Abstract
Assembly of a sequence-specific RNA–protein complex on the 3′ untranslated region (3′UTR) of human α-globin mRNA (α-complex) correlates with mRNA stabilization. Here we map a limited segment of the α-globin 3′UTR that is both necessary and sufficient for α-complex formation. The sequence of this binding region identifies three additional, highly stable mRNAs that share closely related, pyrimidine-rich cis-motifs in their respective 3′UTRs. Each mRNA assembles a sequence-specific ribonucleoprotein complex at this conserved region. These complexes are structurally related, and each contains a 39-kDa cytoplasmic poly(C) binding protein previously demonstrated to be essential to formation of the α-complex. These observations indicate the existence of a general determinant for stabilization of eukaryotic mRNAs.
mRNA stability plays an important role in the regulation of eukaryotic gene expression (1). Stabilities of specific eukaryotic mRNAs can vary markedly (2). Short-lived mRNAs, including those encoding cell cycle control factors and protooncogenes (1), have half-lives measured in minutes. Several of these mRNAs carry multiple iterations of AU-rich elements in their 3′ untranslated regions (3′UTR) that appear to contribute to rapid mRNA turnover (3). At the opposite end of the spectrum are very stable mRNAs. These mRNAs tend to encode proteins that accumulate in high levels in terminally differentiated cells. The existence of shared stability determinants for these stable mRNAs has not been explored.
Globin mRNAs provide a model system in which to study the determinants of mRNA stabilization. Our previous studies identified a discontinuous, pyrimidine-rich cis-acting stability element in the 3′UTR of human α-globin mRNA and a corresponding set of cytoplasmic proteins that form a RNA-protein complex (α-complex) at this site (4–6). The assembly of this complex is tightly linked to the long half-life of the α-globin mRNA (6). These studies also characterized a poly(C) binding protein as an essential component of the α-complex (αCP; refs. 6, 7). The protein components of the α-complex are found in a wide spectrum of tissues and cell lines (8), suggesting that they have a broader function than simply the stabilization of human α-globin mRNA. This possibility is tested in the present study.
MATERIALS AND METHODS
Cell Extracts.
Human erythroleukemia cells (K562), human epithelioid carcinoma cells (HeLa), and rat adrenal pheochromocytoma cells (PC-12) were cultured in standard conditions (ref. 9, PC-12), (ref. 6, K562 and HeLa). Preparation of cytosolic extracts (S100) was as previously described (6).
Electrophoretic Mobility-Shift Assays (EMSAs).
RNA oligonucleotides were synthesized by the University of Pennsylvania Nucleic Acid Core Facility and were 5′-end labeled using T4 polynucleotide kinase (NEB, Beverly, MA) and [γ-32P]ATP (Amersham). All labeled oligonucleotides were gel purified on 12% denaturing gels (10). DNA template for synthesis of the α-globin 3′UTR RNA probe was generated by PCR (6). The DNA template for the α-globin 3′UTR lacking the 20-nt binding site (α-Δmin) was constructed by PCR-mediated deletion (11) using inside primers α3 (5′-GCCCGCTGGGCCTTCCTTGCAC) and α4 (5′-CCGGTGCAAGGAAGGCCCAGC) and outside primers described by Wang et al. (6). Internally labeled RNA probes were synthesized by in vitro transcription with T7 polymerase (MAXIScript T7 RNA polymerase kit; Ambion) in the presence of [α-32P]CTP (Amersham). EMSA analysis was carried out as detailed (6). Briefly, for the in vitro transcribed α-3′UTR, 0.005 μg of labeled RNA was mixed with 30 μg of K562 S100 extract in a total volume of 10 μl at room temperature for 30 min followed by the addition of 1 μl of RNase A+T1 mix (1 unit/μl RNase T1, 10 μg/ml RNase A) and incubated for an additional 10 min at room temperature. The ribonucleoprotein (RNP) complex was electrophoresed on a 5% native acrylamide gel. Unlabeled competitor RNAs were synthesized by MEGAScript T7 RNA polymerase system (Ambion). Poly(C) and poly(CT) competitions were carried out by adding 0.1 μg of RNA (Sigma) to the RNP assembly mixture before the addition of the target RNA. For EMSA with synthetic oligos, 0.005 μg of oligonucleotide (≈20,000 cpm) was mixed with 30 μg of K562 S100 extract, and then incubated and gel-analyzed as detailed above with the exception that the RNase A+T1 step was omitted.
