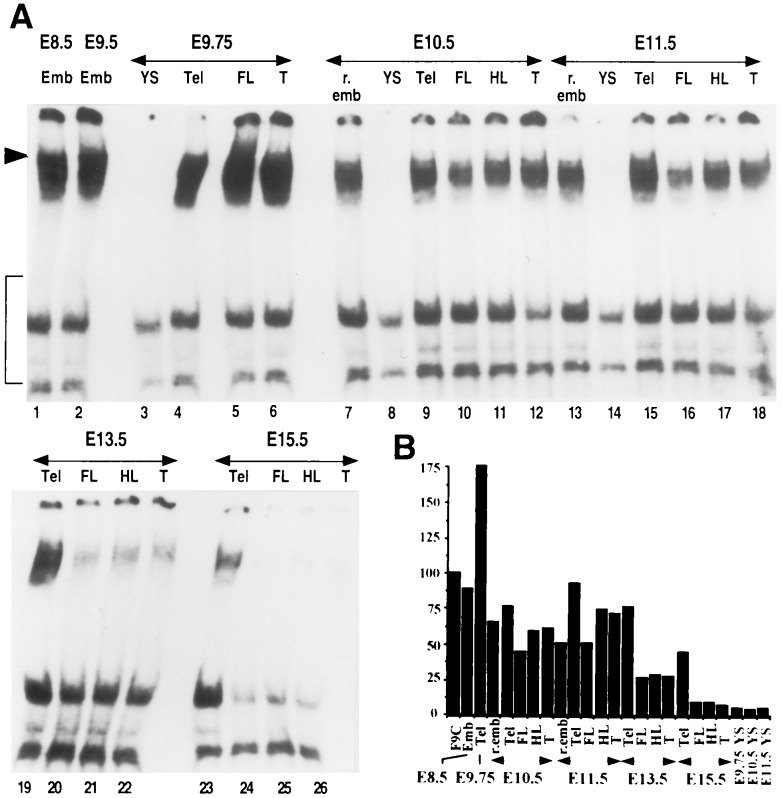
An HSE-binding activity was constitutively present in unshocked embryos throughout postimplantation development (Fig. 1A). This activity was detected at very high levels in E8.5 and E9.5 embryos and peaked at E9.75 (Fig. 1A, lanes 1–6). At E9.75, quantification of the specific shifted complex indicated that it was 2-fold higher in embryonic telencephalon than in F9 control cells (Fig. 1B). It slowly decreased in E10.5 and E11.5 embryos, in which a slightly higher level was detected in the telencephalon than elsewhere (70% of that of F9 cells; Fig. 1A, lanes 7–12 and 13–18). Between E13.5 and E15.5, the activity rapidly disappeared in the limbs and tail bud (Fig. 1A, lanes 20–22 and 24–26) reaching 30% at E13.5 and 8% at E15.5 (Fig. 1B). In contrast, a constitutive HSE-binding activity was still present in the telencephalon at a high level (70% of F9 control cells) until E15.5 when it began to decrease (Fig. 1A, lanes 19 and 23; Fig. 1B). Extraembryonic tissues such as the yolk sac never displayed any constitutive activity at any stage investigated. To verify that these variations in DNA-binding were not due to protein degradation, we used oligonucleotides that can bind the members of the SP1 family (18). Indeed, SP1-like DNA-binding activity in the embryo displayed a constant pattern during this period of development, even in yolk sac extracts where no HSE-binding activity was detected (data not shown).
Figure 1.
Detection of constitutive HSE-binding activity by gel shift assay in postimplantation embryos. Emb, entire embryo at E8.5 or E9.5; r.emb, remaining undissected part of the embryo; YS, yolk sac; Tel, telencephalic vesicles; FL, forelimbs; HL, hindlimbs; T, tail bud. (A) Extracts (27 μg) from dissected structures were incubated with 32P-labeled double-stranded HSE2 oligonucleotide for gel shift assay analysis. Arrowhead points to the specific shifted complexes. Nonspecific complexes are bracketed. Free DNA is not shown. (B) Results of PhosphorImager quantification of the specific shifted complexes. The signal intensities of embryo extracts were reported as the percentage of the density value in F9 control cell extracts.
Therefore, as seen in early embryos, we could detect a constitutive HSE-binding activity in unshocked postimplantation embryos. This activity decreased after E10.5 and almost disappeared by E15.5, except in the telencephalon. This constitutive activity was not limited to the telencephalic part of the brain, but rather to the central nervous system (CNS), since a similar activity was also detected in mesencephalon and spinal cord of E15.5 embryos (data not shown).
HSF2 Is the Major Component of the HSE-Binding Activity Detected in Postimplantation Embryos.
To determine whether the activity detected in postimplantation embryos was related to HSF1 or, as in early embryos (12), to HSF2, we used polyclonal antibodies raised against mouse HSF1 or HSF2 to disturb the signal and/or electrophoretic migration of the complex (Fig. 2). At the dilution used in this experiment, no cross-reactivity of anti-HSF1 antibodies with HSF2 or anti-HSF2 antibodies with HSF1 was observed (12).
