In the past several decades, the discovery and detailed characterization of the various subcellular organelles has revealed the intricate workings of the cell. It is of great interest then, that one of the most prominent structures at the plasma membrane, the caveola, has remained the most enigmatic organelle found in cells. Together with their marker proteins, the caveolins, caveolae are present in large numbers in a variety of cell types. Although they were initially proposed to act as mere conduits for cellular uptake, akin to clathrin-coated vesicles, it is now clear that caveolae and caveolins have pleiotropic functions that modulate numerous cellular processes. Many proposed physiological roles for caveolae are controversial, making a rigorous analysis of their function imperative. A giant leap in this regard is the recent series of reports on the phenotypic characterization of caveolin-deficient mice. As loss of caveolin expression causes a concomitant loss in morphologically identifiable caveolae, such animals allow for the first definitive studies of the role of these organelles in various cells in vivo. In this review, we will discuss the known and proposed functions of caveolae in the context of these caveolin-deficient animal models, give a synopsis of the rapidly emerging themes in the field, and discuss the relevance of caveolae to the understanding of human disease.
Caveolar structure
During the pioneering era of cellular ultrastructure, the rapid discovery of various subcellular organelles led to an explosion in many research areas that constitute what is today known as modern cell biology. The discovery of caveolae and the ensuing research on their function were among the fruits of this endeavor. Because of their resemblance to “little caves” on transmission electron microscopy (TEM), Yamada first coined the term caveolae intracellulare in 1955 to describe conspicuous, 50-100 nm invaginations of the plasma membrane found in gallbladder epithelial cells (1). A few years earlier, Palade had also described the presence of such structures in vascular endothelium, although he favored the term plasmalemmal vesicles (2) to refer to the newly discovered cellular organelle.
Caveolae occur both as invaginations of the plasma membrane proper and as vesicles residing close to the membrane. They are found in many cells types but are notably abundant in fibroblasts, adipocytes, endothelial cells, type I pneumocytes, epithelial cells, and smooth and striated muscle cells. Caveolae are distinct in morphology from the more electron-dense and much larger (about 200-300 nm) clathrin-coated pits. As both types of membrane invaginations are found together in many cell types, caveolae likely serve other functions beyond providing conduits for endocytosis.
Biochemically, caveolar microdomains are very specialized membrane entities. Research in the last decade has provided strong evidence that membranes co-exist in two predominant forms: i) the “liquid-disordered” state — composed of loosely packed phospholipids with relatively rapid lateral diffusion; and ii) the “liquid-ordered” state — composed of an aggregation of cholesterol and sphingolipids — where the bilayer is more rigid and lipids are more confined in their movements. Due to their non-conventional lipid composition, these “liquid-ordered” membranes, termed “lipid rafts,” are biochemically distinct from the surrounding phospholipid bilayer (3). Interestingly, caveolae show a similar enrichment in cholesterol and sphingolipids and have traditionally been considered a specialized form of lipid raft (i.e., an invaginated/vesicular form). What distinguishes caveolae as a subset of these specialized microdomains is the presence of a peculiar family of membrane proteins known as the caveolins.
Caveolin-1 (Cav-1), a 21 to 24 kDa protein that decorates the cytoplasmic surface of caveolae, was the original member of this family and serves as the primary structural component of caveolae (4). In fact, along with cholesterol and sphingolipids, Cav-1 appears to play a direct role in caveolar biogenesis. The two other members of the caveolin family, caveolin-2 (Cav-2) and caveolin-3 (Cav-3), also target exclusively to caveolar microdomains (5, 6). The expression levels of Cav-1, -2, and -3 fully correlate with the presence of caveolae in those tissues. Interestingly, the expression patterns of Cav-1 and -2 are largely distinct from that of Cav-3; whereas adipocytes, endothelial cells, pneumocytes, and fibroblasts have the highest levels of Cav-1 and -2, Cav-3 expression is limited to muscle cells of all types (cardiac, skeletal, and smooth muscle cells) (6, 7). The three proteins overlap in their expression in a few cell types, including smooth muscle cells. Since their discovery, the caveolins have been shown to be intimately involved in all aspects of caveolar function.
Studies of Cav-1-deficient mice
We and others have recently reported the generation of mice with a targeted disruption of the caveolin-1 (Cav1) locus (8, 9). Here, we will consider several of the pivotal observations made in these mice which have expanded our understanding of caveolar function (see Table 1 for a summary). Perhaps the biggest initial surprise in the characterization of these mice was the realization that loss of a major cellular organelle imparts no developmental abnormality or lethality. In fact, cursory examination of the mice into adulthood and beyond reveals no overt phenotypic abnormalities, except for a tendency to remain smaller than their wild-type counterparts (8).
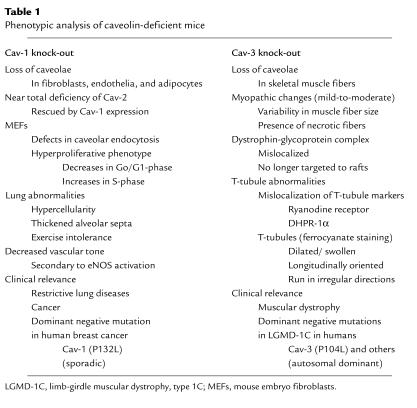
Table 1.