Mapping of the α-Globin mRNA-Protected Region.
Excised gel slices containing RNP complexes were placed in elution buffer overnight (0.5 M ammonium acetate/0.1% SDS/1 mM EDTA, pH 8.0/10% methanol) and the harvested RNA was concentrated by ethanol precipitation (10). Oligonucleotide-directed RNase H cleavage was carried out according to standard protocol (12) and resolved on a 12% acrylamide denaturing gel. The antisense DNA oligonucleotide used to target RNase H cleavage at the 3′ end of the mRNA spanned nucleotides 61–75 (5′-GGGCCGGTGCAAGGA-3′). A set of molecular weight markers (Molecular Weight Marker V, Boehringer Mannheim) was used to calculate the fragment sizes.
Isolation and Analysis of RNP Complexes.
Gel slices containing RNP complexes were excised from the EMSA gel under autoradiographic guidance, and the proteins were harvested in modified elution buffer (1% Triton X-100/20 mM Hepes, pH 7.6/1 mM EDTA/100 mM NaCl/2 mM DTT/20 μg/ml BSA/0.1 mM phenylmethylsulfonyl fluoride; ref. 13) for 4 h at 37°C and precipitated with ice-cold 50% acetone. Precipitated proteins were washed once with ice-cold 80% acetone and resuspended in SDS-loading buffer (14). Proteins were separated by SDS/12.5% PAGE (14) and electroblotted onto a nitrocellulose membrane in buffer containing 25 mM Tris, 192 mM glycine, and 15% methanol. Filter blocking and antibody binding were performed as described (15). The polyclonal anti-αCP antibody was raised in chickens (Cocalico Biologicals, Reamstown, PA) against bacterially expressed and gel-purified αCP2 (M. Kiledjian and S.A.L., unpublished data). Antibody was purified from chicken egg yolks as described (16). Primary antibody for Western analysis was used at 1:10,000 dilution followed by secondary antibody (horseradish peroxidase-conjugated goat anti-chicken IgG; Accurate Chemical, Westbury, NY) used at 1:7,000 dilution. Antibody complexes were detected using the ECL system (Amersham).
RESULTS AND DISCUSSION
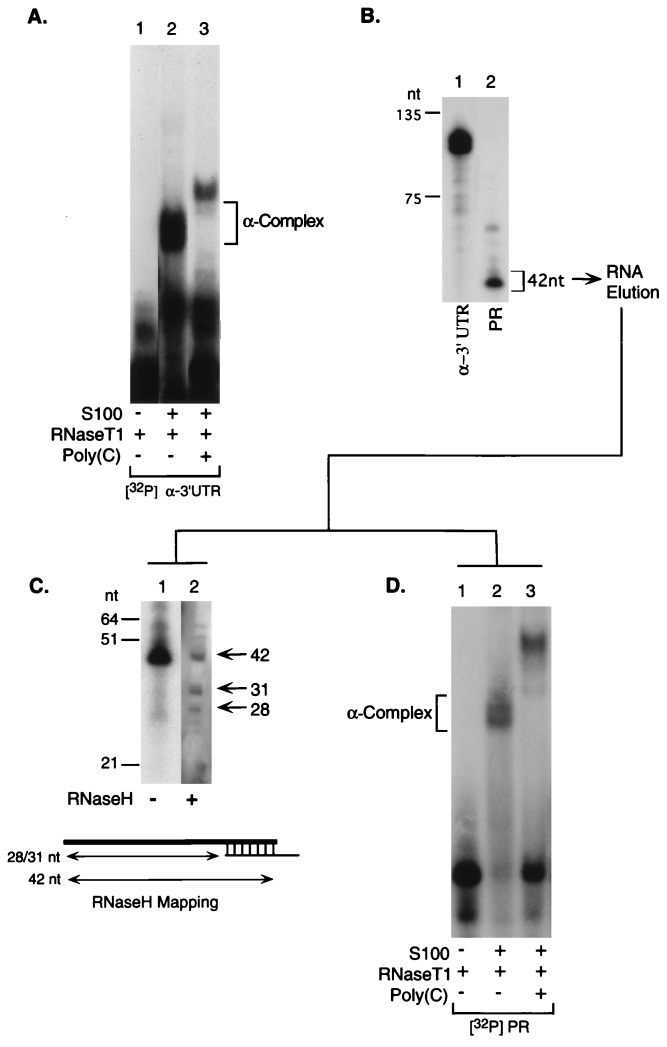
A pyrimidine-rich cis element within the 3′UTR of human α-globin forms a sequence-specific RNP complex that is linked to mRNA stabilization (α-complex; ref. 6). The proteins that constitute the α-complex are highly conserved from mouse to human (6, 7, 17) and are found in a wide spectrum of tissues and cell lines (6, 8). This suggests that the function(s) of the RNP complex containing these proteins may extend beyond stabilization of the erythroid-specific α-globin mRNA. We hypothesized that RNP complexes related to the α-complex might play a general role in mRNA stabilization. To facilitate a search for mRNAs that share stabilization