Figure 2.
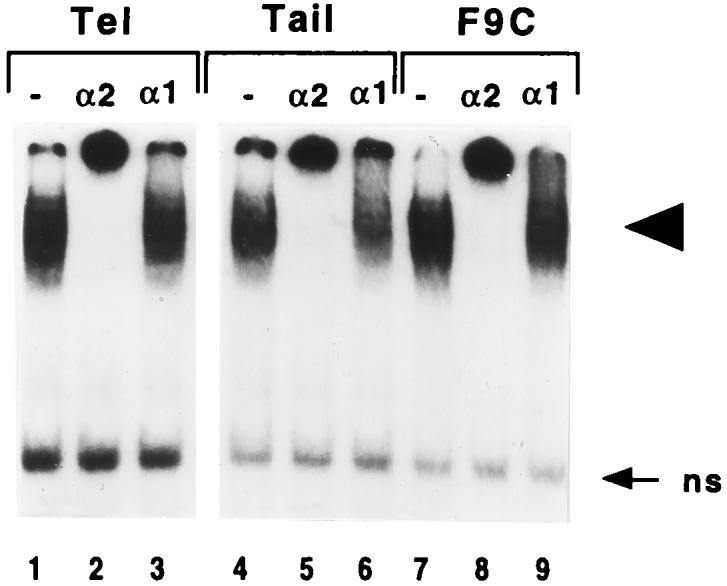
Effect of polyclonal antibodies on specific HSE complexes formed with extracts from postimplantation embryos. Extracts (27 μg) from the telencephalic or tail parts of E11.5 embryos or from F9 control cells (F9C) were preincubated with polyclonal antibodies raised against HSF1 (α1) or HSF2 (α2) or without antibody (−), before incubation with the 32P-labeled double-stranded HSE2 oligonucleotide and gel shift assay analysis. Arrowhead points to the specific complexes, and arrow points to nonspecific complexes (ns).
As in F9 control cells (Fig. 2, lanes 7–9), the HSE complexes detected in extracts from the telencephalic or tail parts of E11.5 embryos were markedly altered by preincubation with anti-HSF2 antibodies, whereas anti-HSF1 antibodies used at the same dilution did not disturb the migration (Fig. 2, compare lanes 2–3 and 5–6 to lanes 1 and 4). Similar alterations were obtained with the other structures. We further verified that heat-shocked tissues from postimplantation embryos (i.e., forelimbs from E13.5 embryos) did still contain a heat-induced activity that cross-reacts with anti-HSF1 antibodies, as found in F9 heat-shocked cells (data not shown).
These results demonstrate, as in blastocysts, that the constitutive HSE-binding activity present in unshocked postimplantation embryos is essentially due to HSF2 and not to a heat-inducible HSF1-like factor that would have been activated during tissue manipulations.
High Levels of HSF2 DNA-Binding Activity Are Correlated with High Levels of HSF2 Protein.
In Western blot analyses. HSF2 protein appears in one or two immunologically reactive bands, corresponding to the two splicing isoforms previously described (10, 23). Higher levels of HSF2 protein were detected at stages with high constitutive DNA-binding activity, namely E8.5, E9.5, and E10.5 (Fig. 3, lanes 2–9). Similarly, high levels of HSF2 protein were found in the telencephalon at E13.5 and E15.5 where high HSF2 DNA-binding activity persists (Fig. 3, lanes 10–14). Likewise, in forelimbs, hindlimbs, and tail, HSF2 protein levels were markedly lower, in correlation with the very low HSF2 DNA-binding activity (Fig. 3, compare lanes 11–13 and 15–17 to lanes 10 and 14). The yolk sac appeared to be devoid of HSF2 protein (Fig. 3, lane 5), consistent with the lack of HSF2 DNA-binding activity.
Figure 3.
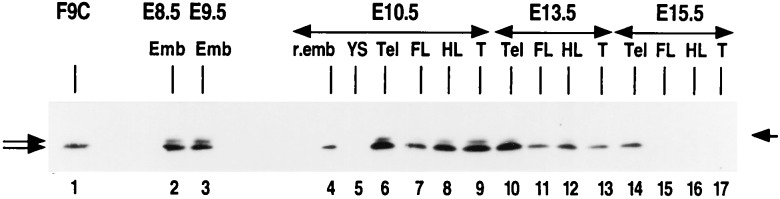
Western blot analysis of HSF2 distribution in postimplantation embryos. F9C, extracts from F9 control cells. Left arrows indicate both HSF2 splicing isoforms; the right arrow indicates the 80-kDa molecular mass marker.
ISH Analysis Reveals that HSF2 Is Specifically Expressed in Restricted Areas of the Nervous System.