Phenotypic analysis of caveolin-deficient mice
Cav-1 and caveolar biogenesis
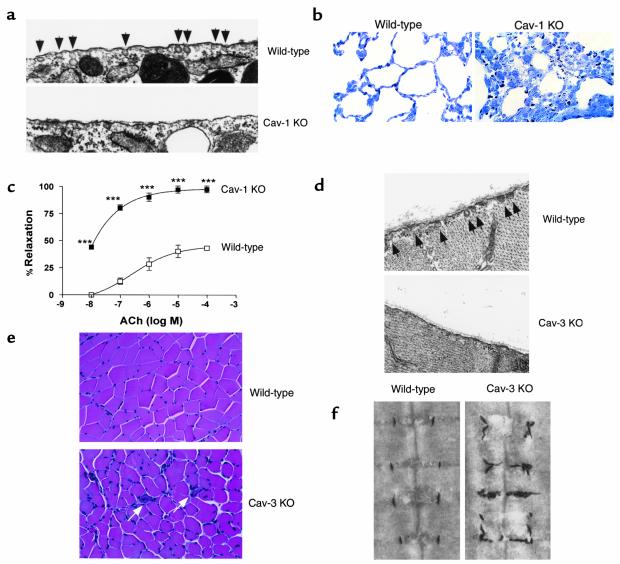
The caveolins appear to mediate the formation of caveolae, as judged by the phenotype of cells in which the expression of these proteins is altered experimentally. In cells such as lymphocytes and transformed fibroblasts, which do not express caveolins, and lack any identifiable caveolae, the de novo production of Cav-1 is sufficient to drive the formation of 50-100 nm invaginations (reviewed in ref. 10). Conversely, anti-sense–mediated down regulation of Cav-1 causes a loss of caveolae in fibroblasts (11). The availability of Cav-1–null mice has recently allowed a rigorous test of this model. These mice show a complete ablation of mophologically identifiable caveolae in endothelia and fat (Figure 1a), firmly establishing that Cav-1 is required for caveolar invagination in these tissues. Primary cells derived from Cav-1–null mice likewise lack caveolae (8, 9).
Figure 1.
Some important phenotypes of Cav-1 and Cav-3–deficient mice. (a) A deficiency in Cav-1 is sufficient to completely disrupt caveolae formation. Tissues thus far examined include endothelial cells (8, 9), adipocytes (Razani and Lisanti, unpublished observations), and mouse embryonic fibroblasts (MEFs) (8). The transmission electron micrographs shown are from near-confluent MEFs (8). (b) Cav-1–deficient mice show lung abnormalities, with constricted alveolar spaces, thickened septa, and hyper-cellularity. One-micron sections of lung parenchyma were cut, stained with Toluidine-Blue, and images were acquired with a 60× objective. Left panel, wild-type mice (WT); Right panel, Cav-1–null mice (KO) (8). (c) Cav-1–deficient mice show hyper-responsiveness to endothelium/nitric oxide (NO)-dependent vasodilation. Note that acetylcholine (Ach)-induced relaxation (a NO-dependent phenomenon) of the Cav-1–null aortic rings was clearly potentiated by the loss of Cav-1 expression. Concentration-dependent relaxation induced by acetylcholine (Ach; expressed as log of molar concentration) in aortas pre-constricted with 10 μM phenylephrine (PE) from wild-type (WT; open squares) and Cav-1–null (KO; black squares) mice. ***P < 0.0001 vs. WT (8). (d) A deficiency in Cav-3 is sufficient to completely disrupt the formation of sarcolemmal caveolae. The transmission electron micrographs shown are from hindlimb skeletal muscle fibers imaged at 25,000× magnification (32). (e) Cav-3–deficient mice show skeletal muscle abnormalities, with variability in muscle fiber size, the presence of some necrotic fibers, and mononuclear infiltrates. Muscle tissue sections from wild-type (left panel) and Cav-3 KO mice (right panel) were cut and stained with hematoxylin and eosin (H&E). (f) Cav-3 protein expression is required for the development of a mature and highly organized T-tubule system. Cav-3 KO T-tubules are dilated and run in irregular directions. Skeletal muscle tissue samples from wild-type (left panel) and Cav-3 KO (right panel) were subjected to T-tubule system staining using potassium ferrocyanate, which gives T-tubules an electron-dense (black) appearance (32).
Although the mechanism of Cav-1’s association with and transformation of the plasma membrane into flask-shaped caveolae remains unknown, the interaction of Cav-1 with lipids likely contributes to this process. Cav-1 binds the two primary components of lipid rafts, cholesterol and sphingolipids, both in vitro and in vivo (10). Given the ability of cholesterol-binding agents such as cyclodextrin and nystatin to flatten caveolae and disassemble their caveolin-enriched coats (4), the association between cholesterol and caveolins appears pivotal for vesicle biogenesis and may be sufficient to explain the cellular phenotype of Cav-1–deficient animals. However, studies to date have not examined possible perturbations in the cholesterol content of the plasma membrane or lipid rafts in Cav-1–null tissues. The absence of Cav-1 could also indirectly lead to the loss of caveolae by destabilizing the arrangement of cholesterol in lipid rafts. Additionally, there is expanding literature on the importance of Cav-1 for intracellular cholesterol transport (reviewed in ref. 12). It will be interesting to know whether a lack of Cav-1 imparts any deficiencies in the ability of cells to maintain intracellular cholesterol trafficking and homeostasis.
Interactions of Cav-1 with the other caveolins
Cav-1 is known to have very intricate interactions with its closely related family member, Cav-2. All known terminally differentiated tissues that express Cav-1 also express Cav-2. In addition, after its synthesis in the endoplasmic reticulum (ER), Cav-1 has been shown to either homo-oligomerize or hetero-oligomerize with Cav-2 into a unit of 14 to 16 individual molecules (7, 10). Cav-2 localization is also highly dependent on Cav-1; the Cav-2 protein is retained intracellularly within the Golgi compartment in the absence of Cav-1 expression (13, 14). Down-regulation of Cav-1, however, does not appear to affect Cav-2 levels, as reduction of Cav-1 levels observed in many tumor cell-lines or elicited by anti-sense strategies has no effect on Cav-2 expression (11, 15). Intriguingly, however, in all Cav-1–null tissues examined, the expression of Cav-2 is severely reduced (8, 9). The reduction in Cav-2 levels is not transcriptional, as its mRNA levels remain unperturbed.