determinants, we first identified a minimal segment within the α-globin 3′UTR sufficient for α-complex assembly. When incubated with S100 extract isolated from a human erythroleukemic cell line (K562), the α-globin 3′UTR forms an RNP complex sensitive to poly(C) competition (α-complex, Fig. 1A; 6). A 42-nt RNA segment is protected from RNase digestion by the α-complex (Fig. 1B). The termini of the 42-nt protected region were determined by oligonucleotide-directed RNase H cleavage (detailed in Fig. 1C; ref. 12). This protected region was found to be sufficient for reassembly of the α-complex (Fig. 1D).
Figure 1.
The human α-complex protects a 42-nt segment of the 3′UTR that is sufficient for α-complex assembly. (A) The α-complex was assembled in vitro and resolved on a nondenaturing polyacrylamide gel. Lane 1, [32P]α-3′UTR digested with RNaseT1; lane 2, [32P]α-3′UTR incubated with S100 extract from human erythroleukemia cells (K562) before RNaseT1 digestion; lane 3, [32P]α-3′UTR incubated with S100 in the presence of poly(C) competitor before RNaseT1 digestion. The position of the α-complex is indicated. The structure of the more slowly migrating alternative complex formed in the presence of the poly(C) competitor, in which the 39-kDa αCP is replaced by a 48-kDa poly(CU)-binding protein, is detailed elsewhere (17). (B) A gel slice containing the α-complex was excised from the gel shown in A, lane 2, the RNA was eluted, electrophoresed on 6% denaturing gel, and autoradiographed. Lane 1, full-length undigested [32P]α-3′UTR; lane 2, RNA protected by the α-complex (42-nt protected region; PR). (C) The 3′ end of the 32P-labeled protected fragment was mapped by site-directed RNaseH cleavage. The protected region mRNA was cleaved at a site specified by a hybridized antisense oligonucleotide, and the span of the resulting fragment were determined. Based on this data the termini of the 42-bp protected region were determined. Lane 1, protected region eluted from B, lane 2; lane 2, RNA segment eluted from B and subjected to site-directed RNase H cleavage. The size marker (nt) is indicated to the left. The diagram depicts the position of the antisense oligonucleotide that targeted the RNase H cleavage and indicates the sizes of the observed bands. (D) The RNA protected region isolated from an excised gel slice (B, lane 2) was incubated with S100 extract and was electrophoresed as in A. Lane 1, the eluted [32P]PR RNA digested with RNase T1 in the absence of S100; lane 2, [32P]PR incubated in S100 before digestion with RNase T1; lane 3, poly(C) competitor added to the S100 extract before incubation with the [32P]PR and RNase digestion.