ISH analyses were performed to examine the correlation between patterns of HSF2 mRNA expression and protein activity. As the mHSF1 and mHSF2 genes share some highly conserved sequences (4), the specificity of the detection was ensured by limiting the antisense riboprobe to the 3′ region of HSF2 cDNA, thereby excluding cross-hybridization of the HSF2 riboprobe with HSF1 mRNAs.
The analysis was performed at three time points of embryogenesis: E9.5, E12.5, and E15.5 (see above) to encompass HSF2 DNA-binding activity.
The aim of this study was not to give a complete description of HSF2 gene expression during embryogenesis. Rather, we intended to analyze whether the high levels of HSF2 protein and HSF2 DNA-binding activity detected earlier were correlated with the level of HSF2 mRNA transcripts, and whether the expression of the HSF2 gene was uniform inside the tissues where the protein was abundant (especially the CNS). Thick sections were thus examined.
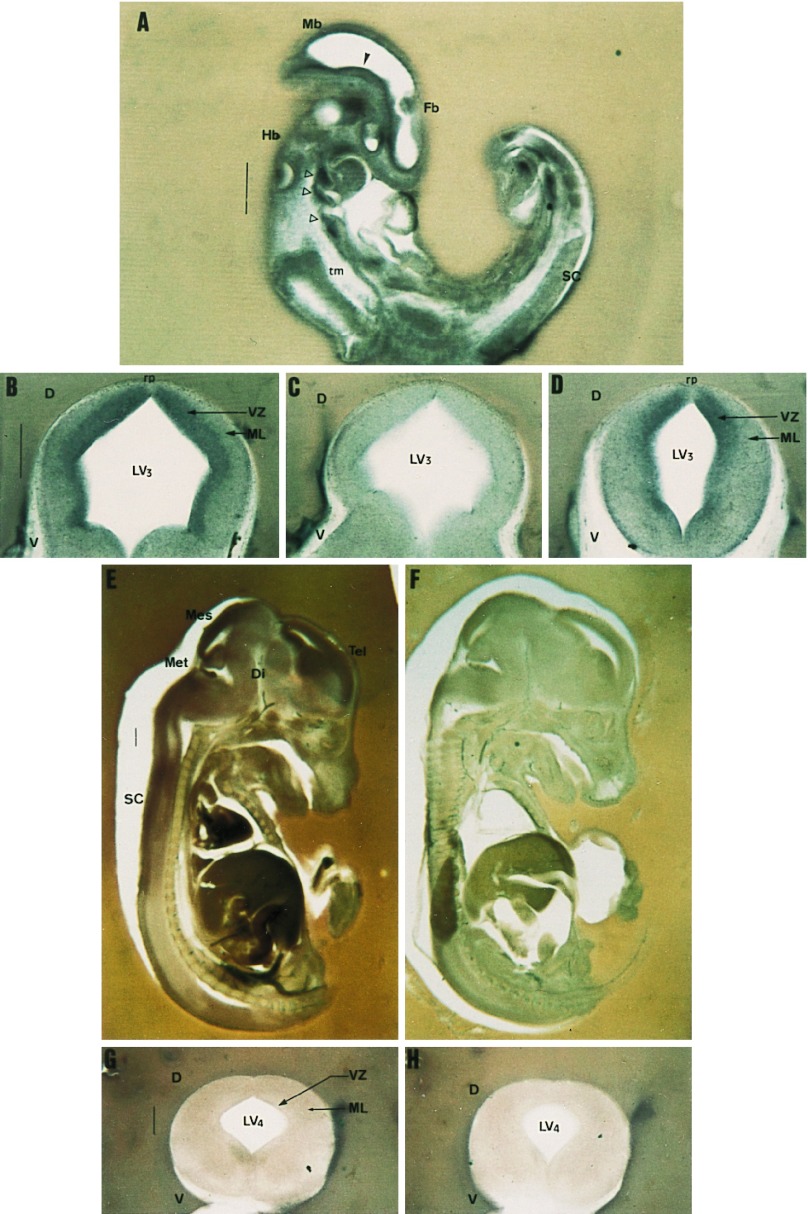
E9.5 embryos were labeled ubiquitously (Fig. 4A), apart from trunk mesenchyme, where the signal was not higher than background. Most of the nervous system displayed HSF2 expression. Particularly prominent was the neuroepithelial labeling of the head (Fig. 4A, closed arrowhead), delineating the roof as well as the floor of telencephalic through mesencephalic vesicles. Besides, branchial regions displayed a signal as strong as the neural tube (Fig. 4A, open arrowheads).
Figure 4.
