Abstract
Pax-6 in vertebrates and its homolog eyeless in Drosophila are known to be essential for eye development. Here we investigate the role of Pax-6 in eye development in another major systematic group, molluscs. We demonstrate that alternatively spliced RNAs derived from a single Pax-6 gene in the squid (Loligo opalescens) are expressed in the embryonic eye, olfactory organ, brain, and arms. Despite significant sequence differences between squid Pax-6 and Drosophila eyeless in the region outside the paired- and homeodomains, squid Pax-6 is able to induce the formation of ectopic eyes in Drosophila. Our results support the idea that Pax-6 related genes are necessary for eye and olfactory system formation throughout the animal kingdom.
Keywords: Pax-6, olfactory organ, evolution mollusc, crystallin
Eyes of very diverse type and structure ranging from simple light-sensitive receptors to complex image-forming eyes can be found in the animal kingdom (1–3). Most of the major animal groups comprise species with a simple eye spot. A more elaborate optical system can be found in only six of the animal phyla; these, however, contribute about 96% of the known species (2). Different explanations for the diversity of eyes have been proposed. The morphological differences of the various eyes have been considered as evidence that they did not share a common ancestor and thus are polyphyletic in origin. Indeed, it has been estimated that photoreceptors may have evolved independently 40–60 times (1). An alternative view suggesting a common evolutionary origin of the various eye types has also been proposed (4). Recent data based on the demonstration that the paired domain/homeodomain transcription factor, Pax-6/eyeless, has a critical role in eye development in vertebrates (5–10) and Drosophila (11) support the idea of a monophyletic origin of the eyes.
Heterozygous mutations in Pax-6 of vertebrates are associated with a variety of eye diseases, including aniridia in human and Small eye (Sey) in rodents (5, 6, 8). In homozygotes, Pax-6 mutations are lethal due to a complete absence of eyes and nose and severe defects in brain formation (see ref. 12 for a review). The curtailing of normal eyeless expression in the eye primordia of Drosophila leads to a reduction or complete absence of the compound eyes (11, 13). Targeted expression of Drosophila eyeless or mouse Pax-6 in various imaginal disc primordia of Drosophila results in supernumerary eyes (14). On the basis of these results it was proposed that eyeless/Pax-6 is the master control gene for eye morphogenesis (11, 14, 15). Taken together these data suggest that two types of image-forming eyes, complex eyes of vertebrates and compound eyes of arthropods, share—at least partially—developmental pathways. Furthermore, this suggests that the last common ancestor of these organisms at the protostome–deuterostome divergence possessed eyes in which a Pax-6 gene was already active (16).
Molluscs represent a third phylum in which highly complex eyes are present. Cephalopod molluscs (squid, octopus, cuttlefish) possess a well developed nervous system and are highly intelligent (17). The complex eyes of cephalopod molluscs and vertebrates have been considered a classical example of convergent evolution (18). The eyes in these two systematic groups are remarkably similar in general appearance and organization but they are formed by different mechanisms during development and differ in many details.
A possible strategy used to evolve complex image-forming eyes from the primitive eyes present in the last common ancestor is the use of similar developmental mechanisms with the same or closely related transcription factors. If this assumption is correct, one would expect that in cephalopod molluscs a Pax-6 homolog is involved in visual system development as it is in Drosophila and vertebrates. In this report, we present evidence corroborating this prediction by way of the structural and functional characterization of a Pax-6 homolog of the squid, Loligo opalescens.
MATERIALS AND METHODS
Squid Collection.
Squids (L. opalescens) were collected at the Hopkins Marine Station, Stanford University (Pacific Grove, CA). For in situ hybridization, embryos were fixed overnight at 4°C in 3.7% formaldehyde, 0.1 M Mops (pH 7.2), 2 mM EGTA, and 1 mM MgSO4, rinsed once in 0.9% NaCl, transferred to 90% methanol, and stored at −20°C before hybridization. Embryos used for RNA isolation were frozen at −70°C.
RNA Isolation and RNA Blot Analysis.
Total RNA from indicated tissues was isolated by the acidic guanidinium thiocyanate-phenol-chloroform extraction method (RNazol B; Tel-Test, Friendswood, TX). Poly(A)+ RNA was purified from total RNA by using the Dynabeads mRNA purification kit (Dynal, Oslo). For Northern blot experiments, about 2 μg of poly(A)+ RNA from the indicated tissues were separated by agarose gel electrophoresis, transferred to Duralon UV membrane (Stratagene), and hybridized with 32P-labeled PCR fragment (positions 70–361) in QuickHyb as recommended by the manufacturer (Stratagene). Squid β-actin PCR probe was used as a control.
Cloning and Characterization of the Squid Pax-6 cDNA.