Razani et al. went on to dissect these effects in primary mouse embryonic fibroblasts (MEFs), finding that Cav-2 is indeed trapped in the Golgi compartment and that its expression can be rescued — and its localization returned to the plasma membrane — following re-introduction of Cav-1 (8). Furthermore, proteasomal inhibitors (but not lysosomal inhibitors) can restore the expression of Cav-2 to nearly wild-type levels, indicating that in the absence of Cav-1, Cav-2 is most likely mis-folded during transport through the ER/Golgi and is degraded by the proteasomal apparatus.
Cav-1 and cellular proliferation
Based on several lines of evidence, Cav-1 has been considered a candidate tumor suppressor (16). Cav-1 mRNA and protein levels are down-regulated during cell transformation of cultured NIH-3T3 cells, in transgenic mouse models of breast cancer, and in cell lines derived from human tumors (reviewed in ref. 17). Recombinant expression of Cav-1 in transformed NIH-3T3 cells or cell lines derived from human cancers suppresses their transformed phenotype, e.g. anchorage-dependent growth in soft agar is abrogated (17). Targeted downregulation of Cav-1 expression by a vector-based antisense approach promotes anchorage-independent cell growth in soft agar, drives tumorigenesis in nude mice, and hyper-activates the p42/44 MAP kinase cascade in NIH-3T3 cells (11). An analogous gene silencing approach in C. elegans, RNA interference, also yields a phenotype highly reminiscent of hyperactivated Ras signaling (18).
Traditionally, characterization of mice with a targeted deletion of a candidate oncogene or tumor suppressor requires an analysis of the growth properties of primary mouse embryonic fibroblasts (MEFs). Razani et al. therefore used this strategy to assess the effects of Cav-1 on cellular proliferation and the cell cycle (8). Consistent with a role for Cav-1 as a tumor suppressor, Cav-1–null MEFs had more active cell cycle profiles, with increases in the proportion of cells in the S-phase. Phenotypically, this higher activity led to increased cellular proliferation with a nearly 3-fold increase in cell number over a 10-day period. These Cav-1–null active cell cycle profiles could be rescued via re-expression of Cav-1 in the knockout MEFs. In an attempt to pinpoint the molecular components driving this proliferation, Razani et al. looked at the p42/44 MAP kinase cascade, which they found, surprisingly, to be unaffected in the knockout cells (8). Therefore, in primary cells, the loss of Cav-1 predisposes cells to increased proliferation independent of the major pathway previously thought to be affected by Cav-1. Future studies will have to look at the expression of an array of cell cycle proteins and their relative activity in the hope of elucidating the underlying basis for the increased proliferation in Cav-1–null cells.
The altered cell cycle profiles observed in Cav-1–deficient primary cells is important in light of the recent discovery of mutations in the CAV1 gene in a large proportion of invasive human breast carcinomas (19). Although Cav-1–deficient mice do not appear to develop tumors even in old age, they will nonetheless be extremely useful in assessing the exacerbation of cancer phenotypes in the large array of tumor-prone mice currently available.
Cav-1 and endocytosis
Although endocytosis has traditionally been associated with clathrin-coated vesicle transport (20), it has become clear that certain endocytic events exclusively occur via caveolae. Albumin, the cholera and tetanus toxins, certain GPI-anchored proteins, and even some viruses and bacteria seem to target caveolae for internalization (21–23). Razani et al. have tested this assertion by incubating primary MEFs with fluorophore-conjugated albumin. At all time-points tested, Cav-1–null MEFs failed to show surface labeling or internalization of albumin, while transfection of Cav-1 into the null cells restored this uptake (8). Conversely, the uptake of fluorophore-conjugated transferrin, a clathrin-mediated event, was unaffected in both wild-type and knockout cells. These results provide clear evidence for the importance of the caveolae membrane system in at least some endocytic events. As these studies were conducted in primary cultured cells, it will be important to test these ideas in vivo as well.
In an attempt at in vivo analysis of endocytosis, Drab et al. measured the albumin concentration in cerebrospinal fluid (CSF), a condition that has been thought to depend on caveola-mediated transport of albumin from the blood, but found it to be normal (9). Recently however, Lisanti and colleagues have reexamined this question using more direct in vivo assays of endocytosis (24). When Cav-1–null mice were perfused with gold-conjugated albumin and their lungs imaged by transmission electron microscopy, all labeled albumin remained in the blood vessel lumen, suggesting that no endocytosis had occurred. In contrast, gold-conjugated albumin was concentrated and internalized by lung endothelial cell caveolae in wild-type mice. Uptake of radio-iodinated albumin into aortic ring segments, a still more sensitive and quantitative measure, was also undetectable in Cav-1–deficient tissue, whereas wild-type ring segments showed robust uptake that was time- and temperature-dependent and could be competed by unlabeled “cold” albumin. Therefore, it appears that caveolae can act as important portals for the endocytosis of major proteins such as albumin. However, the fact that Cav-1–null mice remain viable suggests that other pathways must at least partially compensate for the absence of caveolae in vivo.