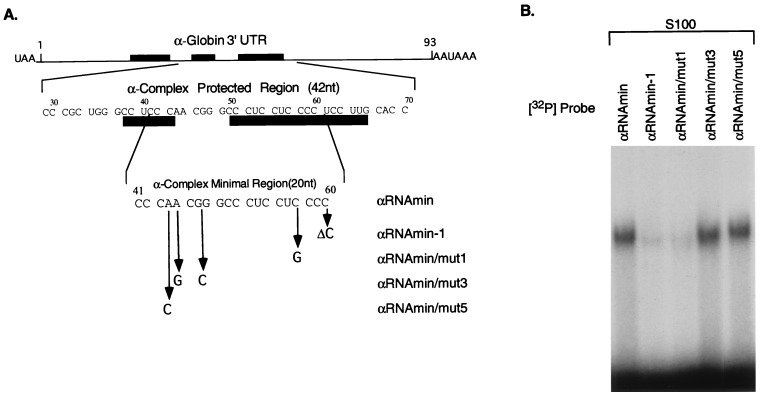
The minimal binding site was mapped in greater detail. A series of synthetic oligonucleotides representing portions of the 42-nt protected region were tested for their abilities to assemble the α-complex (data not shown). A 20-nt segment was capable of forming the α-complex (αRNAmin; Fig. 2A). Deletion of a single base from either the 5′ or 3′ terminus results in a substantial loss of complex formation (αRNAmin−1; Fig. 2 A and B; 5′ deletion not shown). Furthermore, deletion of this 20-nt minimal binding site from the full α-3′UTR results in a complete loss of α-complex formation (result not shown; see αΔmin mutation, below). These data indicate that the 20-nt segment, αRNAmin, is both necessary and sufficient for α-complex formation.
Figure 2.
A 20-nt segment of α-globin 3′UTR is sufficient for α-complex assembly. (A) The diagram represents the α-globin mRNA 3′UTR. The first sequence below the diagram is that of the 42-nt region of the 3′UTR protected from RNase digestion by the α-complex (protected region). The filled rectangles indicate positions of the pyrimidine-rich sequence elements of the α-globin stability complex (6). Numbers above the sequence indicate nucleotide position starting from the first nucleotide after the translational stop signal UAA. The second sequence represents the minimal binding site of the α-complex (αRNAmin) as determined by terminal deletion mapping (data not shown). Mutations in αRNAmin that were tested for their effects on α-complex assembly are indicated below this sequence. (B) RNP complex assembly on the αRNAmin and on four derivative mutant sequences. Gel mobility-shift assays were performed on equal amounts (documented by analysis by denaturing gel electrophoresis; data not shown) of each of the indicated mutant RNA segments (detailed in A) using S100 extract from K562 cells.
Domains within the α-globin 3′UTR region that contribute to mRNA stability have been mapped by linker-scanning mutagenesis (αH mutations; ref. 5). Mutations corresponding to two of these sets of base substitutions that overlay the minimal binding site were introduced into the αRNAmin sequence and tested for their effects on α-complex formation (Fig. 2 A and B). Mutant αRNAmin/mut1 (related to destabilizing mutant αH19) fails to form the α-complex, whereas αRNAmin/mut3 and αRNAmin/mut5 (both related to nondestabilizing mutant αH15) assemble the α-complex with the same efficiency as the wild-type sequence. Thus, the effects of mutations in the minimal binding site on α-complex formation parallel their respective effects on complex formation in the context of the full-length 3′UTR.
The 20-nt αRNAmin overlies two of three polypyrimidine segments with the 3′UTR that are important for α-globin RNA stability (Fig. 2A; ref. 5). Interestingly, mutations that lie outside the αRNAmin and thus do not appear to be involved in the RNP proteins binding also can block complex formation (6). Computer modeling of the α-3′UTR RNA secondary structure using foldrna program (GCG) indicates that these flanking mutations have the potential to alter the predicted secondary structure of α-3′UTR; in the wild-type α-3′UTR, the αRNAmin region is predicted to be single-stranded while inhibitory mutations introduce secondary structure (data not shown). These results indicate that there are two classes of mutations within α-3′UTR that block α-complex formation: those directly affecting binding of RNP proteins (such as mutations within αRNAmin) and those introducing foreign secondary structure(s) into this region, which may hinder access to the RNP binding site.