An initial Pax-6 PCR fragment was obtained with primers 5′-CCGCTCGAGGGITG(T/C)GTITC(G/A/T/C)AA-3′ (5′ oligonucleotide corresponding to the conserved GCVSK sequence in the paired domain, positions 210–223 in Fig. 1) and 5′-GTATCTAGAGTC(A/C/G/T)CG(A/G/T)AT(T/C)TCCCA-3′ (3′ oligonucleotide corresponding to the WEIRD sequence in the paired domain, positions 357–371 in Fig. 1) using genomic DNA of the squid Loligo vulgaris as a template. The 5′ and 3′ primers had XhoI and XbaI restriction sites, respectively, which are underlined. The PCR was conducted in two steps: 5 cycles at low stringency with ramping (1 min at 94°C, in 1 min to 37°C, 1 min at 37°C, in 2.5 min to 72°C, 1 min at 72°C, and in 1 min to 94°C) and then 30 cycles without ramping (30 sec at 94°C, 30 sec at 50°C, 30 sec at 72°C) with final extension at 72°C for 2 min. A 162-bp fragment of the Pax-6 gene, identified by low stringency Southern hybridization using a mouse Pax-6 cDNA as a probe, was isolated, cloned in Bluescript KS+, and sequenced. This genomic fragment was used to screen a squid Ommastrephes sloani pacificus genomic library (5 × 105 plaques) made in λEMBL3 (19). Four independent plaques were isolated. A 5-kb EcoRI restriction fragment hybridizing with a Pax-6-specific probe was identified, cloned, and sequenced. It contained an exon sequence (347 bp) encoding part of the paired domain (see Fig. 1) and its flanking intron sequences. The full-length Pax-6 coding sequence was obtained by PCR using primers 5′-AAGATTCTCGGACGGTACTATGA-3′ (5′ primer, positions 222–244 in Fig. 1) and 5′-CCATTT(C/T)GCTC(G/T)TC(G/T)GTT(A/T)GA(A/G)AACCA-3′ (3′primer, positions 873–899 in Fig. 1) and then 5′- and 3′-rapid amplification of cDNA ends (RACE) (BRL/GIBCO kits) using squid L. opalescens embryonic cDNA as a template. The cDNA for RACE reactions was synthesized using poly(A)+ RNA isolated from squid embryos as a template. cDNA2 was obtained in 5′ RACE reaction using primers 5′-CTCATAGTACCGTCCGAGAATCTT-3′ (positions 222–245) and 5′-GAGTCGAGTCTGGAAGGGGA-3′ (positions 116–135).
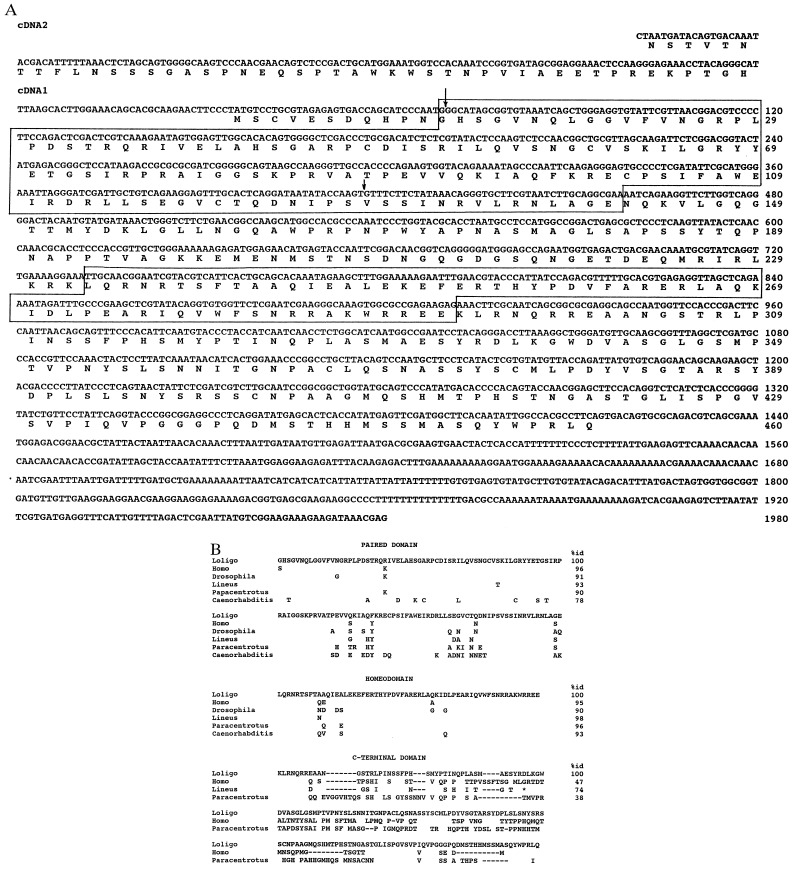
Figure 1.
(A) Nucleotide and deduced amino acid sequences of the squid L. opalescens embryonic Pax-6 cDNA. The paired domain and homeodomain are boxed. Two arrows indicate the positions of two known introns in the L. opalescens Pax-6 gene. (B) Comparison of the amino acid sequences between paired-, homeo- and C-terminal domains of vertebrate and invertebrate species. The squid sequence is shown in full; for other sequences only differing amino acids are shown. A - indicates gaps that were introduced to maximize similarities in the C-terminal domain; an * marks the end of the known nemertine sequence. The percent identities are shown for the squid sequence.
In Situ Hybridization.