Cav-1 in the lung
Because two of its major cellular constituents, endothelial cells and type I pneumocytes, are rich in Cav-1, the lung possesses an extraordinary abundance of caveolae (7, 25); so the finding of an overt histological abnormality in Cav-1–null lungs is no surprise. Nevertheless, the nature of the pulmonary phenotype of these animals was somewhat unexpected. In Cav-1–deficient mice, the alveolar spaces appear significantly smaller or constricted, with thickened alveolar septa and marked hyper-cellularity (Figure 1b) (8, 9). Staining with the endothelial marker VEGF-R (Flk-1) shows that endothelial cells are significantly increased in number as compared to wild-type animals. Complementary studies with other cellular markers specific to Type I and II pneumocytes, and alveolar macrophages have not yet been reported. However, Razani et al. stained lung sections with Ki67, a marker of cellular proliferation, and found dramatically increased staining in Cav-1–null lungs (8). In light of the hyper-proliferation observed in primary MEFs, these increases in cell number could be due to an aberration in cell cycle checkpoints. However, because no hypercellularity is observed in several other Cav-1–null tissues, alternative explanations of this lung phenotype should be considered. For example, ineffective lung function (due to faulty endothelial cells or pneumocytes) could also impart higher levels of stress to the Cav-1–null lung parenchyma and induce a compensatory increase in cell number.
The histopathologic features of Cav-1–deficient mice are highly reminiscent of a well-known array of disorders. In the so-called restrictive lung diseases, including idiopathic pulmonary fibrosis and acute respiratory distress syndrome, increases in cell number and thickening of the alveolar septae cause dysfunctions of pulmonary gas exchange and overall lung capacity. For this reason, both Razani et al. and Drab et al. have also assessed the lung function of these mice via a swimming test. As expected for animals with a restrictive type disorder, Cav-1–null mice are easily fatigued and markedly exercise intolerant in swimming chambers (8, 9). A more rigorous assessment of lung physiology is required, including an analysis of lung function via whole-body plethysmography and measurements of arterial blood gases.
The two reports also looked at the extracellular matrix composition in Cav-1–null lungs. Although both groups indicated increases in matrix deposition, they did not agree on its composition. The study of Drab et al. noted enhanced fibrosis (in the form of trichrome-stainable collagen) (9), whereas Razani et al. noted enhanced reticulin-staining (indicative of increased basement membrane deposition) with little or no fibrosis (8). It is unlikely that this discrepancy is due to differences in the strains of mice used by the two groups, since both examined F1-generation C57BL/6-129/Svj hybrids. Differences in the interpretation of histological samples may explain the disagreement on this point.
The vascular physiology of Cav-1–deficient mice
A major proposed function of caveolae is the compartmentalization and regulation of signal transduction (26). Biochemical and morphological experiments have shown that a variety of signaling molecules are concentrated within these plasma membrane microdomains and that Cav-1 plays a negative regulatory role in signaling by functioning as a direct inhibitor of these signaling molecules (reviewed in ref. 17). Indeed, there is mounting evidence for the physiological significance of one such functional interaction, that between endothelial nitric oxide synthase (eNOS) and Cav-1. eNOS targets to caveolae (27, 28) in a palmitoylation-dependent manner (28). Interestingly, although localization to caveolae is required for maximal eNOS activity, Cav-1 can inhibit this activity by directly interacting with eNOS (28, 29). Recently, Bucci et al. (30) showed that a cell-permeable peptide harboring the scaffolding domain of Cav-1, a region of the molecule implicated in the inhibition of eNOS signaling, can abrogate nitric oxide-mediated vascular permeability and vasodilatory responses in vivo. These data provided the first functional evidence of a role for Cav-1 in NO-dependent processes and vascular responses.
Following up on this finding, Razani et al. and Drab et al. measured the tension in isolated aortic rings in response to vasoconstrictive and vasodilatory stimuli (8, 9). Both groups found that Cav-1–null aortas exhibit a blunted response to vasoconstrictive agents, such as phenylephrine. More importantly, there was a marked increase in acetylcholine (ACh)-induced vasorelaxation even at extremely low concentrations of ACh (Figure 1c). As ACh acts in the endothelium primarily as an activator of eNOS, these results strongly suggest that the absence of Cav-1 leads to constitutive eNOS activity and to the rapid loss of vascular tone. To further bolster the eNOS hypothesis, Razani et al. also showed that addition of the potent eNOS inhibitor, L-NAME, could restore the tension in Cav-1–null aortic rings to wild-type levels (8). Drab et al., taking another approach, assessed the functionality of the nitric oxide pathway in primary cultured vascular smooth muscle cells and demonstrated a several-fold increase in NO and its downstream mediator, cGMP, in the Cav-1–null cells (9). Extending their studies to include an assessment of general vascular tone in Cav-1–deficient mice, Drab et al. also measured the contractility of isolated arteries in response to various vasoconstrictive agents. As expected, they found a blunted response in the Cav-1–null vessels (9). Taken together, these results show an obvious role for Cav-1 and caveolae in normal vascular responses. In the future it will be important to see if Cav-1–null mice have altered blood pressures either at rest or under stress. As much of this analysis has already been conducted on eNOS transgenic and knockout mice, it will be interesting to see how mice with a deficiency in a modulator of eNOS will behave.
Studies of Cav-3–deficient mice
Cav-3 is most closely related to Cav-1 based on protein sequence homology; Cav-1 and Cav-3 are about 65% identical and about 85% similar. However, Cav-3 mRNA and protein are expressed predominantly in muscle tissue types (heart, diaphragm, skeletal, and smooth muscle) (31). The expression of Cav-3 is induced during the differentiation of skeletal myoblasts in culture and Cav-3 is localized to the muscle cell plasma membrane (sarcolemma).