The complex formed by the minimal binding domain appears to be the same as that formed on the intact 3′UTR, and both complexes are affected in a parallel manner by informative mutations (Fig. 2 A and B). It should be noted that when α-complex formation on the full-length α-globin 3′UTR is blocked by excess of cold poly(C) competitor an alternative complex is formed (Fig. 1A). It has been shown that in this alternative complex the αCP is replaced by poly(CU)-binding protein (6, 17). This alternative complex also assembles on 42-nt RNA segment (protected region, Fig. 1D) but it is not observed when the 20-nt minimal binding site is used (data not shown; also see Fig. 3B). This suggests that while the 20-nt binding site is sufficient to form an αCP-dependent complex, bases outside this region, encompassed by the larger 42-nt protected segment, are needed for binding of the poly(CU)-binding protein-containing alternative complex.
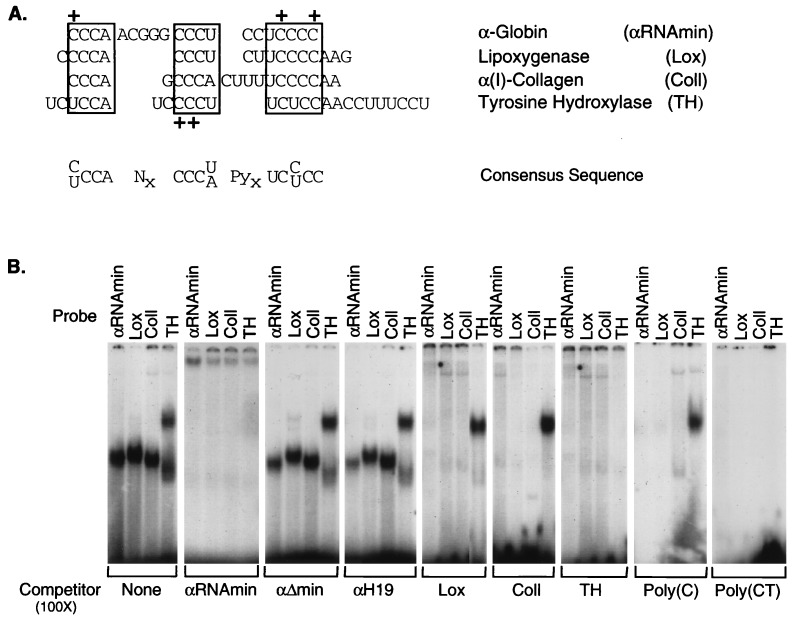
Figure 3.
Structurally related RNP complexes form at pyrimidine-rich segments of 3′UTRs from four stable mRNAs. (A) Sequence alignment of pyrimidine-rich segments within the 3′UTRs of human α-globin (αRNAmin), rabbit 15-lipoxygenase (Lox; ref. 19), human α(I)-collagen (ColI), and rat tyrosine hydroxylase (TH; ref. 18). The sequences shared by all four UTRs are boxed. Gaps are introduced to maximize alignment. A consensus sequence is shown under the sequence alignment. + denotes the positions of mutations within α-globin 3′UTR (ref. 6 and present data) or tyrosine hydroxylase 3′UTR (18) that block RNP complex formation. (B) Gel shift analysis of RNP complex assembly on each of the four 3′UTRs. RNP complexes were assembled and resolved by nondenaturing PAGE as in Fig. 1. The 32P-labeled RNA segment used as a probe in the gel shifts is noted above the lanes, and the unlabeled competitor sequence is noted below each gel. αΔmin and αH19 are two mutant α-3′UTR sequences unable to assemble the α-complex (detailed in text).
We next used the αRNAmin sequence to search the GenBank database (blast program; ref. 20) for related sequence segments in other mRNAs. Only three mRNAs that contain sequences similar to that of αRNAmin were identified: α(I)-collagen, 15-lipoxygenase, and tyrosine hydroxylase (Fig. 3A). Remarkably, sequence homology with all three mRNAs was within their respective 3′UTRs. All three of these mRNAs are highly stable, and their corresponding 3′UTR segments have been noted to be of functional importance (see below). Each 3′UTR contains a pyrimidine-rich cluster conforming to the consensus sequence (C/U)CCANxCCC(U/A)PyxUC(C/U)CC (Fig. 3A). Substitutions of cytosine bases within this segment of the α-globin RNA (Fig. 2, ref. 6) or TH RNA segment (18) block RNP complex formation (summarized in Fig. 3A). These data support the hypothesis that sequences within or immediately adjacent to these clusters are important for RNA-protein interactions.