Whole mount in situ hybridization was conducted as described (20). Digoxigenin labeling of the probes and detection were conducted using the Genius RNA labeling and detection kit (Boehringer Mannheim). Specimens were photographed on Kodak Ektachrome 160 film with a Nikon SMZ-U microscope. Whole mount hybridized embryos were embedded in JB-4 glycol methacrylate resin and sectioned with a glass knife. Sections were photographed on Kodak Ektachrome film with a Zeiss Axiophot microscope.
Ectopic Expression of Squid Pax-6 cDNA in Drosophila.
The full-length squid Pax-6 cDNA1 (positions 2–1510 in Fig. 1) was first cloned as a NotI fragment in the Bluescript KS+ plasmid and then recloned in the GAL-UAS plasmid, pUAST (21). Orientation of the insert was confirmed by restriction mapping, and flies were transformed as described (14). Ectopic induction of squid Pax-6 was accomplished by crossing the UAS-Pax-6 transformants to the MS1096, dppblink, and E132 GAL4 expressing lines, and subsequent analysis of the ectopic eyes was performed as described (14).
RESULTS
Isolation and Characterization of Squid Pax-6 cDNAs.
A squid Pax-6 cDNA was cloned using a PCR approach (see Materials and Methods, Fig. 1). There is a single Pax-6 gene in the squid genome as judged by Southern blot hybridization with several probes corresponding to different parts of the squid Pax-6 cDNA (data not shown). By 5′-RACE reactions, we identified two variants of the squid Pax-6 cDNA differing by their 5′ exon(s), as was previously observed for the quail Pax-6 and Drosophila eyeless cDNAs (11, 22). The squid cDNA1 has an ORF encoding 460 amino acids and contains a potential initiator methionine codon 11 codons upstream of the encoded paired domain. The deduced protein has a molecular mass of 50,568 Da. cDNA2 does not encode methionine residues within a stretch specifying 44 amino acid residues upstream of the paired domain, which also lacks methionine. Although we do not know at present whether there is an initiator methionine codon further upstream in cDNA2, it could be that cDNA2 encodes a variant Pax-6 protein containing a homeodomain but not a paired domain as was observed previously for quail and Caenorhabditis elegans Pax-6 (22–25).
Comparison of squid Pax-6 with that from other species (refs. 7, 11, 22, and 24–27; see Fig. 1B) confirmed the high degree of conservation of this protein. The highest overall deduced amino acid sequence identities between Pax-6 of the squid and that of other species is 78% with a nemertine, 67% with vertebrates, and 63% with a sea urchin. In the region of the paired domain, squid Pax-6 shows 91–95% identity with its vertebrate, Drosophila, nemertine, and sea urchin homologs, and 78% identity with C. elegans Pax-6-related proteins; the homeodomain of squid Pax-6 shows 90–98% identities with Pax-6 from these different species. Sequences C− terminal of the homeodomain are also conserved but to a lesser extent with amino acid sequence identities of 74%, 47% and 38% between Pax-6 of squid and nemertine, human, and sea urchin, respectively. This region has been called the PST domain and shown to possess a potent transactivator function (26, 28, 29). The squid Pax-6 linker region between the paired and homeodomain shows 51%, 42%, and 36% identity with nemertine, sea urchin, and human sequences, respectively. Regions upstream of the paired domain show no similarities at all.
At present the positions of only two introns have been identified in the squid Pax-6 gene; one is located in codon 1 and the other between codons 116 and 117 within the region encoding the paired domain (Fig. 1). They coincide exactly with the positions of introns in the Pax-6 gene of vertebrates, nemertine, Drosophila, and C. elegans (7, 11, 22, 24, 25, 27). The second intron found in the paired box of vertebrates, which is used for alternative splicing of an additional small exon (28), is missing in the squid as in all other invertebrate species analyzed so far. In Drosophila and C. elegans additional splice sites have been found in the 3′ part of the paired domain that are absent in squid and nemertine.
Squid Pax-6 Transcripts.