HSF2 ISH during postimplantation development. Sections 100–250 μm thick of E9.5 (A), E12.5 (B–D), and E15.5 (E–H) embryos were hybridized with digoxygenin-labeled antisense or sense (C, F, and H) HSF2 probes. (A) Parasagittal section of E9.5 embryo; HSF2 is expressed throughout the embryo except in the trunk mesenchyme. Arrowheads point to structures where HSF2 is highly expressed (see text). (B–D) Coronal sections of E12.5 dissected neural tube at the level of the mesencephalon (B and C) or spinal cord (D). The expression domain maps to proliferative regions of the neural tube. Note that the roofplate region (rp) is not stained. (E and F) Sagittal sections of E15.5 embryos display markedly decayed HSF2 expression. Weak staining is only detectable in the different developing encephalic vesicles and spinal cord. (G and H) Coronal sections of E15.5 neural tubes (at the level of spinal cord) show diffuse low labeling. D, dorsal; Di, diencephalon; Fb, forebrain; Hb, hindbrain; LV3, mesencephalic vesicle; LV4, fourth ventricle; Mb, midbrain; Mes, mesencephalon; Met, metencephalon; ML, mantle layer; rp, roofplate; SC, spinal cord; Tel, telencephalon; tm, trunk mesenchyme; V, ventral; VZ, ventricular zone. (Bar = 500 μm.)
At E12.5, HSF2 expression became more restricted to the nervous system. Dissected neural tubes displayed a homogenous expression profile along the antero-posterior axis. Specifically, coronal sections of the midbrain revealed a strong signal surrounding the ventricle (Fig. 4B compared with Fig. 4C). This domain fits with the proliferating ventricular layer. Likewise, at a more posterior level, the most highly labeled domain of the spinal cord was the ependymal layer which also contains mitotically active cells and separates the lumen of the ventricle from the mantle layer composed of postmitotic cells (Fig. 4D). Close examination of the roof of the neural tube showed no signal in a thin triangular shaped domain, which roughly corresponds to the roofplate (Fig. 4 B and D).
In contrast to previous stages, HSF2 mRNA levels declined in E15.5 embryos (Fig. 4E). Compared with the control (Fig. 4F), they remained detectable and were significantly higher in the neural tube, labeling all encephalic vesicles, with a lower intensity in the diencephalon and anterior mesencephalon. The labeling on coronal sections from different parts of the brain as well as from spinal cord was weak (compare Fig. 4G to Fig. 4H) and it clearly appeared to be no longer restricted to the neuroepithelium as previously observed at E12.5.
Variations in the Luciferase Reporter Activity Driven by the hsp70.1 Promoter Do Not Correlate with Variations in HSF2 DNA-Binding Activity.
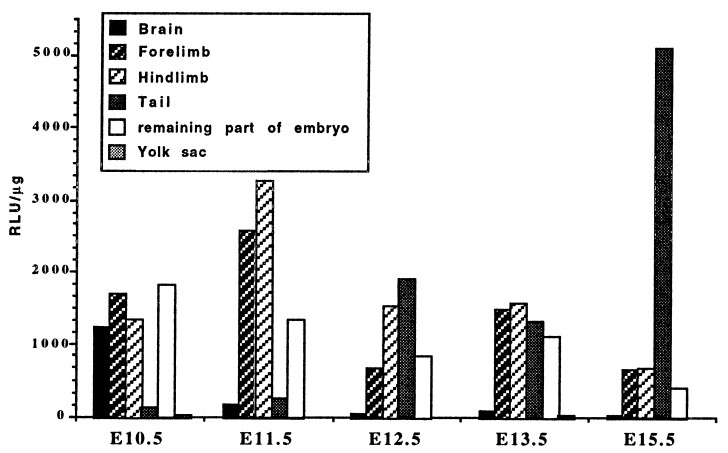
Until now, it was admitted that the main targets of the HSFs were the promoters of the heat shock genes. A transgenic line has been generated by Thompson et al. (21) carrying a luciferase reporter gene under the control of the inducible hsp70.1 gene promoter. The pattern of luciferase activity in the transgenic mouse has been shown to be faithfully correlated with that of the endogenous HSP70 protein during preimplantation development and at adult stages (21, 22). We thus determined the luciferase specific activity on tissue extracts from E8.5 to E15.5 transgenic embryos (Fig. 5). The general pattern of luciferase activity in whole embryos was roughly similar to the HSF2 DNA binding activity profile: an increase after implantation with a maximum at E10.5 and a progressive decrease during the second part of embryogenesis (data not shown). Moreover, the luciferase activity was very low in the yolk sac as compared to the embryo (Fig. 5), in parallel to what was observed for HSF2 activity.
Figure 5.
Luciferase activity in tissues of HSP70.1-luciferase transgenic embryos. Each value is the mean of the measurements performed with three embryos. The variations between embryos are <20%.
However, a closer examination reveals quantitative and qualitative differences between HSF2 DNA-binding and luciferase activities. The increase in luciferase activity between implantation and E10.5 was sharp (data not shown) whereas the increase in DNA-binding activity was smoother. In particular, HSF2 activity persisted in the CNS longer than in other tissues, whereas luciferase activity disappeared from this tissue before decreasing in the others (Fig. 5). Luciferase activity was highest in the limb buds at E11.5, in contrast to HSF2. Moreover, the luciferase activity in the tailbud increased regularly and significantly during embryogenesis, in agreement with the high luciferase activity found in the tail of the adult transgenic mice (21).