The shared pyrimidine-rich sequences suggest that these four 3′UTR segments might subserve a common function. The effect of the cis-element in the α-globin 3′UTR on mRNA stability in vivo correlates with its ability to assemble the α-complex in vitro (6). Therefore, each of the three additional pyrimidine-rich sequences was tested for their ability to bind proteins. All four RNA segments form a sequence-specific RNP complex (Fig. 3B). Each complex is self-competed by its respective 3′UTR segment and can be crosscompeted by each of the other RNAs. In addition, formation of each complex is blocked by poly(C) or poly(CT) competitor, but not by poly(A), poly(U), or poly(G). This pattern of competition has been previously demonstrated for the α-globin (6) and the tyrosine hydroxylase RNP complexes (9). In contrast, complex formation is not competed by a mutant α-globin 3′UTR that is itself unable to form the α-complex (αH19, ref. 5), nor by the α-globin 3′UTR from which the 20-nt minimal binding site has been deleted (α-Δmin). These results suggest that the α-globin, α(I)-collagen, 15-lipoxygenase, and tyrosine hydroxylase 3′UTRs form sequence-specific RNP complexes that share one or more protein components.
Unlike the other three mRNA segments, the tyrosine hydroxylase sequence forms two complexes. The appearance of the upper band is distinct from the others by the criterion of the competition studies (Fig. 3B). In addition, its intensity shows a high degree of interexperimental variation (data not shown). Since the work of Czyzyk-Krzeska et al. (9) identified a single poly(C)-sensitive tyrosine hydroxylase complex in hypoxia-induced cell extracts, the appearance of the alternative (upper) complex may reflect a shift in the protein composition of extracts isolated at different physiological states.
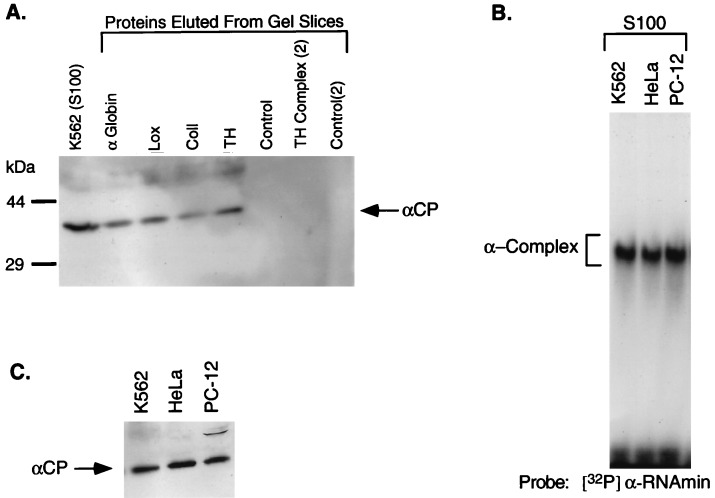
It has been previously demonstrated that the poly(C) binding activity in the α-complex is encoded by two closely related 39-kDa proteins, αCP1 and αCP2 (7). Before their structural characterization, these two proteins were referred to by the general term “poly(C)-binding proteins” (6). The αCP1 and αCP2 proteins may be interchangeable in the formation of the α-complex. The poly(C) sensitivity of all four RNP complexes (Fig. 3B) suggests that the αCP proteins might be a common component. To test this possibility, RNP complexes were resolved on a nondenaturing polyacrylamide gel, and proteins were eluted from the appropriate gel slice, resolved by SDS/PAGE, and analyzed by Western blotting using a polyclonal anti-αCP antiserum. Gel slices from corresponding regions of a control lane (S100 without added added RNA) were analyzed in parallel. αCP was detected in each of the four RNP complexes (Fig. 4A). The absence of this protein from the control lanes suggests that it is specifically recruited to this region of the gel by its incorporation into each RNP complex. These results demonstrate that one or both of the αCP proteins are involved in formation of the α-globin, α(I)-collagen, 15-lipoxygenase, and tyrosine hydroxylase 3′UTR RNP complexes.