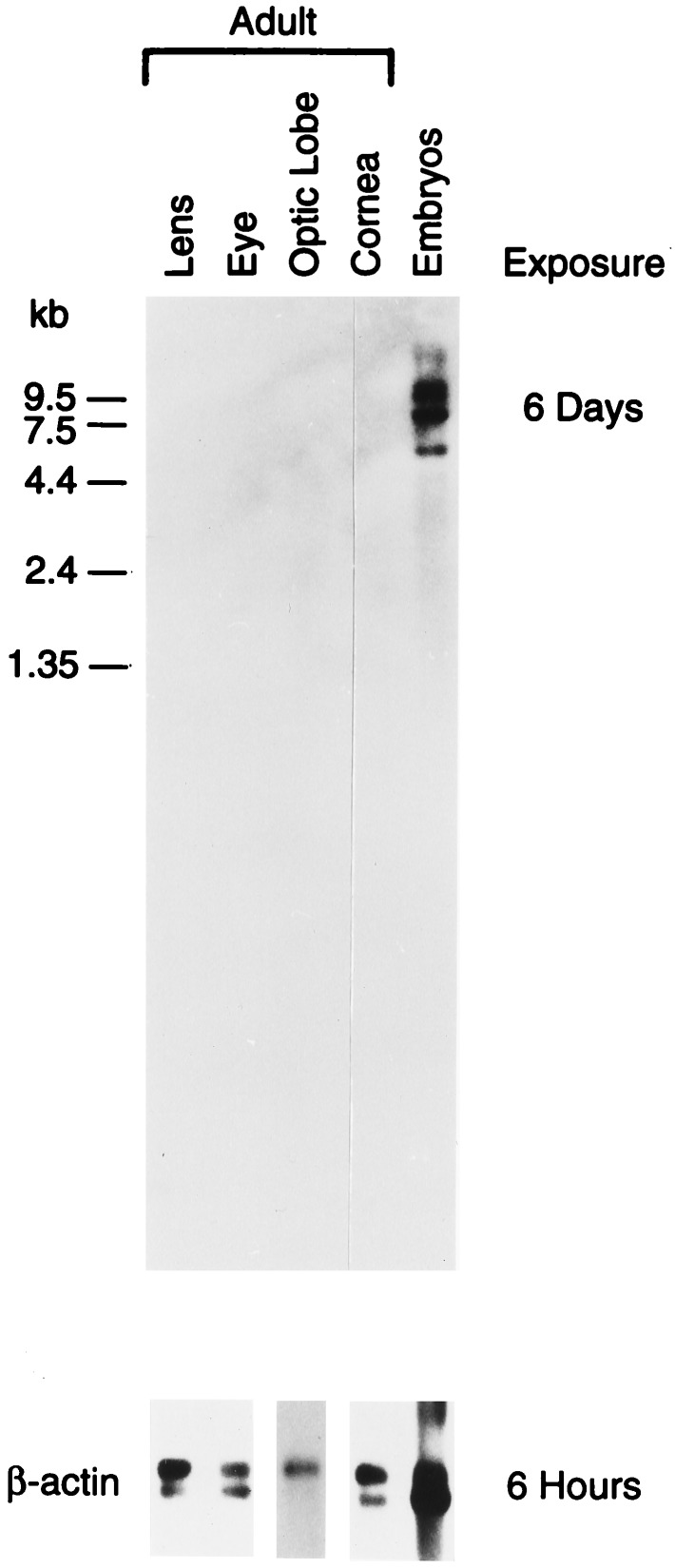
To estimate the size of the squid Pax-6 mRNA and investigate its expression profile, we conducted Northern blot hybridization tests using available squid adult tissues and embryos (Fig. 2). No hybridization signal was observed in the RNAs from adult tissues. However, a low level of expression in adult optic lobe and cornea was demonstrated by PCR with Pax-6-specific primers (data not shown). In contrast to the results obtained with adult tissues, four bands of hybridization indicating Pax-6 RNAs with a length of ≈5.6, 8, 11, and 12 kb were detected in the squid embryonic material. Because there appears to be only one Pax-6 gene in the squid, these data suggest that the squid Pax-6 primary transcript is alternatively spliced. The longest available cDNAs (1980 and 2000 nt) are much shorter than the smallest mRNA, suggesting that significant parts of 5′- and/or 3′-untranslated regions are missing from our clones. The squid Pax-6 mRNAs are longer than their 2.7–3 kb homologs in vertebrates (7). The sea urchin Pax-6 mRNA is at least 5 kb long judging from the cDNA sequencing data (26).
Figure 2.
Northern blot analysis of the squid Pax-6 gene in different adult tissues and embryos.
Developmental Expression Pattern of Squid Pax-6.
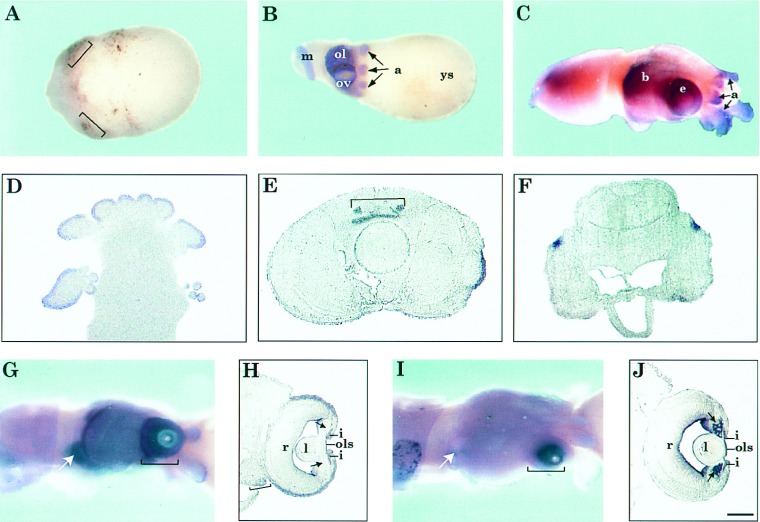
To investigate Pax-6 expression at early stages of squid development and obtain more detailed information on the localization of Pax-6 mRNA in the embryos, we conducted whole mount in situ hybridization experiments. We could not detect expression of Pax-6 at Arnold stage 15 (Naef stage VI) and earlier (not shown) (17, 30, 31). The first traces of Pax-6 expression were detected at Arnold stages 16–17 (Naef stages VII–VIII) in the region of the rudimentary eye primordia (Fig. 3A). At these early developmental stages the squid embryos consist of three components: two layers of cells (ectoderm and mes-endoderm) comprising the actual embryo, an inner syncytial epithelium (transitory formation, referred to as “yolk syncytium”), and a central mass of yolk (32). The retina anlage becomes evident at Arnold stage 17 (Naef stages VII–VIII) but it is still not enclosed by the ectodermal fold. At Arnold stages 19–20 (Naef stages VIII–IX), when the optic vesicle is formed by internalization of the eye placode, Pax-6 expression was increased and clearly seen in the regions of the developing eyes, optic lobe, arms, and mantle (Fig. 3B). The expression of Pax-6 in the arms and mantle of the squid embryo indicates that this transcription factor has a role in the development of these tissues, an observation that deserves further study. Differentiation of the lens primordia and formation of the iris fold begin after Arnold stages 19–20. At Arnold stages 26–28 (Naef stages XIV–XVIII), Pax-6 expression was detected predominantly in the eyes, chemosensory olfactory organ (33–35), arms, and brain (Fig. 3 C and G). In the olfactory organ, arms, and suckers, Pax-6 was expressed mainly in the outer layers (Fig. 3D) that are rich in nerve endings, while in the brain expression was detected mainly in the cerebral ganglion (Fig. 3 E and F). In the eye, Pax-6 expression was detected at the surface of the eye, the developing iris and the anterior part of the anterior lens segment (Fig. 3 G and H). We could not detect any expression in the inner lentigenic cells that give rise to the lens posterior segment or in the retina. Pax-6 mRNA is also present in the fold of tissue posterior