Immunocytochemical Localization of HSPs Does Not Reveal Any Obvious Correlation Between the Levels of HSF2 and HSPs.
The putative correlation between the pattern of HSF2 DNA-binding activity and that of HSP synthesis (other than HSP70.1) was investigated by Western blot analysis and immunocytochemistry.
First, Western blot analyses were performed on E10.5 to E15.5 extracts. No obvious variations in the total amount of HSP70 family members were observed among embryonic tissues and stages (data not shown). For the HSP90 family members, slight variations appeared among tissues, but to a lesser extent than those of HSF2. The only significant parallel observed between HSF2 and HSP90 protein levels is that HSP90 was more abundant in the CNS (data not shown).
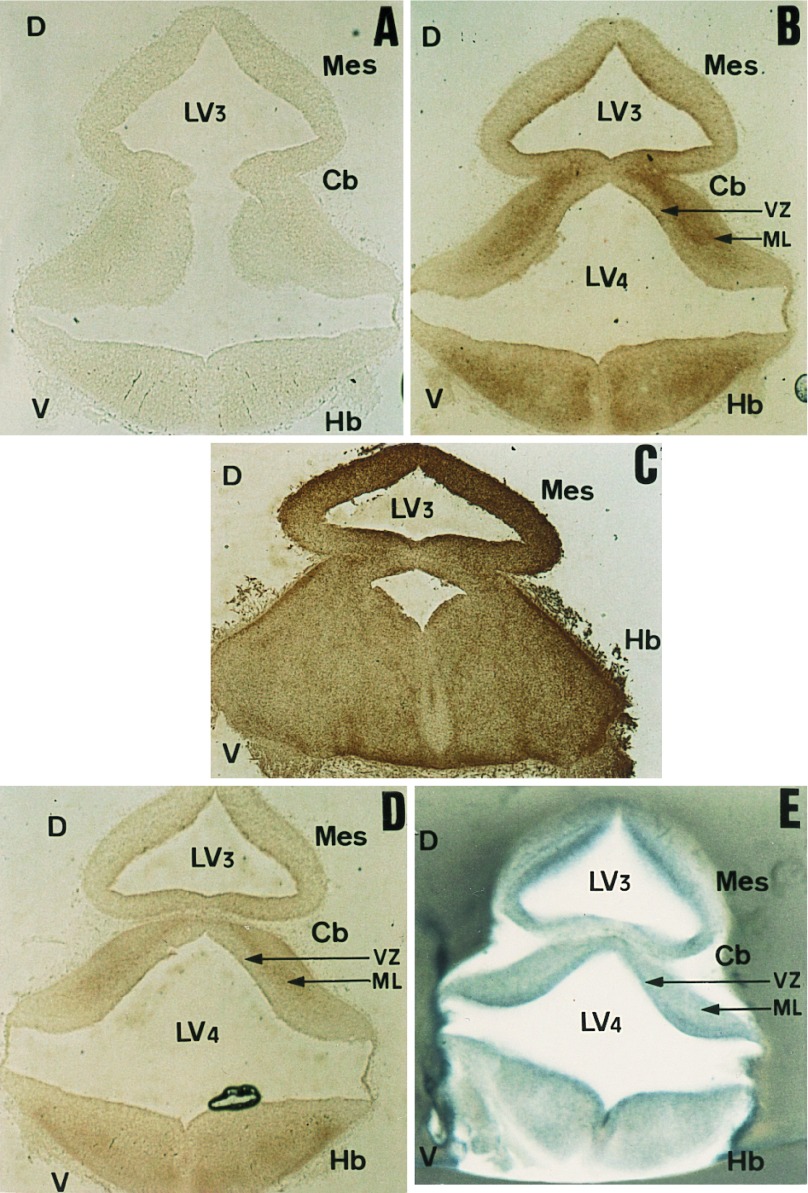
Second, the distribution of HSPs was analyzed by immunocytochemistry in the CNS where both HSF2 expression and activity were high. This analysis was performed on the same embryonic stages as for ISH—i.e., E9.5, E12.5, and E15.5. Five antibodies raised against HSP25, HSP70, HSC70, HSP90α, and HSP90β allowed us to distinguish between the stress-inducible and constitutive forms of these proteins. We found that the constitutive forms of the HSPs were the first to be expressed in the CNS (as early as E9.5). At E9.5, these forms appeared to be preferentially expressed in cephalic mesenchyme components, which give rise to the peripheral nervous system, rather than in the neural tube (M.L. and M.M., unpublished results). At E12.5, the inducible forms of HSP70 and HSP90 were still almost undetectable (Fig. 6A). The cognate HSC70 expression occurred at different levels of the mantle layer—e.g., in the hindbrain and cerebellum (Fig. 6B). The labeling seems to map postmitotic rather than mitotically active cells of the neural tube. Some staining was also detected in the ventral part of the diencephalon (presumptive hypothalamus). HSP90β, as described earlier, was ubiquitously expressed along the neural tube (Fig. 6C). Anti-HSP25 also labeled the mantle layer of the neural tube (Fig. 6D), but to a lesser extent than HSC70 (compare with Fig. 6B). Therefore, it clearly appears that the expression of each constitutive HSP is different and unrelated to the abundance of HSF2 mRNA (as shown in Fig. 6E). This discrepancy remains true at E15.5, but extends to the five proteins studied.