Figure 4.
αCP is a component of the RNP complexes formed on 3′UTR segments of each of four stable mRNAs. (A) RNP complexes were formed on each of the four 3′UTR segments in vitro, resolved by nondenaturing PAGE (as in Fig. 3B), and gel slices containing each of the RNP complexes were then excised. Proteins were eluted from each gel slice, concentrated by acetone precipitation, resolved by SDS/12.5% PAGE, and electrotransferred to a nitrocellulose membrane. The blot was incubated with a chicken anti-αCP antibody, and immune complexes were visualized with a secondary antibody using ECL (Amersham). The identity of each sample is shown above the lanes. TH complex (2) corresponds to the more slowly migrating of the two tyrosine hydroxylase complexes. As controls, proteins were eluted from positions on a lane containing S100 extract (no added RNA) corresponding to each RNP complex (Control) as well as from a region of the lane corresponding to the upper TH RNP complex [Control (2)]. (B) RNP complexes were formed on the α-globin 3′UTR segment (αRNAmin) with S100 extracts from three different cell types: K562, HeLa, and PC-12. The resultant complexes were electrophoresed as in Fig. 1. The identity of each S100 extract is indicated above the lanes. (C) Western blot analysis of S100 extracts from K562, HeLa, and PC-12 cells. The immunoreactive αCP is indicated.
The observation that α-globin, tyrosine hydroxylase, 15-lipoxygenase, and α(I)-collagen mRNAs have distinct patterns of tissue specific expression suggests that the distribution of the entire set of protein components of the four RNP complexes has a comparably wide distribution. Consistent with this observation we find that S100 extracts from rat PC-12 pheochromocytoma cells, human K562 leukemic erythroblasts, and HeLa cells all form identically migrating RNP complexes on each of the four 3′UTR segments (Fig. 4B and data not shown). In addition, αCP can be directly demonstrated in the S100 extracts of all three cell types (Fig. 4C). Therefore, the protein constituents of RNP complexes that form on each of the 3′UTRs are conserved across species (rat to human) as well as being expressed in a variety of cell types.
The mRNAs analyzed in this study all have been previously characterized by their stability. α-Globin and 15-lipoxygenase mRNAs both are expressed during erythroid cell differentiation (19, 21). The high-level stability of α-globin mRNA is well established and is essential to hemoglobin synthesis (22). Although the 3′UTR of rabbit 15-lipoxygenase mRNA has been most carefully studied for its contribution to translational repression (21), this mRNA is maintained in stable form for several days posttranscription before translational activation in reticulocytes (17, 23). The 5′-proximal portion of the 15-lipoxygenase 3′UTR contains 10 repeats of a 19-nt pyrimidine-rich repeat that forms a poly(C)-sensitive RNP complex (LOX-BP; ref. 19). A single representative 19-nt repeat appears to be sufficient for assembly of this complex (Fig. 3B). It is possible that this complex may mediate stability as well as control translational activity of the 15-lipoxygenase mRNA. The stability of tyrosine hydroxylase mRNA, which encodes the rate-limiting enzyme in catacholamine synthesis, was characterized during oxygen starvation of rat adrenal pheochromocytoma PC-12 cells (9, 18). A 27-nt pyrimidine-rich sequence within the 3′UTR of tyrosine hydroxylase serves as a binding site of a hypoxia-inducible RNP complex; the activity of complex formation increases in parallel with the hypoxia-induced increase in mRNA stability from a half-life of 10 to 30 h (18). This complex is poly(C) sensitive and can be inhibited by mutations within the pyrimidine-rich element. α(I)-collagen mRNA appears to be stable in a variety of cell types (24). This property can be further accentuated in response to pathophysiologic stimuli and the extended half-life is paralleled by induction of RNP complex formation at the respective 3′UTR pyrimidine-rich element (David Brenner, personal communication). Thus the four mRNAs characterized in this report form related RNP complexes in their 3′UTRs, yet demonstrate unique aspects of their stability phenotypes and may contain distinct subsets of proteins.