to the eye (see bracket, Fig. 3H) which will move forward in development to surround the eyeball and form the cornea (17). Pax-6 mRNA staining is not yet seen on the developing corneal fold above the eye in this section. To eliminate the possibility of poor probe penetration we removed the eye, cut it open and hybridized it as whole mount. The pattern of hybridization was indistinguishable from that obtained with intact embryos (data not shown). Whole mount hybridization using two mixed antisense probes from S-crystallins (Lops12 and Lops7; refs. 36 and 37) that belong to the major squid lens and cornea-specific proteins and are synthesized in the lentigenic cells of the embryos (38), showed as expected expression in the inner and outer lentigenic cells (Fig. 3 I and J).
Figure 3.
Expression of Pax-6 during L. opalescens embryonic development. (A–C, G, and I) Whole mount in situ hybridization of squid embryos using Pax-6 (A–C and G) and S-crystallin (I) antisense riboprobes. Sense probes were used as controls (data not shown). (D–F, H, and J) Frontal, 10-μm plastic sections of embryos hybridized as whole mount with Pax-6 (D–F and H) and S-crystallin (J) antisense probes (20). (A) Stage 17 embryo. Brackets indicate the eye primordia where first traces of Pax-6 expression are detected. (B) Stage 20 embryo. Arrows point to the arms. (C) Stage 27 embryo. The yolk sac was removed. Arrows point to the arms. (D–F) Sections of stage 27 embryo. (D) Section through the arms and suckers. (E) Section through the brain adjacent to the posterior edge of the eyes. Bracket indicates the cerebral ganglion. (F) Section through the posterior end of the brain. (G) Stage 27 embryo. Bracket indicates eye and arrow indicates olfactory organ, where expression of Pax-6 is detected. (H) Section through the eye of an embryo as in G. Brackets indicate areas from which the cornea will develop as a fold from the edge of forward growing arms and arrows point to lentigenic cells which do not express Pax-6. (I) Stage 27 embryo, hybridized with S-crystallin probe. Bracket indicates eye where expression of S-crystallins is detected. Note that there is no expression in the olfactory organ (arrow). (J) Section through the eye of an embryo as in I. Brackets and arrows are as in H. Label at the inner edge of the eye chamber is background staining also observed with the sense probe. (Bar in J = 300 μm in A and B, 150 μm in C, 100 μm in D–F, 200 μm in G and I, and 50 μm in H and J.) Abbreviations: a, arms; b, brain; e, eye; ep, eye primordia; i, iris; l, lens; m, mantle; ol, optic lobe; ols, outer lens segment; ov, optic vesicle; r, retina; ys, yolk sac.
Ectopic Expression of Squid Pax-6 in Drosophila.
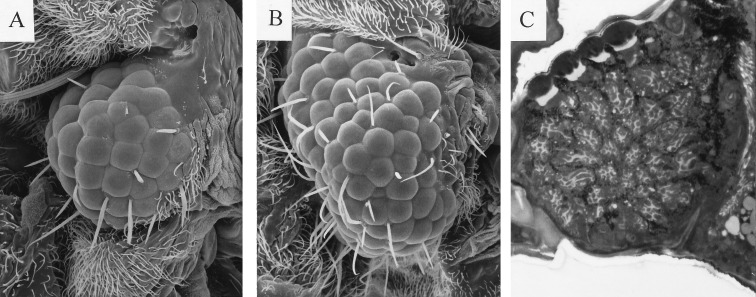
Functional studies of Pax-6 in squid are complicated due to the absence of mutants, transgenic methods, and appropriate cell lines. Thus, as a first approach for investigating the functional role of squid Pax-6, we decided to test whether it can induce ectopic eyes in Drosophila. We used the GAL4 system (21) to target squid Pax-6 cDNA1 expression to various imaginal discs. The results demonstrated (Fig. 4) that squid Pax-6 is able to induce ectopic Drosophila eyes on wings, antennae, and legs, as was previously demonstrated for Drosophila eyeless and mouse Pax-6 (14). All Drosophila eye-specific structures including cornea, pigment cells, cone cells and photoreceptors with rhabdomeres were formed in the ectopic eyes induced by squid Pax-6 cDNA.