Figure 6.
Comparative distribution profiles of the major HSPs and HSF2 gene expression within the CNS of E12.5 embryos. (A–E) Immunological detection of HSPs on coronal cryostat sections of E12.5 dissected neural tubes at level of midbrain. Sections were incubated with anti-HSP90α (A), anti-HSC70 (B), anti-HSP90β (C), or anti-HSP25 (D). (E) ISH with digoxygenin-labeled HSF2 antisense riboprobe on a 200-μm-thick coronal section, performed at the level of midbrain Cb, cerebellum; other abbreviations are identical to those in Fig. 4.
DISCUSSION
The mechanisms leading to the developmental regulation of heat shock gene expression in vertebrates are poorly characterized. It has been suggested that HSF2 might be the form of heat shock transcription factor involved in this control (16, 24). To test this hypothesis and to characterize the mechanisms involved in HSF2 activation, we examined both the expression and DNA-binding activity of this factor during mouse development.
HSE-Binding Activity Profile During Mouse Development.
Gel shift assays revealed a constitutive HSE-binding activity from E8.5 to E15.5. This activity could be obtained without any stresses and was similar to that previously found in preimplantation embryos at the morula and blastocyst stages. In both studies (ref. 12 and this study), it can be attributed to HSF2. It cannot be ruled out that another HSF, still unknown but immunologically related to HSF2, could account for such an activity in so far as there are three HSFs in chicken (2). However, two sets of data argue for an involvement of HSF2. First, the constitutive activity displayed thermosensitivity properties similar to those of in vitro-synthesized HSF2 (M.R., M.L., V.M., and M.M., unpublished results). Second, the ISH analyses presented here show that HSF2 mRNA levels were well correlated with that of the constitutive activity.
Therefore, as far as DNA-binding activity is concerned, HSF2 is active during most of development and can thus be considered as an embryonic factor. HSF2 DNA-binding activity increased until E9.75, when it peaked. It was then maintained at more or less similar levels until E10.5 and progressively decreased to E13.5. At this stage, a dramatic decrease was observed in the whole embryo, except in the CNS where it remained high until E15.5.
Besides, HSF2 activity was present in almost all embryonic tissues, but never in extraembryonic ones, such as the yolk sac. The absence of protein degradation was verified by silver staining and Western blot analysis, showing that other proteins (i.e., HSP70 and HSP90) were detectable (data not shown). Moreover, an SP1-like binding activity could be observed in the same extracts, showing that the absence of HSF2 DNA-binding activity was not an artefact but probably specific of HSF2.
Developmental Control of HSF2.
Comparison of gel shift assay and Western blot analyses showed a strong positive correlation between constitutive HSE-binding activity (due to HSF2) and HSF2 protein distribution. Embryonic stages and tissues displaying high levels of HSF2 DNA-binding activity (i.e., the telencephalic part of the brain at E13.5 and E15.5) also showed the highest levels of HSF2 protein (Figs. 1 and 3). Moreover, when HSF2 was inactive in the gel shift assay (i.e., in the yolk sac at any stage and in the limbs at E15.5), the protein could not be detected by Western blot analysis. This suggests that HSF2 is not expressed in these tissues, at least not at detectable levels.
ISH analysis also showed a good correlation between HSF2 mRNA distribution and HSF2 activity. Indeed, maximal HSF2 mRNA expression took place around mid-gestation (i.e., E9.5; Fig. 4A), in parallel with HSF2 DNA-binding activity which peaked at E9.75. Therefore, we can conclude that variations in HSF2 DNA-binding activity during mouse postimplantation development are mainly due to developmentally regulated HSF2 gene expression. However, we cannot conclude whether the mechanism controlling HSF2 expression is transcriptional or posttranscriptional by mRNA stabilization. Constitutive HSF2 activity persisted later than HSF2 transcription (30% of the DNA-binding activity still detected at E15.5 versus much lower HSF2 mRNA levels) suggesting that the half-life of HSF2 protein is rather long, which was confirmed by immunocytochemical analysis (see note).
HSF2 mRNA levels were significantly higher in the CNS than elsewhere, eventually becoming restricted to it as development progressed. In chicken, Northern blot and ISH analyses revealed that HSF2 is also expressed at higher levels in the developing brain [as well as in the intestine (2)]. At E12.5 we showed that the ISH signal is preferentially localized in the ventricular or ependymal layers of the neural tube, corresponding to the proliferative cell-rich zones, in contrast to postmitotic cells. This might be correlated with a role for this factor in neural proliferation and would be reminiscent of the role of a related rat factor, BF-1, during neural cell proliferation in the telencephalon (25). Like BF-1, HSF structurally belongs to the HNF3/forkhead multigene family (26, 27).