In order to study the function of Cav-3 in a whole animal model and gain insights into the molecular perturbations that occur in its absence, we and others have recently generated Cav3–/– mice using standard homologous recombination techniques (32, 33). As with Cav-1–deficient mice, these mice are both viable and fertile, with no overt abnormalities, although they lack caveolae in cells that normally express this protein. Although Cav-3 had been thought to be a major factor in caveolar biogenesis, most of the evidence was indirect and relied on structural comparisons with Cav-1. The recent data confirm that a deficiency in Cav-3 protein expression leads to a loss of sarcolemmal caveolae (Figure 1d) (32, 33) and indicate that this protein is required for caveolar biogenesis in skeletal muscle cells in vivo. Table 1 provides a summary of the features of Cav3–/– mice that offer insights into the roles of caveolae in these and other cell types.
Cav-3 and muscular dystrophy
Based purely on its high homology with Cav-1 and specific localization to muscle caveolae, it is tempting to think of Cav-3 simply as a specialized caveolin carrying out the repertoire of Cav-1’s functions selectively in muscle. To a certain extent this has been corroborated, with numerous studies demonstrating the compartmentalization of signaling molecules in muscle caveolae and the ability of Cav-3 to modulate muscle cell behavior (34). However, caveolae and Cav-3 seem to also have specialized roles in muscle tissue, quite distinct from that of Cav-1. In particular, Cav-3 forms a complex with dystrophin and its associated glycoproteins, cytoskeletal components of the caveola-rich muscle cell plasma membrane (31).
A major advance in understanding the physiological role of Cav-3 in muscle came when Minetti and colleagues identified a novel autosomal dominant form of limb-girdle muscular dystrophy (LGMD-1C) caused by mutations within the coding sequence of the human CAV3 gene (3p25) (35). Patients with this disease show several signs of muscle pathology: calf hypertrophy, muscle cramps, and mild-to-moderate proximal muscle weakness. However, histology reveals only moderate myopathic changes. These mutations within the CAV3 gene are responsible for the generation of unstable Cav-3 aggregates, which undergo ubiquitination and proteosomal degradation (36). As such, LGMD-1C mutations lead to a reduction of about 95% in Cav-3 protein expression. Since the initial description of LGMD-1C, several other mutations leading to decreases in Cav-3 levels have also been implicated in other reported cases of muscular dystrophy (37, 38).
The generation of Cav-3–deficient mice marked a perfect opportunity to observe the recapitulation of this type of muscular dystrophy in an animal model. Interestingly, two groups found that the mice show no overt clinical symptoms even into old age (32, 33). In light of the fact that patients with LGMD-1C are mildly symptomatic, this unexpected finding probably indicates that the deleterious effects of human CAV3 dominant negative mutations extend beyond the loss of Cav-3 function. Histological analysis of the skeletal muscle tissue from Cav-3–null mice revealed mild myopathic changes, with variability in the size of muscle fibers, necrotic fibers, and some mononuclear cell infiltration (Figure 1e) (32, 33). Hagiwara et al. (30) also found the highest degree of muscle degeneration in the soleus and gastrocnemius muscles, possibly explaining the calf symptomatology seen in the LGMD-1C patients.
Very recently, Sunada et al. have reported the phenotype of a muscle-specific transgenic mouse expressing a mutation (P104L) found in LGMD-1C patients (39). In contrast to the mild myopathic phenotype observed in the Cav-3–null mice and LGMD-1C patients, these transgenic mice are severely affected both histologically and clinically, and they show poor growth and hind limb paralysis. The discrepancy between this phenotype and the mild dystrophy seen in the knockout mice and in LGMD-1C patients is probably explained by non-specific over-expression toxicity in the muscle tissue.
As several components of the dystrophin complex are concentrated in caveolae and in complex with Cav-3, the studies of Galbiati et al. and Hagiwara et al. also examined possible perturbations in the localization or expression of these proteins. Using immunohistochemical staining of muscle sections, both groups found similar sarcolemmal staining and expression for an array of dystrophin and dystrophin-associated proteins (32, 33). However, via biochemical fractionation of the muscle cell plasma membrane, Galbiati et al. found that dystrophin and several of its associated proteins are excluded from lipid raft microdomains/caveolae in Cav-3–deficient mice (32). Thus, the caveolar localization of the dystrophin complex is probably critical for the maintenance of skeletal muscle structural integrity. In future studies, it will be important to dissect the interactions between components of the dystrophin complex in Cav-3 null mice. In this way, it is hoped that one can elucidate, at a molecular level, alterations in the cytoskeletal architecture of muscle tissue that are characteristic of muscular dystrophy.
Cav-3 and T-tubules
Although Cav-3 is localized to sarcolemmal caveolae in the fully differentiated muscle, it has been shown to be transiently associated with the developing T-tubule system during skeletal muscle maturation (40). Additionally, antisense-mediated downregulation of Cav-3 in a skeletal myoblast cell-line, C2C12 cells, is sufficient to abrogate myoblast fusion and myotube formation (41). Galbiati et al. therefore tested the integrity of the T-tubule system in Cav-3–null mice. Immuno-staining with specific antibodies to two well-characterized T-tubule markers, dihydropyridine receptor-1α and ryanodine receptor, showed diffuse and disorganized labeling, suggestive of abnormalities in the T-tubule system (32). Furthermore, ultrastructural visualization of Cav-3–null skeletal muscle, via ferrocyanate staining, showed dilated and irregularly-oriented T-tubules (Figure 1f). In the future, analysis of primary cultured muscle cells from the developing embryo will be important in assessing the progress of T-tubule formation in the absence of Cav-3. In this way, it should be possible to assess the role of Cav-3 in T-tubule maturation and myoblast differentiation. Indeed, this work might have possible implications in the pathogenesis LGMD-1C in humans. T-tubule abnormalities have also recently been observed in patients with this form of the disorder (42).