Figure 4.
The ectopic expression of L. opalescens Pax-6 cDNA1 by means of the GAL4 system (21) induces the formation of ectopic eye in Drosophila. (A and B) Scanning electron micrograph of an ectopic eye induced on the wing by misexpression of L. opalescens Pax-6 cDNA1 (A) and Drosophila eyeless (B). (C) Section through a L. opalescens Pax-6-induced ectopic eye stained with Azur II and methylene blue.
DISCUSSION
We have isolated cDNAs corresponding to the squid Pax-6 gene. Squid Pax-6 shows the highest overall amino acid identity (78%) with Pax-6 from nemertines, consistent with nemertines being coelomate animals and molluscs and nemertines having a close evolutionary relationship (27, 39). Despite the fact that only the paired- and homeodomain regions of Pax-6/eyeless are well conserved between squid and Drosophila, squid Pax-6 can induce the formation of ectopic eyes in Drosophila as was previously demonstrated for Drosophila eyeless and mouse Pax-6 (14). It has been shown for vertebrate Pax-6 that the C-terminal PST domain is necessary for function of the protein (28, 29). Although squid Pax-6 and Drosophila eyeless C-terminal domains show appreciable differences in sequence and have different lengths (168 and 368 amino acids, respectively), both are proline-, serine- and threonine-rich (33% and 37%, respectively). It seems reasonable to propose that structural features such as secondary or tertiary folding, rather than direct similarity in amino acid sequences, are responsible for the common functional properties of squid Pax-6 and Drosophila eyeless, as was proposed for the C-terminal domains of vertebrate Pax-3 and Drosophila Gooseberry and Paired proteins (40).
The expression of squid Pax-6 in the developing brain, olfactory organ, and eyes shows parallels and differences with the expression pattern of Pax-6 in vertebrates, Drosophila, and nemertines. Despite remarkable similarity in general appearance of the eye in vertebrates and cephalopods, they are formed differently in the course of development. In the squid, the eye develops from a thickened ectodermal monolayer, which forms a multilayered oval mitotic placode on the dorsal surface of the head lobe. The eye vesicle develops by the internalization of this placode (17). The developing eye vesicle of squids is sealed off by the three layers of the primary eye fold: outer and inner ectodermal layers and a layer of mesoderm that separates the ectodermal layers. The outer ectodermal layer will produce the iris and outer lens segment, while the inner ectodermal layer will give rise to the inner lens segment. The lens is formed by projection of lentigenic processes and consists of two parts separated by a septum (41). The cornea has a very different ectodermal origin from the rest of the eye in the squid and is formed as a new skin fold from the edge of the forward growing arms (42). In vertebrates, the optic vesicle appears as an evagination of the diencephalon and the lens develops from the overlying ectoderm (see refs. 43 and 44 for reviews). During eye development in vertebrates, Pax-6 expression occurs in the optic vesicle, the overlaying surface ectoderm, and, at later stages of development, in the developing retina, iris/ciliary body, lens, and cornea (7, 9, 45). In Drosophila, eyeless is expressed in the eye portion of the eye–antennal imaginal disc, with higher expression being observed in the undifferentiated cells anterior to the morphogenetic furrow than behind the furrow where cells start to differentiate (11). Thus, both in vertebrates and Drosophila Pax-6/eyeless is involved in the formation of a morphogenetic field that will give rise to the eye. Explantation and transplantation experiments with squid eye rudiments from Arnold stages 15–17 (Naef stages VI–VIII) demonstrated that they possess the ability for autonomous eye differentiation (46), and our data are consistent with the idea that Pax-6 is necessary for establishing the “eye field,” and that eye determination in squid is primarily a process intrinsic to the differentiation capacities of the blastoderm (46).
Our data also show that there are some notable differences between the expression pattern of Pax-6 in the developing eyes of squid and vertebrates, and that the expression pattern of squid Pax-6 in the developing eye is more similar to that of eyeless in Drosophila than of Pax-6 in vertebrates. eyeless is actively expressed in undifferentiated precursor cells of the eye imaginal disks in Drosophila and its expression decreases significantly in differentiating cells posterior to the morphogenetic furrow (11). In vertebrates, Pax-6 is expressed in presumptive neural retina and in amacrine and ganglion cells at later stages of retina development (7, 9, 47). In the squid, expression of Pax-6 was not detected in differentiating retina cells (Fig. 3H). Moreover, in vertebrates Pax-6 is expressed in the developing lens (7, 9, 45) and has been implicated in transcription of many crystallin genes (see ref. 48 for a review). However, we did not find squid Pax-6 mRNA in the inner lentigenic cells, a major site of S-crystallin synthesis (38, 49). Nonetheless, Pax-6 may still be involved in the regulation of S-crystallin synthesis in the developing anterior segment of the squid lens, where expression of both Pax-6 and two S-crystallin genes (Lops7 and -12) were observed. We have noted potential Pax-6 binding sites in the promoters of several squid S-crystallin genes (data not shown). Thus, a possible role of Pax-6 in crystallin gene regulation in the squid remains to be established. Together, our results suggest that squid Pax-6 is not as critical for later stages of retina and lens development as it is in vertebrates.