In Search of Functions for HSF2 During Mouse Development.
The very specific distribution of HSF2 suggests a role for this factor during mouse development. In early embryos, HSF2 is present and active at stages with high levels of HSP expression (15). However, experiments performed with the transgenic mouse line harboring the luciferase reporter gene under the control of the hsp70.1 promoter demonstrated that the transcriptional activity of this heat shock gene promoter was not correlated with the level of HSF2 during the mid-part of embryogenesis. Both quantitatively and qualitatively, luciferase activity was far from being parallel to HSF2 DNA-binding activity. HSF2 remained high in the brain after day 15.5, whereas luciferase activity decreased in the CNS before decreasing in the other parts of the embryo.
Immunocytochemical analysis was used to examine the expression of other HSPs during development in the CNS, where HSF2 was demonstrated to be preferentially localized. Analysis of all the major HSPs showed that the constitutive forms were more or less ubiquitously expressed in the CNS at all stages, with the constitutive HSP90β being the most abundant. Expression of inducible HSPs was not detected before E15.5, consistent with previous results showing a delayed constitutive expression of inducible forms of HSPs in the embryo (28). We found no obvious correlations between the expression patterns of the major HSPs and that of HSF2. As development progressed from mid to late ontogeny, HSF2 expression was down-regulated while expression of the HSPs was increasing. Furthermore, at similar stages, the expression profiles were differently distributed. For example, at E12.5, the HSF2 hybridization signal was clearly localized in the ventricular layer of the neural tube (Fig. 6E). By comparison, HSP staining was different: HSP90β was ubiquitously distributed (Fig. 6C), while HSC70 (Fig. 6B) and, to a lower extent HSP25 (Fig. 6D), were localized in the mantle layer of the neural tube. Therefore, HSF2 does not seem to be responsible by itself for constitutive HSP expression, at least in the CNS. However, we cannot exclude that, in other tissues, HSF2 might be directly involved in the control of HSPs expression, nor that its action is positively or negatively modulated by other transcription factors, which would preclude a clear vision of the transcriptional role of HSF2.
At this point, we would like to emphasize the insights provided by our new data on potential HSF2 targets and functions, compared with what was already known from the in vitro model of embryonal carcinoma and ES cells. In embryonal carcinoma cells, the occurrence of high transcription rates for hsc70 and hsp90 genes remains controversial (13, 17, 28–30). The fact that HSF2 does not occupy the hsp70.1 promoter (13) is therefore not so surprising and does not preclude that HSF2 might not be involved in HSP gene activation during development. In contrast, the data provided by the in vivo analysis of the transgenic hsp70.1-luciferase embryos showed a clear transcriptional activation of the hsp70.1 gene during development. This gene was therefore a good putative target candidate for HSF2. However, it turned out that neither the hsp70.1 gene activation profile nor the distribution of any other HSP so far tested is compatible with the hypothetic activation of the corresponding genes by HSF2 in postimplantation development.
These results contrast with those obtained on mouse testis in which the HSF2 expression profile is well correlated with that of an HSP gene specifically expressed in this tissue, hsp70.2 (11). It was recently shown that mHSF2 is subjected to posttranscriptional regulation (10, 23). Alternative splicing yields two HSF2 mRNA and protein isoforms, each of which can be detected in specific tissues. The largest isoform is predominant in testis and the smallest in many other tissues. We show that during mouse development, as in adult mice, the smallest isoform is quantitatively the major one (see Fig. 3). Both isoforms have distinct transcriptional activities, the largest being the most transcriptionally active (23). Therefore, the HSF2 isoform that is expressed during development may not be transcriptionally active enough to regulate HSP expression. Alternatively, this isoform might activate the transcription of genes different from heat shock genes or interact with other transcription factor to antagonize or stimulate their transcriptional properties.
We believe this study provides radically new insights in the search for HSF2 target genes and opens the way to a quest for other functions for this “orphan” factor.
Acknowledgments
We are grateful to V. Zimarino for the gift of HSF cDNA clones and M. Catelli for the gift of anti-HSP90 antibodies. We are indebted to J.-P. Renard, Y. Mercier, and J.-F. Oudin for the gift of the transgenic line and their help in obtaining the embryos and luciferase assays. We thank R. Sousa-Yeh for critical reading of this manuscript, M. Wassef’s lab for help with ISH, Y. Gitton for support and helpful comments, and E. Christians for helpful discussions. This work was supported by a grant from the Association pour la Recherche sur le Cancer (Grant 6505), by ACC from the Ministère de la Recherche and by the Ecole Normale Supérieure.