Caveolin mutations in human disease
LGMDs are a clinically and genetically heterogeneous group of myopathies and include both autosomal dominant and recessive forms. Because mutations in CAV3 underlie the autosomal dominant LGMD-1C (35), it is interesting to consider the pathogenesis of this disorder in light of the genetic data on caveolins in mice and other organisms. The two best studied human disease mutations in CAV3 cause either a Pro→Leu substitution at amino acid position 104 (P104L) or a deletion of three amino acids at positions 63-65 (ΔTFT) (35). In each case, LGMD-1C patients have a reduction of about 95% in Cav-3 levels. It is now known that these mutants act in a dominant-negative fashion by oligomerizing with wild-type Cav-3 and directing their proteosomal degradation (43). Recently, several reports have identified other dominant-negative mutations in the coding sequence of CAV3 that lead to the protein’s displacement from plasma membrane caveolae or to loss of its expression in muscle tissue. Herrmann et al. identified a missense mutation (A45T) as the culprit in a patient with LGMD-1C (38). Carbone et al. identified a sporadic mutation in Cav-3 (R26Q) in two patients with elevated creatinine kinase levels (hyperCKemia) that leads to a decrease of about 65% in Cav-3 expression (44). Betz et al. identified several of these mutations (R26Q, A45T, P104L) in a group of families with Rippling muscle disease, an autosomal dominant disorder with hyper-irritable muscles (45). Taken together, these finding clearly show that disease-causing mutations in Cav-3 are for the most part in residues that cause a dominant-negative phenotype, one in which the Cav-3 mutant disrupts the normal functioning of the wild-type Cav-3 protein.
Interestingly, most of the disease-causing mutations in CAV3 occur at one of the 12 residues that are conserved over metazoans from worms to humans (Figure 2). One might therefore predict that corresponding mutations in Cav-1 and -2 would have similarly deleterious effects. Indeed, a recent report indicates that the CAV1 gene is mutated in up to 16% of human breast cancer samples examined and that the CAV1 mutation P132L, which is analogous to the CAV3 P104L mutation described above, occurs in these breast tumors (Figure 2). Expression of recombinant Cav-1 harboring the P132L mutation is sufficient to transform NIH-3T3 cells (19). As similar results have been previously obtained using an anti-sense approach to ablate Cav-1 expression (11), it appears that this mutation behaves in a dominant-negative fashion. This first description of a mutation in the CAV1 gene is sure to be followed by the identification of many similar dominant-negative mutations in other human diseases.
Figure 2.
Dominant-negative disease-causing mutations in the human caveolin genes (Cav-1 and Cav-3). Sequence alignment of all the known caveolins indicates that most of the disease-causing mutations found in human caveolin-3 (R26Q, ΔTFT, and P104L; LGMD-1C) and human caveolin-1 (P132L; breast cancer) coincide with one of the twelve residues (demarcated by boxes) that are conserved in all caveolins from worms (C. elegans) to humans. Interestingly, many of these mutations seem to act in a dominant-negative fashion by mis-localizing or causing the degradation of the wild-type caveolin proteins. Note that the mutations, Cav-1 (P132L) and Cav-3 (P104L), are analogous (see asterisk). LGMD-1C, limb-girdle muscular dystrophy, type 1c.
Future directions
The generation and characterization of Cav-1 and -3 deficient mice are important in many respects. At present, we have learned much from these initial phenotypic analyses. These mice manifest some interesting pathophysiological changes, with clearly abnormal lungs and a hypotonic vasculature (in the case of a Cav-1 deficiency) and mild-to-moderately dystrophic muscle tissue (in the case of a Cav-3 deficiency). The generation of these mice also marks the first time caveolar function can be directly assessed in vivo.
Due to the sheer volume of the relevant literature, not all of their possible roles could be assessed in these initial studies. Still, it is intriguing that even in the absence of these prominent membrane entities in such critical sites as the lungs, the heart, and the skeletal musculature, mutant animals can live and function relatively normally. Moreover, some of the most widely anticipated phenotypes in these mice were not immediately realized. Thus, despite Cav-1’s intricate role in cholesterol trafficking and homeostasis and its proposed roles in atherosclerosis, Cav-1–null mice do not appear to have altered serum cholesterol levels (9) (our unpublished observations) or overt signs of atherosclerotic lesions/plaque formation (our unpublished observations). Obviously, more rigorous analyses need to be conducted, including lipid influx/efflux experiments, placement of Cav-1–null mouse cohorts on atherogenic diets, and crossing Cav-1–null mice with hyper-cholestrolemic mouse models. In addition, although Cav-1 suppresses several oncogenic pathways and is clearly down-regulated in numerous tumors, Cav-1–null mice do not present with a higher incidence of carcinomas (8). However, they do show dramatic signs of hyper-proliferation both in vitro and in vivo (8, 9), indicating that full onset tumorigenesis might require the synergistic targeting of Cav-1 and other classically known tumor-suppressors. In this regard, treatment of these mice with carcinogens and/or crossing Cav-1–null mice with certain tumor-prone mice will be an important next step.
In 1994, Lisanti and colleagues proposed that the caveolar compartmentalization of signaling molecules has distinct advantages as it provides a mechanism for the regulation of subsequent signaling events and explains cross-talk between different signaling pathways, an idea known as the “caveolae signaling hypothesis” (26). In direct support of this notion, Cav-1–null mice clearly show hyper-activation of eNOS signaling, as indicated by hypotonic vessel responses (8, 9). This finding provides direct verification of only one of a series of signaling pathways proposed to be regulated by Cav-1 (10). Assessments of the rates, efficiency, and functionality of these and other signaling pathways in the caveolin-null setting will be essential in further establishing the broad physiological relevance of this hypothesis.