The expression of Pax-6 in the developing vertebrate nose and olfactory bulbs (7), nemertine cerebral (chemosensory) organ (27), and squid olfactory organ suggests that also these structures may have a common ancestral origin. It is noteworthy that development of the olfactory and visual systems has a number of common features. In vertebrates, both the visual and olfactory systems are ectodermally derived and depend upon determinative interactions with particular regions of the brain (43). Of special interest here is that expression of Pax-6 is critical for the development of both sensory systems (9). In Drosophila, the olfactory and visual organs derive from the same eye–antennal imaginal disc; moreover, both systems express a number of identical genes during development (for example, irregular chiasm C-roughest for cell death, lozenge for pattern formation, retinal degeneration B for maintenance and physiology, norpA for phototransduction and odorant reception; see ref. 50 for a review). Further experiments comparing the roles of Pax-6 in the developing olfactory and visual systems seem warranted.
It is not clear at present whether squid Pax-6 initiates the development of ectopic eyes in Drosophila directly or indirectly by activating eyeless. Moreover, it is not known how many common genes acting downstream of Pax-6 are involved in the cascade leading to eye development in different systematic groups. At least two other candidate genes have been identified. One is eyes absent which is necessary for Drosophila eye development (51); two homologous genes for eyes absent are expressed in vertebrate lens and retina (52), and one homolog was recently identified in the squid (S.I.T., unpublished data). The homeobox gene, sine oculis, is also essential for eye development in Drosophila (53) and acts after eyes absent (54). The sine oculis homolog, Six-3, is expressed in the vertebrate eye (55). sine oculis homologs have not been identified yet in the squid.
Our data support the idea that morphologically distinct eyes of different species have arisen through elaboration of a common conserved Pax-6-dependent mechanism (11, 14, 15) that is operative at early stages of eye development and that the anatomical differences among eyes arose later in evolution. Consequently, we believe that eyes in cephalopods and vertebrates have a common evolutionary origin and are products of parallel rather than convergent evolution (56).
Acknowledgments
We thank Dr. J. Marthy for help with the isolation of squid spermatophores for DNA isolation and Drs. W. Gilly and M. Perri for help with collection and fixing of squid embryos. We thank Drs. W. Gilly, P. Grant, and J. West for help with analysis of the results of in situ hybridization; A. Cvekl for pointing out the potential Pax-6 binding sites in S-crystallin promoters, and Frederick Biomedical Supercomputing Center, Frederick Cancer Research and Development Center (Frederick, MD) for allocation of computing time and staff support. This work has been supported by grants from the Swiss National Science Foundation and the Kantons of Basel To W.G., from the Janggen-Pöhn Stiftung to G.H., and from the Collen Foundation and the Sandoz Foundation to P.C.
ABBREVIATION
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Salvini-Plawen L V, Mayr E. Evol Biol. 1977;10:207–263. [Google Scholar]
- 2.Land M F, Fernald R D. Annu Rev Neurosci. 1992;15:1–29. doi: 10.1146/annurev.ne.15.030192.000245. [DOI] [PubMed] [Google Scholar]
- 3.Wolken J J. Light Detectors, Photoreceptors, and Imaging Systems in Nature. New York: Oxford Univ. Press; 1995. [Google Scholar]
- 4.Eakin R M. Am Zool. 1979;19:647–653. [Google Scholar]
- 5.Ton C C T, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, Royer-Pokora B, Collins F, Swaroop A, Strong L C, Sounders G F. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 6.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 7.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 8.Matsuo T, Osumi-Yamashita N, Noji S, Ohuchi H, Koyama E, Myokai F, Matsuo N, Taniguchi S, Doi H, Iseki S. Nat Genet. 1993;3:299–304. doi: 10.1038/ng0493-299. [DOI] [PubMed] [Google Scholar]
- 9.Grindley J C, Davidson D R, Hill R E. Development (Cambridge, UK) 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 10.Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie N D. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 11.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 12.Hanson I, van Heyningen V. Trends Genet. 1995;11:268–272. doi: 10.1016/s0168-9525(00)89073-3. [DOI] [PubMed] [Google Scholar]
- 13.Lindsley D, Zimm G. The Genome of Drosophila melanogaster. New York: Academic; 1992. [Google Scholar]