ABBREVIATIONS
- HSP
heat shock protein
- HSF
heat shock factor
- HSE
heat shock element
- E
embryonic day
- ISH
in situ hybridization
- CNS
central nervous system
Note
. We have now confirmed the preferential expression of HSF2 protein in mitotically active neuronal cells by preliminary immunocytochemistry with anti-HSF2 polyclonal antibodies (M.R., M.M., and V.M., unpublished results)
Note Added in Proof
A new HSF, HSF4, has been described in humans (31), but no embryological data are available.
References
- 1.Pauli D, Tissières A. In: Developmental Expression of the Heat Shock Genes in Drosophila melanogaster. Morimoto R I, Tissières A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 361–378. [Google Scholar]
- 2.Nakai A, Morimoto R I. Mol Cell Biol. 1993;13:1983–1997. doi: 10.1128/mcb.13.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabindran S K, Giorgi G, Clos J, Wu C. Proc Natl Acad Sci USA. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarge K D, Zimarino V, Holm K, Wu C, Morimoto R I. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- 5.Schuetz T J, Gallo G J, Sheldon L, Tempst P, Kingston R E. Proc Natl Acad Sci USA. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christians E, Michel M, Adenot P, Mezger M, Rallu M, Morange M, Renard J P. Mol Cell Biol. 1997;17:778–788. doi: 10.1128/mcb.17.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bevilacqua A, Kinnunen L H, Bevilacqua S, Mangia F. Development (Cambridge, UK) 1995;121:113–122. [Google Scholar]
- 8.Sistonen L, Sarge K D, Phillips B, Abravaya K, Morimoto R I. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sistonen L, Sarge K D, Morimoto R I. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiorenza M T, Farkas T, Dissing M, Kolding D, Zimarino V. Nucleic Acids Res. 1995;23:467–474. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarge K D, Park-Sarge O K, Kirby J D, Mayo K E, Morimoto R I. Biol Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- 12.Mezger V, Rallu M, Morimoto R I, Morange M, Renard J P. Dev Biol. 1994;166:819–822. doi: 10.1006/dbio.1994.1361. [DOI] [PubMed] [Google Scholar]
- 13.Murphy S P, Gorzowski J J, Sarge K D, Phillips B. Mol Cell Biol. 1994;14:5309–5317. doi: 10.1128/mcb.14.8.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mezger V, Renard J P, Christians E, Morange M. Dev Biol. 1994;165:627–638. doi: 10.1006/dbio.1994.1281. [DOI] [PubMed] [Google Scholar]
- 15.Mezger V, Legagneux V, Babinet C, Morange M, Bensaude O. In: Heat Shock Protein Synthesis in Preimplantation Mouse Embryos and Embryonal Carcinoma Cells. Hightower H, Nover L, editors. Berlin: Springer; 1991. pp. 153–166. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto R I, Jurivich D A, Kroeger P E, Mathur S K, Murphy S P, Nakai A, Sarge K, Abravaya K, Sistonen L T. In: Regulation of Heat Shock Gene Transcription by a Family of Heat Shock Factors. Morimoto R I, Tissières A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 417–445. [Google Scholar]
- 17.Mezger V, Bensaude O, Morange M. Mol Cell Biol. 1989;9:3888–3896. doi: 10.1128/mcb.9.9.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nardelli J, Gibson T J, Vesque C, Charnay P. Nature (London) 1991;349:175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- 19.Sarge K D, Murphy S P, Morimoto R I. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson D G. In: Whole Mount in Situ Hybridization of Vertebrate Embryos. Wilkinson D G, editor. New York: Oxford Univ. Press; 1992. pp. 75–83. [Google Scholar]
- 21.Thompson E M, Christians E, Stinnakre M G, Renard J P. Mol Cell Biol. 1994;14:4694–4703. doi: 10.1128/mcb.14.7.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christians E, Campion E, Thompson E M, Renard J P. Development (Cambridge, UK) 1995;121:113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- 23.Goodson M L, Park-Sarge O K, Sarge K D. Mol Cell Biol. 1995;15:5288–5293. doi: 10.1128/mcb.15.10.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 25.Xuan S, Baptista C A, Balas G, Tao W, Soares V C, Lai E. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 26.Harrison C J, Bohm A A, Nelson H C M. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- 27.Vuister G W, Kim S J, Wu C, Bax A. Biochemistry. 1994;33:10–16. doi: 10.1021/bi00167a002. [DOI] [PubMed] [Google Scholar]
- 28.Kothary R, Perry M D, Moran L A, Rossant J. Dev Biol. 1987;121:342–348. doi: 10.1016/0012-1606(87)90170-9. [DOI] [PubMed] [Google Scholar]
- 29.Giebel L B, Dworniczak B P, Bautz E K F. Dev Biol. 1988;125:200–207. doi: 10.1016/0012-1606(88)90073-5. [DOI] [PubMed] [Google Scholar]
- 30.Legagneux V, Mezger V, Quélard C, Barnier J V, Bensaude O, Morange M. Differentiation (Berlin) 1989;41:42–48. doi: 10.1111/j.1432-0436.1989.tb00730.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto R, Nagata K. Mol Cell, Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]