In conclusion, we demonstrate that four mRNAs characterized by their high stabilities each assemble a 3′UTR RNP complex containing shared cis- and trans-components. Cis-acting polypyrimidine clusters have been either directly or indirectly implicated in the stabilization of these mRNAs; the trans-acting protein constituents of these complexes include a shared 39-kDa poly(C)-binding protein, αCP, that is abundant and ubiquitous. The RNP complex, or set of related RNP complexes described in this report, therefore may constitute a general determinant of mRNA stabilization.
Acknowledgments
We thank Mike Kiledjian for his efforts in generating the anti-CP antiserum, David Brenner for sharing his unpublished data, and Rabah Iratni, Francisco Velazquez, and members of the Liebhaber laboratory for helpful discussions. This work was supported in part by a grant from the National Institutes of Health (HL38632). S.A.L. is an Investigator in the Howard Hughes Medical Institute.
ABBREVIATION
- 3′UTR
3′ untranslated region
- RNP
ribonucleoprotein
- EMSA
electrophoretic mobility-shift assay
References
- 1.Ross J. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belasco J. In: Control of mRNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. [Google Scholar]
- 3.Chen C-Y A, Shyu A-B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 4.Weiss I M, Liebhaber S A. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss I M, Liebhaber S A. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Mol Cell Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiledjian M, Wang X, Liebhaber S A. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aasheim H C, Loukianova T, Deggerdal A, Smeland E B. Nucleic Acids Res. 1994;22:959–964. doi: 10.1093/nar/22.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czyzyk-Krzeska M F, Dominski Z, Kole R, Millhorn D E. J Biol Chem. 1994;269:9940–9945. [PubMed] [Google Scholar]
- 10.Maxam A M, Gilbert W. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higuchi R. In: PCR Protocols: A Guide to Methods and Applications. Innis M, Gelfand D, Sninsky J, White T, editors. San Diego: Academic; 1990. [Google Scholar]
- 12.Keller W, Crouch R. Proc Natl Acad Sci USA. 1972;69:3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ossipow V, Laemmli U K, Schibler U. Nucleic Acids Res. 1993;21:6040–6041. doi: 10.1093/nar/21.25.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hames B D. In: Gel Electrophoresis of Proteins: A Practical Approach. Hames B D, Rickwood D, editors. Oxford: IRL; 1981. [Google Scholar]
- 15.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 16.McLaren R D, Prosser C G, Grieve R C J, Borissenko M J. Immunol Methods. 1994;177:175–184. doi: 10.1016/0022-1759(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Liebhaber S A. EMBO J. 1996;15:5040–5051. [PMC free article] [PubMed] [Google Scholar]
- 18.Czyzyk-Krzeska M F, Beresh J E. J Biol Chem. 1996;271:3293–3299. doi: 10.1074/jbc.271.6.3293. [DOI] [PubMed] [Google Scholar]
- 19.Ostareck-Lederer A, Ostareck D H, Standart N, Thiele B J. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Meyers W W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Lodish H F, Small B. Cell. 1976;7:59–69. doi: 10.1016/0092-8674(76)90255-5. [DOI] [PubMed] [Google Scholar]
- 22.Liebhaber S A, Wang X, Kiledjian M, Weiss I. In: Molecular Biology of Hemoglobin Switching. Stamatoyannopoulis G, editor. Vol. 1. Andover U.K.: Intercept; 1995. pp. 375–389. [Google Scholar]
- 23.Hohne M, Thiele B-J, Prehn S, Giessmann E, Nack B, Rapoport S M. Biomed Biochim Acta. 1988;47:75–78. [PubMed] [Google Scholar]
- 24.Maatta A, Ekholm E, Penttinen R P. Biochim Biophys Acta. 1995;1260:294–300. doi: 10.1016/0167-4781(94)00207-j. [DOI] [PubMed] [Google Scholar]