It should also be noted that the generation of Cav-1 and Cav-3–null mice does not indicate an absolute deficiency in caveolar formation. In Cav-1–null mice, skeletal and cardiac muscle cells still form caveolae due to unperturbed Cav-3 expression (our unpublished observations); conversely, in Cav-3–null mice, numerous non-muscle tissues and cell types, including endothelial cells, still form caveolae due to unperturbed Cav-1 expression (32). Although there is no known compensatory upregulation by any of the caveolins in these mutant mice, whole-organism ablation of caveolae will come only by inter-crossing Cav-1 and -3–null mice, to generate Cav-1/Cav-3 double knock-out mice. Such a dual-deficiency will also be important in the study of smooth muscle cells and other cell types that co-express Cav-1 and Cav-3. In their initial characterization of Cav-1–null mice, Drab et al. noted that smooth muscle cells totally lacked caveolae (as data not shown) and showed abnormal calcium and eNOS signaling. This finding is difficult to reconcile, given the fact that Cav-3 should still be expressed in this cell type. Perhaps Cav-1 and Cav-3 serve different functions in caveolar biogenesis in smooth muscle cells. Thus, the dual ablation of both genes will be needed to gain a complete understanding of caveolar function in smooth muscle cells and whole animals.
Clearly, the recent reports described above mark the beginning of a renaissance in caveolae research. The thorough characterization of caveolin-deficient mouse models will be extremely important in understanding the molecular mechanisms of certain human diseases. Based on the growing list of phenotypes in these mice, they provide the best available organismal model for the study of caveolar function in relation to human pathophysiology.
Acknowledgments
This work was supported by grants from the National Institutes of Health, the Muscular Dystrophy Association, the American Heart Association, and the Komen Breast Cancer Foundation, as well as a Hirschl/Weil-Caulier Career Scientist Award (all to M.P.L.). B.R. was supported by NIH Medical Scientist Training Grant T32-GM07288.
References
- 1.Yamada E. The fine structure of the gall bladder epithelium of the mouse. J Biophys Biochem Cytol. 1955;1:445–458. doi: 10.1083/jcb.1.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palade GE. Fine structure of blood capillaries. J Appl Phys. 1953;24:1424–1436. [Google Scholar]
- 3.Galbiati F, Razani B, Lisanti MP. Emerging themes in lipid rafts and caveolae. Cell. 2001;106:403–411. doi: 10.1016/s0092-8674(01)00472-x. [DOI] [PubMed] [Google Scholar]
- 4.Rothberg KG, et al. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 5.Scherer PE, et al. Identification, sequence and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci USA. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Z-L. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J Biol Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- 7.Scherer PE, et al. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J Biol Chem. 1997;272:29337–29346. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- 8.Razani B, et al. Caveolin-1 null mice are viable, but show evidence of hyper- proliferative and vascular abnormalities. J Biol Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- 9.Drab M, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 10.Smart EJ, et al. Caveolins, liquid-ordered domains, and signal transduction. Mol Cell Biol. 1999;19:7289–7304. doi: 10.1128/mcb.19.11.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galbiati F, et al. Targeted down-regulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J. 1998;17:6633–6648. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fielding CJ, Fielding PE. Cholesterol and caveolae: structural and functional relationships. Biochim Biophys Acta. 2000;1529:210–222. doi: 10.1016/s1388-1981(00)00150-5. [DOI] [PubMed] [Google Scholar]
- 13.Parolini I, et al. Expression of caveolin-1 is required for the transport of caveolin-2 to the plasma membrane. Retention of caveolin-2 at the level of the Golgi complex. J Biol Chem. 1999;274:25718–25725. doi: 10.1074/jbc.274.36.25718. [DOI] [PubMed] [Google Scholar]
- 14.Mora R, et al. Caveolin-2 localizes to the Golgi complex but redistributes to plasma membrane, caveolae, and rafts when co-expressed with caveolin-1. J Biol Chem. 1999;274:25708–25717. doi: 10.1074/jbc.274.36.25708. [DOI] [PubMed] [Google Scholar]
- 15.Engelman JA, et al. Recombinant expression of caveolin-1 in oncogenically transformed cells abrogates anchorage-independent growth. J Biol Chem. 1997;272:16374–16381. doi: 10.1074/jbc.272.26.16374. [DOI] [PubMed] [Google Scholar]
- 16.Razani B, Schlegel A, Liu J, Lisanti MP. Caveolin-1, a putative tumour suppressor gene. Biochem Soc Trans. 2001;29:494–499. doi: 10.1042/bst0290494. [DOI] [PubMed] [Google Scholar]
- 17.Razani B, Schlegel A, Lisanti MP. Caveolin proteins in signaling, oncogenic transformation and muscular dystrophy. J Cell Sci. 2000;113:2103–2109. doi: 10.1242/jcs.113.12.2103. [DOI] [PubMed] [Google Scholar]
- 18.Scheel J, Srinivasan J, Honnert U, Henske A, Kurzchalia TV. Involvement of caveolin-1 in meiotic cell-cycle progression in Caenorhabditis elegans. Nat Cell Biol. 1999;1:127–129. doi: 10.1038/10100. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi K, et al. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001;61:2361–2364. [PubMed] [Google Scholar]