- 14.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 15.Halder G, Callaerts P, Gehring W J. Curr Opin Genet Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 16.Valentine J W, Erwin D H, Jablonski D. Dev Biol. 1996;173:373–381. doi: 10.1006/dbio.1996.0033. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert D L, Adelman W J, Arnold J M, editors. Squid as Experimental Animals. New York: Plenum; 1990. [Google Scholar]
- 18.Packard A. Biol Rev. 1972;47:241–307. [Google Scholar]
- 19.Tomarev S I, Zinovieva R D, Piatigorsky J. J Biol Chem. 1992;267:8604–8612. [PubMed] [Google Scholar]
- 20.Wilkinson D G. In: In Situ Hybridization: A Practical Approach. Wilkinson D G, editor. Oxford: IRL; 1993. pp. 74–83. [Google Scholar]
- 21.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 22.Dozier C, Carriere C, Grevin D, Martin P, Quatannens B, Stehelin D, Saule S. Cell Growth Differ. 1993;4:281–289. [PubMed] [Google Scholar]
- 23.Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chisholm A D, Horvitz H R. Nature (London) 1995;377:52–55. doi: 10.1038/377052a0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Emmons S W. Nature (London) 1995;377:55–59. doi: 10.1038/377055a0. [DOI] [PubMed] [Google Scholar]
- 26.Czerny T, Busslinger M. Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loosli F, Kmita-Cunisse M, Gehring W J. Proc Natl Acad Sci USA. 1996;93:2658–2663. doi: 10.1073/pnas.93.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D, Maas R L. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 29.Carriere C, Plaza S, Caboche J, Dozier C, Bailly M, Martin P, Saule S. Cell Growth Differ. 1995;6:1531–1540. [PubMed] [Google Scholar]
- 30.Naef A. Die Cephalopoden. Vol. 35. Naples, Italy: Fauna Flora Golfo Napoli; 1928. (monographia). [Google Scholar]
- 31.Segawa S, Yang W T, Marthy H-J, Hanlon R T. Veliger. 1988;30:230–243. [Google Scholar]
- 32.Marthy, H.-J. (1985) in Cellular and Molecular Control of Direct Cell Interaction, NATO ASI Series A , ed. Marthy, H.-J. (Plenum, New York), pp. 159–197.
- 33.Emery D G. Tissue Cell. 1975;7:357–367. doi: 10.1016/0040-8166(75)90011-7. [DOI] [PubMed] [Google Scholar]
- 34.Gilly Wm F, Lucero M T. J Exp Biol. 1992;162:209–229. [Google Scholar]
- 35.Budelmann B U. Mar Freshwater Behav Physiol. 1996;27:59–75. [Google Scholar]
- 36.Tomarev S I, Chung S, Piatigorsky J. J Mol Evol. 1995;41:1048–1056. doi: 10.1007/BF00173186. [DOI] [PubMed] [Google Scholar]
- 37.Tomarev S I, Piatigorsky J. Eur J Biochem. 1996;235:449–465. doi: 10.1111/j.1432-1033.1996.00449.x. [DOI] [PubMed] [Google Scholar]
- 38.West J A, Sivak J G, Pasternak J, Piatigorsky J. Dev Dyn. 1994;199:85–92. doi: 10.1002/aja.1001990202. [DOI] [PubMed] [Google Scholar]
- 39.Turbeville J M, Field K G, Raff A R. Mol Biol Evol. 1992;9:235–249. doi: 10.1093/oxfordjournals.molbev.a040716. [DOI] [PubMed] [Google Scholar]
- 40.Xue L, Noll M. EMBO J. 1996;15:3722–3731. [PMC free article] [PubMed] [Google Scholar]
- 41.Willekens B, Vrensen G, Jacob T, Duncan G. Tissue Cell. 1984;16:941–950. doi: 10.1016/0040-8166(84)90073-9. [DOI] [PubMed] [Google Scholar]
- 42.Arnold J M. J Exp Zool. 1984;232:187–195. doi: 10.1002/jez.1402320206. [DOI] [PubMed] [Google Scholar]
- 43.Grainger R M. Trends Genet. 1992;8:349–355. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- 44.Saha M S, Servetnick M, Grainger R M. Curr Opin Genet Dev. 1992;2:582–588. doi: 10.1016/s0959-437x(05)80176-5. [DOI] [PubMed] [Google Scholar]
- 45.Li H-S, Yang J-M, Jacobson R D, Pasko D, Sundin O. Dev Biol. 1994;162:181–194. doi: 10.1006/dbio.1994.1077. [DOI] [PubMed] [Google Scholar]
- 46.Marthy H-J. J Embryol Exp Morphol. 1973;29:347–361. [PubMed] [Google Scholar]
- 47.Hitchcock P F, Nacdonald R E, VanDeRyt J T, Wilson S W. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Cvekl A, Piatigorsky J. BioEssays. 1996;18:621–631. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 49.Brahma S K. J Embryol Exp Morphol. 1978;46:111–118. [PubMed] [Google Scholar]
- 50.Gaines P, Carlson J R. Braz J Med Biol Res. 1995;28:161–167. [PubMed] [Google Scholar]
- 51.Bonini N M, Leiserson M, Benzer S. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 52.Mishima N, Tomarev S I. Invest Ophthalmol Visual Sci. 1996;37:S150. [PubMed] [Google Scholar]
- 53.Cheyette B N R, Green P J, Matrin K, Garren H, Hartenstein V, Zipursky S L. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 54.Thomas B J, Zipursky S L. Trends Cell Biol. 1994;4:389–394. doi: 10.1016/0962-8924(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 55.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development (Cambridge, UK) 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 56.Zuckerkandl E. J Mol Evol. 1994;39:661–678. doi: 10.1007/BF00160412. [DOI] [PubMed] [Google Scholar]