- 20.Takei K, Haucke V. Clathrin-mediated endocytosis: membrane factors pull the trigger. Trends Cell Biol. 2001;11:385–391. doi: 10.1016/s0962-8924(01)02082-7. [DOI] [PubMed] [Google Scholar]
- 21.Montesano R, Roth J, Robert A, Orci L. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins. Nature. 1982;296:651–653. doi: 10.1038/296651a0. [DOI] [PubMed] [Google Scholar]
- 22.Schnitzer JE, Oh P, Pinney E, Allard J. Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules. J Cell Biol. 1994;127:1217–1232. doi: 10.1083/jcb.127.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin JS, Gao Z, Abraham SN. Involvement of cellular caveolae in bacterial entry into mast cells. Science. 2000;289:785–788. doi: 10.1126/science.289.5480.785. [DOI] [PubMed] [Google Scholar]
- 24.Schubert, W., et al. 2001. Caveolae-deficient endothelial cells show defects in the uptake and transport of albumin in vivo. J. Biol. Chem. In press. [DOI] [PubMed]
- 25.Lisanti MP, et al. Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: implications for human disease. J Cell Biol. 1994;126:111–126. doi: 10.1083/jcb.126.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisanti MP, Scherer P, Tang Z-L, Sargiacomo M. Caveolae, caveolin and caveolin-rich membrane domains: a signalling hypothesis. Trends Cell Biol. 1994;4:231–235. doi: 10.1016/0962-8924(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 27.Shaul PW, et al. Acylation targets endothelial nitric-oxide synthase to plasmalemmal caveolae. J Biol Chem. 1996;271:6518–6522. doi: 10.1074/jbc.271.11.6518. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Cardena G, Fan R, Stern D, Liu J, Sessa WC. Endothelial nitric oxide synthase is regulated by tyrsosine phosphorylation and interacts with caveolin-1. J Biol Chem. 1996;271:27237–27240. doi: 10.1074/jbc.271.44.27237. [DOI] [PubMed] [Google Scholar]
- 29.Michel JB, Feron O, Sacks D, Michel T. Reciprocal regulation of endothelial nitric-oxide synthase by calcium-calmodulin and caveolin. J Biol Chem. 1997;272:15583–15586. doi: 10.1074/jbc.272.25.15583. [DOI] [PubMed] [Google Scholar]
- 30.Bucci M, et al. In vivo delivery of the caveolin-1 scaffolding domain inhibits nitric oxide synthesis and reduces inflammation. Nat Med. 2000;6:1362–1367. doi: 10.1038/82176. [DOI] [PubMed] [Google Scholar]
- 31.Song KS, et al. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J Biol Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- 32.Galbiati F, et al. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and T-tubule abnormalities. J Biol Chem. 2001;276:21425–21433. doi: 10.1074/jbc.M100828200. [DOI] [PubMed] [Google Scholar]
- 33.Hagiwara Y, et al. Caveolin-3 deficiency causes muscle degeneration in mice. Hum Mol Genet. 2000;9:3047–3054. doi: 10.1093/hmg/9.20.3047. [DOI] [PubMed] [Google Scholar]
- 34.Galbiati F, Razani B, Lisanti MP. Caveolae and caveolin-3 in muscular dystrophy. Trends Mol Med. 2001;7:435–441. doi: 10.1016/s1471-4914(01)02105-0. [DOI] [PubMed] [Google Scholar]
- 35.Minetti C, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 36.Galbiati F, Volonte D, Minetti C, Chu JB, Lisanti MP. Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the golgi complex. J Biol Chem. 1999;274:25632–25641. doi: 10.1074/jbc.274.36.25632. [DOI] [PubMed] [Google Scholar]
- 37.McNally EM, et al. Caveolin-3 in muscular dystrophy. Hum Mol Genet. 1998;7:871–877. doi: 10.1093/hmg/7.5.871. [DOI] [PubMed] [Google Scholar]
- 38.Herrmann R, et al. Dissociation of the dystroglycan complex in caveolin-3–deficient limb girdle muscular dystrophy. Hum Mol Genet. 2000;9:2335–2340. doi: 10.1093/oxfordjournals.hmg.a018926. [DOI] [PubMed] [Google Scholar]
- 39.Sunada Y, et al. Transgenic mice expressing mutant caveolin-3 show severe myopathy associated with increased nNOS activity. Hum Mol Genet. 2001;10:173–178. doi: 10.1093/hmg/10.3.173. [DOI] [PubMed] [Google Scholar]
- 40.Parton RG, Way M, Zorzi N, Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galbiati F, Volonte D, Engelman JA, Scherer PE, Lisanti MP. Targeted down-regulation of caveolin-3 is sufficient to inhibit myotube formation in differentiating C2C12 myoblasts. Transient activation of p38 mitogen-activated protein kinase is required for induction of caveolin-3 expression and subsequent myotube formation. J Biol Chem. 1999;274:30315–30321. doi: 10.1074/jbc.274.42.30315. [DOI] [PubMed] [Google Scholar]
- 42.Minetti, C., et al. 2001. Impairment of caveolae formation and T-system disorganization in human muscular dystrophy with caveolin-3 deficiency. Am. J. Pathol. In press. [DOI] [PMC free article] [PubMed]
- 43.Galbiati F, Volonte D, Minetti C, Bregman DB, Lisanti MP. Limb-girdle muscular dystrophy (LGMD-1C) mutants of caveolin-3 undergo ubiquitination and proteasomal degradation. Treatment with proteasomal inhibitors blocks the dominant negative effect of LGMD-1C mutants and rescues wild-type caveolin-3. J Biol Chem. 2000;275:37702–37711. doi: 10.1074/jbc.M006657200. [DOI] [PubMed] [Google Scholar]
- 44.Carbone I, et al. Mutation in the CAV3 gene causes partial caveolin-3 deficiency and hyperCKemia. Neurology. 2000;54:1373–1376. doi: 10.1212/wnl.54.6.1373. [DOI] [PubMed] [Google Scholar]
- 45.Betz RC, et al. Mutations in CAV3 cause mechanical hyperirritability of skeletal muscle in rippling muscle disease. Nat Genet. 2001;28:218–219. doi: 10.1038/90050. [DOI] [PubMed] [Google Scholar]